Abstract
The accurate mapping of clones derived from genomic regions containing complex arrangements of repeated elements presents special problems for DNA sequencers. Recent advances in the automation of optical mapping have enabled us to map a set of 16 BAC clones derived from the DAZ locus of the human Y chromosome long arm, a locus in which the entire DAZ gene as well as subsections within the gene copies have been duplicated. High-resolution optical mapping employing seven enzymes places these clones into two contigs representing four distinct copies of the DAZ gene and highlights a number of differences between individual copies of DAZ.
At the outset of the Human Genome Project the construction of high-density genetic and physical maps of the genome was prominent among the initial goals. As these goals have come to fruition (Cohen et al. 1993; Dib et al. 1996; Deloukas et al. 1998), the primary emphasis has shifted to the determination of the complete nucleotide sequence of the more than 3000 Mb comprising the 24 different human chromosomes (Collins et al. 1998). The predominant approach to sequence acquisition is the shotgun subcloning of fingerprinted DNA clones, generally BAC, PAC, and P1 clones of considerable size and stability (McPherson 1997; Rowen et al. 1997; Sanger Centre and Washington University Genome Sequencing Center 1998), or, alternatively, whole genome shotgun sequencing (Weber and Myers 1997; Venter et al. 1998). Inherent in this approach, however, is the realization and acceptance that a certain percentage of the genome is likely to be excluded from analysis, that is, regions containing large blocks of repetitive sequences that are therefore presumably of low sequence complexity (Collins et al. 1998). These repeat-containing regions include centromeres, telomeres, rDNA clusters, and heterochromatic chromosomal regions composed mainly of satellite DNA, for example, 1qh, 9qh, and Yqh. Unfortunately, the present inability to characterize clones containing repeated DNA sequences adequately may inadvertently result in the exclusion of some gene-containing regions from the final human genome sequence assembly.
The euchromatic portion of the human Y chromosome, until recently, was considered to be comprised of a small number of genes, among them male sex-determining and fertility factors, dispersed throughout a sea of Y-specific repetitive sequences (Foote et al. 1992; Affara et al. 1994). Recently, however, it has become apparent that what were once thought to be Y-chromosome repetitive sequences may actually represent a number of gene families (Lahn and Page 1997). Among these is the DAZ (deleted in azoospermia) gene cluster, deletions of which are associated with decreased fertility resulting from severely decreased or absent mature sperm production (Reijo et al. 1995, 1996). The DAZ locus appears to have arisen from the transposition of an ancestral copy of the DAZ gene DAZL (also known as DAZH [Saxena et al. 1996], DAZLA [Seboun et al. 1997], or SPYGLA [Shan et al. 1996]), located on chromosome 3, to the Y chromosome (Saxena et al. 1996). Coincident or subsequent to this transposition, the entire gene, as well as several regions within the gene, were duplicated. The entire DAZ gene has been duplicated several times, resulting in at least two pairs of head-to-head copies separated by 300–400 kb (Saxena et al. 2000). Internally, a 2.4-kb genomic fragment (the “DAZ repeat”) containing exon 7 is present from 8 to possibly 15 times within the copies of DAZ, and a segment of ∼10 kb containing exons 2 through 6 has also been duplicated one or more times in different copies of DAZ (this paper; Saxena et al. 2000). The complexity of this locus has made the physical mapping of the DAZ region exceedingly difficult. The majority of STS markers derived from this locus are not single-copy, making the creation of clone contigs by PCR analysis virtually impossible.
Optical mapping is a recently developed DNA mapping system in which single DNA molecules are employed in the generation of ordered restriction maps (for review, see Aston et al. 1999). Substrates for optical mapping range from long-range PCR products (Skiadas et al. 1999), cloned DNA including cosmids and BAC clones (Cai et al. 1998; Jing et al. 1998), to genomic DNA from microorganisms (Jing et al. 1999; Lin et al. 1999) as well as from humans. The unique capability of optical mapping to create high-resolution multienzyme ordered restriction maps of DNA molecules affords us the opportunity to accurately characterize cloned DNA templates containing potentially confusing arrangements of repetitive coding and noncoding sequences. Recently, we have introduced increased automation into the optical mapping process, drastically reducing the time required to produce individual restriction maps. This, in turn, allows us to broaden the scope of targets we can analyze. As an example of this increased capacity, we have chosen to demonstrate the power of optical mapping by analyzing a collection of 16 BAC clones known to originate from the DAZ locus by hybridization studies and, through the use of optical mapping, have succeeded in creating contigs that clarify the relationships between the clones and the copies of DAZ from which they are derived. Our results are consistent with the existence of four copies of the DAZ gene, each of which has its own distinctive arrangement of repetitive elements, patterns that, although difficult to discover using other mapping techniques, are immediately obvious through the use of optical mapping.
RESULTS
New map construction algorithms were developed that specifically deal with problems encountered in working with BAC clones as compared with cosmid clones (Anantharaman et al. 1997; Jing et al. 1998). Algorithms were constructed that modeled errors unique to large insert clones; these included increased DNA breakage and an increased number of small DNA map fragments (see Advancements in Map Construction Techniques in the Methods section below). An overview of the complete optical mapping process is depicted in Figure 1.
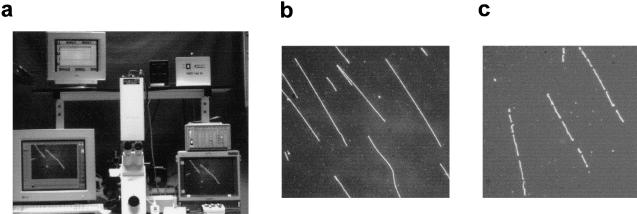
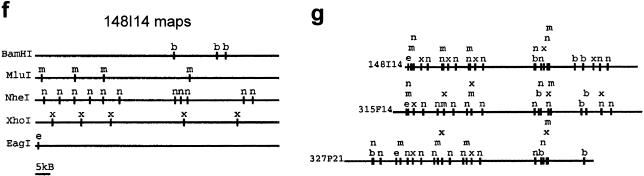
Figure 1.
Overview of optical mapping system. (a) Optical mapping station for image collection, including a Zeiss 135M Axiovert microscope, SIT camera, and Sony monitor for viewing slides and CCD camera (attached underneath microscope), Ludl controller, and Dell and Sun computers for image collection. (b) Uncut BAC DNA deposited on optical mapping surface. (c) BAC DNA (clone 148I14) digested on optical mapping surface with restriction enzyme XhoI. (d) Illustration demonstrating construction of highest probability single-enzyme (XhoI) map (white background) from individual input molecules (gray background) of clone 148I14. (e) Output of automatic map-making program indicating the three most probable XhoI maps of clone 148I14. Statistics provided by the map-making program include the number of molecules used to construct the map, the probability (P) of each map, the percentage of molecules that fit the map (good mols), the fraction of cuts that are false cuts, and the percentage of molecules that show breakage at either the right or left ends (breakage R or L). (f) Single-enzyme maps of clone 148I14. In general, the map with the highest probability is chosen as the final map. (g) Composite map of clone 148I14 is constructed by overlaying the single-enzyme maps after the orientation of single-enzyme maps has been determined. GENTIG then attempts to create a contig of this clone with any other overlapping clones (315F14 and 327P21).
Determination of NotI Sites within Inserts
BAC clones were first characterized with respect to the presence or absence of NotI sites within the human DNA insert. Circular DNA molecules were mounted on surfaces using the peel method and then digested with NotI on the surface (Fig. 2a,b). The vector, pBeloBAC11, contains two NotI restriction sites flanking the cloning site (Kim et al. 1996). None of our clones contains any internal NotI sites, allowing us to do our analysis on NotI-linearized BAC DNA.
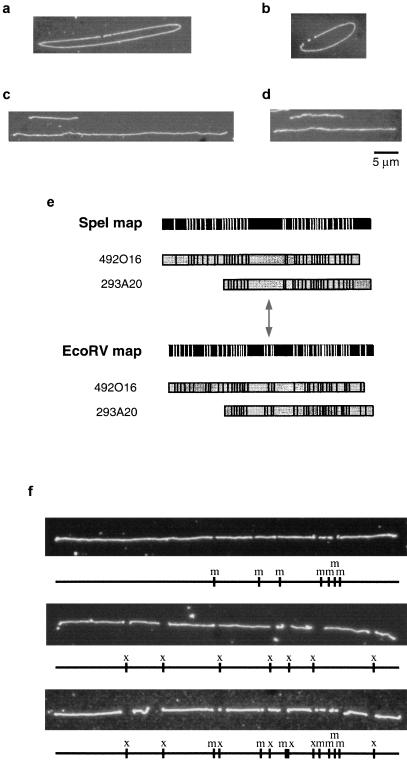
Figure 2.
Optical mapping of BAC clones. (a,b) Digestion of BAC clones 492O16 (a) and 148I14 (b) with NotI. Small fragments represent cloning vector. Note lack of NotI sites within insert. (c,d) Sizing of BAC clones 492O16 (c) and 132B16 (d) using λ DNA as external standard. (e) EcoRV and SpeI maps of BAC clones 492O16 and 293A20. Note symmetrical arrangement of sites surrounding the small cluster of EcoRV fragments and projecting in either direction from the center of the large SpeI fragment, indicated by double-headed arrow. (f) Orientation of single-enzyme maps of clone 492K23 using double digestion approach.
Determination of Clone Size
Next, we used optical methods to determine the sizes of the BAC clones. NotI-linearized BAC DNA was mixed with intact λ DNA, which served as an external size standard. This mixture was mounted on mapping surfaces and digested with EagI, which cuts λ at two sites. In this way we could easily differentiate between λ standards and random-sheared DNA fragments. Between 25 and 50 images containing both BAC and λ DNA were collected for each BAC clone (Fig. 2c, d). The sizes of the BAC clones were calculated automatically based on their relative fluorescence compared to the λ standard. The sizes of the BACs ranged from 83 kb to 229 kb. These results are, for the most part, consistent with pulsed field gel analysis (data not shown). Five clones showed a > 10% difference between pulsed field gel sizes and those derived from optical mapping. Sizing discrepancies are likely because of difficulties in calculating an exact size from pulsed field gels. We have used the optical mapping sizes in our map construction, although the ultimate contig construction was identical regardless of which size estimates were used.
Single-Enzyme Restriction Maps
BAC clones were subjected to restriction enzyme digestion as described in Methods. All clones were mapped with the enzymes BamHI, EagI, MluI, NheI, and XhoI. A total of 75 maps were made for the 16 clones, not including those enzymes for which a particular clone had no sites. Representative images of digested molecules are shown in Figure 2f. In addition, each clone was also digested with the enzymes EcoRV and/or SpeI. These enzymes were known previously to cut within most copies of the DAZ repeat (which contains exon 7) (Saxena et al. 1996), and we used these digestions to provide an overall picture of the location and placement of the DAZ gene within the clones, although these enzyme maps were not incorporated into the final composite maps of the clones.
The single-enzyme maps showed several interesting features of several of the clones. The most noticeable is that two clones (235I11 and 352E14) each contain a cluster of 2.5-kb MluI fragments (Fig. 3). We suspected that these might represent a polymorphism present in the 2.4-kb repeats in one of the copies of the DAZ gene. End-sequencing of shotgun subcloned MluI fragments from these clones and comparison to previously sequenced cosmids known to contain a copy of DAZ (Saxena et al. 1996) proved that these MluI fragments are, in fact, copies of the 2.4-kb repeat (data not shown). Clusters of BamHI fragments are also present in several of the clones (e.g., 343O6, 530K16, 50D17; Fig. 3). These clusters are present in the 3′-flanking regions of various DAZ gene copies (Saxena et al. 1996). Also, the EcoRV and SpeI maps are illuminating in their display of the organization of the DAZ repeats, and, consequently, the DAZ gene. Even without prior knowledge that the DAZ genes exist in head-to-head copies, several of the clones contain an apparently symmetrical organization of EcoRV and SpeI fragments that suggests such an arrangement (Fig. 2e).
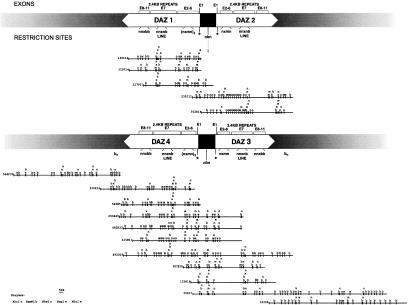
Figure 3.
Y DAZ BAC clone contigs. Contigs created automatically by the GENTIG program. Copies of DAZ have been assigned the names DAZ1–DAZ4. Recent results suggest that DAZ1 lies closest to the Y centromere and DAZ4 to the telomere (Saxena et al. 2000). Above the contigs is a diagram indicating the location of the DAZ genes within the contigs, the approximate positions of exons within the DAZ genes, and the positions of several restriction motifs that are indicative of specific regions of DAZ (see text). The smaller contig is a combination of two separate contigs created by GENTIG and overlapped as described in text. The arrow below the upper diagram indicates an nbn motif where head-to-head copies of DAZ1 and DAZ2 meet as well as where DAZ4 and DAZ3 meet (lower diagram).
Construction of Composite Maps and Contigs
The single-enzyme maps of BAC clone inserts do not contain any directionality. To create composite maps, therefore, individual maps were oriented with respect to each other by performing double digestions on mounted DNA molecules (Fig. 2f). Because double digestions occasionally resulted in a multitude of small fragments, many less than 1 or 2 kb long, which tend to desorb from the surface, double digestions were sometimes scored, not by counting every single expected fragment, but rather by looking for diagnostic fragments that would appear in one orientation rather than the other. Based on these results, composite maps were generated by superimposing all oriented single-enzyme maps for each clone. It is important to emphasize that our composite maps are not based on the fragments from the double digestions but instead on the combination of individual single-enzyme maps. Thus, any errors in sizing fragments derived from digestion with even one enzyme may result in some deviation in the order of very close restriction sites.
However, even given these caveats, the contigs created from the composite maps provide us with a remarkably clear picture of the organization of the DAZ gene cluster. The 16 BAC clones fall into three clusters based on GENTIG output (Fig. 3). Two contigs representing DAZ1 (clones 327P21/315F14/148I14) and DAZ2 (clones 352E14/235I11) have been combined based on independent mapping data (Saxena et al. 2000). The two clones having the unusual MluI site within the DAZ repeat (235I11 and 352E14) constitute one group. In fact, clone 352E14 is completely contained within clone 235I11. Three other clones (148I14, 315F14, and 327P21) comprise another contig. Clones 315F14 and 148I14 are essentially identical, whereas clone 327 extends further in one direction and is truncated at the other end relative to the other two clones in this contig. The first interesting thing to note about these clones is that they clearly do not contain the MluI site in the 2.4-kb DAZ repeats as do 235I11 and 352E14, distinguishing these two copies of DAZ from each other.
More interesting, however, are the various restriction site motifs that become apparent from the composite map contigs. The first is the cluster of restriction sites nmxn (NheI MluI XhoI NheI) at the 5′-end of the DAZ gene (immediately following the EagI site, Fig. 3). It appears that the 148I14/315F14/327P21 contig contains three copies of this particular region. Based on data from our efforts to sequence clone 148I14 (unpubl.), we now know that within this genomic region lie exons 2–6 of the DAZ gene. In addition, we do find that in our shotgun sequencing, we have a three-fold excess of coverage in this region. Thus, it appears that this copy of DAZ, represented by these three clones, in addition to containing the amplified DAZ repeat, also contains three copies of exons 2–6, whereas the 235/352 contig seems to contain only one copy.
The remaining 11 clones form a single contig, containing two copies of the DAZ gene (Fig. 3). Based on the nmxn motif, it appears that one of these (DAZ4) contains two copies of exons 2–6, and the adjoining copy (DAZ3) contains a single copy of exons 2–6. In addition, although some of the 2.4-kb DAZ repeats in DAZ4 (those following the LINE repeat, see below) appear to contain the MluI site, none of the 2.4-kb repeats in DAZ3 seem to contain MluI sites.
Several other motifs appear in all of the copies of DAZ. The cluster bnnxn (BamHI NheI NheI XhoI NheI) is present within the region we expect to contain the 2.4-kb DAZ repeats (Fig. 3). It was already known that in at least one sequenced cosmid clone containing DAZ (63C9, Saxena et al. 1996) a LINE sequence interrupts the 2.4-kb repeat cluster. Restriction site analysis of this sequence demonstrates that the bnnxn motif is derived from the LINE repeat and that this insertion is present in all four copies of DAZ. Also, the cluster bbxnn is present in all DAZ copies, except DAZ2, near the 3′-end of the DAZ gene. This is yet another characteristic way in which this copy of DAZ diverges from the other copies. Clones 343O6 and 566F22, which are located at the ends of this contig, were known, prior to mapping, not to contain any markers derived from within the DAZ gene and were presumed to flank the DAZ genes, which is confirmed by our maps.
Notice should also be taken of the regions where the 5′-ends of the DAZ pairs meet. The EagI sites indicate the 5′-ends of the genes (Saxena et al. 2000; J. Giacalone, unpubl.). But, between the EagI sites is the motif nbn. This is obvious in the large BAC contig (Fig. 3). As clone 352E14 contains a similar cluster of sites between two EagI sites (indicated by arrow under upper diagram in Fig. 3), it is likely that this clone contains the 5′-ends of two adjacent DAZ copies. Because of these and several other overlapping restriction sites and because we believe that there are at least four copies of DAZ arranged in two head-to-head pairs (Saxena et al. 1996, 2000), it would therefore be a legitimate presumption to connect the two small contigs together in this region (Fig. 3). GENTIG, however, would not have formed this larger contig because of insufficient overlap of restriction fragments.
DISCUSSION
The major focus of the Human Genome Project currently is to compile the “complete” sequence by the year 2003, with a working draft by 2001 (Collins et al. 1998). However, it is generally accepted that the term complete signifies that proportion of the genome that is the most straightforward to assemble (Dunham et al. 1999; Hattori et al. 2000). This is essentially a practical consideration, in that there is no obvious way to address the issue of assembling sequence from regions that are rife with repetitive sequences. Unfortunately, in setting aside regions such as centromeres, telomeres, and other heterochromatic regions, there is the risk of overlooking other regions that, although not heterochromatic in nature, will be equally difficult to assemble. A number of loci in the genome associated with disease pathologies—for example, Williams, DiGeorge, Prader Willi/Angelman and Smith-Magenis syndromes—have recently been shown to result from rearrangements of duplicated genomic regions (for review, see Eichler 1998). In addition, recent work indicates that as much as half of the euchromatic region of the human Y chromosome may, in fact, be composed of a number of gene families, whereas previously these regions were suspected of being merely Y-chromosome repetitive sequences (Lahn and Page 1997). They are, in fact, repetitive, but in this case it is genes that are present in multiple copies, as exemplified by the complex DAZ locus on Yq.
A major stumbling block in analysis of these regions is the inability to map clones derived from such loci accurately. The most common and least time-consuming methods of clone analysis—standard PCR, Southern blot analysis, and DNA fingerprinting—do not adequately unravel the complexities of these regions because of the high degree of sequence similarity between the repeated segments. Complete restriction mapping requires multiple single and double digestions, which may still give ambiguous results, as well as partial digestions and end-labeling experiments. Optical mapping is a DNA mapping approach that is ideally suited to such difficult regions. The ability to rapidly produce multienzyme, high-resolution ordered restriction maps of cloned DNA is critical to the analysis of complex regions of the genome exhibiting multiple levels of sequence repetition. The DAZ locus, a region of human Yq implicated in male fertility and proper sperm development, is such a region and provided a perfect substrate to test the efficacy of optical mapping.
At the start of this project, our knowledge of the DAZ gene complex was sketchy. The exact number of copies of DAZ on the Y chromosome was uncertain, although the existence of two head-to-head pairs was postulated. We were also aware that a genomic segment of about 2.4 kb containing exon 7 had been duplicated a number of times. Our optical mapping analysis of a collection of 16 BAC clones derived from the DAZ locus supports the existence of four copies of DAZ (Saxena et al. 2000) as all of the clones fall into three contigs, two containing a single copy of DAZ and one large contig containing two copies. Our most interesting results involve the nature of the intragenic repeated segments. EcoRV and SpeI digestion of all clones confirmed the existence of the multiple copies of the 2.4-kb DAZ repeat in all of these copies of DAZ. The unusual finding was the potential divergence between the 2.4-kb repeat in different copies of the gene. The existence of MluI sites in apparently all of the 2.4-kb repeats in DAZ2 (the 235I11/352E14 contig), in some of the repeats in DAZ4, and seemingly in none of the repeats from either of the other two copies of DAZ leads to speculation on possible evolutionary aspects of these copies of DAZ. Perhaps one of the 2.4-kb repeats within DAZ2 acquired an MluI site and through a gene-conversion-type event the other repeats within this copy of DAZ also acquired the MluI site. The existence of MluI sites in some of the 2.4-kb repeats in DAZ3 may indicate the beginning of the conversion of these repeats.
The application of optical approaches to large-scale genome mapping ultimately requires the ability to fashion a high-throughput, highly automated system with a minimum of user intervention. Optical mapping has made large strides toward fulfilling this mandate. The most significant improvement in our system is the development of an automated map-making function. Previously (Cai et al. 1998) map-making required the use of spreadsheet programs to calculate average fragment lengths, a time-consuming process that is now done automatically. Further refinements have also made optical mapping more efficient, including development of more consistent mapping surfaces (trimethyl/vinyl silanes with acrylamide overlay), the ability to analyze multiple BACs per optical mapping surface, and the automatic determination of clone contigs. Also, automated focusing and image tiling programs have made image acquisition an essentially user-independent task. The rapid evolution of optical mapping and its application to analysis of both cloned and genomic DNA (Jing et al. 1999; Lin et al. 1999) presents us with the possibility of creating, in the near future, complete restriction maps of large genomes or whole clone libraries in a time- and labor-efficient way.
METHODS
Advancements in Map Construction Techniques
Although algorithms have been developed for the construction of optical maps from cosmid clones (Anantharaman et al. 1997; Jing et al. 1998), these same approaches do not work well for BAC clones because of the increased amount of DNA breakage and the presence of large numbers of small DNA fragments. New map construction algorithms were developed that specifically deal with these issues.
Map construction takes place in three stages. First, an interactive tool called “visionade” is used to select high quality molecules from the image and mark the restriction sites. Subsequently, the relative mass of restriction fragments (in order) is automatically calculated from the integrated fluorescence intensity and local illumination conditions (Aston et al. 1999), producing a single molecule map. Typically 50–150 of these single molecule maps are combined to compute an accurate restriction map using a Bayesian model of many sources of potential errors in the single molecule maps. Sizing errors, missing restriction sites, spurious cuts, unknown orientation, and DNA contamination are modeled as described (Anantharaman et al. 1997; Anantharaman and Mishra 1998). In addition, because the large size of BAC clones subjects them to shear-mediated breakage, we now model missing ends of DNA molecules (up to 80% of the ends of all molecules within a group can be missing with up to 40% of the mass of any individual molecule lost). Given these and other statistical models of the mapping process, a finished map is constructed using previously published Bayesian inference techniques (Anantharaman et al. 1997). Single-enzyme maps are oriented with respect to each other using double digestions, and composite maps are created by overlaying all single-enzyme maps for an individual clone. Finally, map overlaps are determined using a program called GENTIG (Anantharaman et al. 1998). Here again, Bayesian inference is employed by GENTIG to model errors in maps that affect construction of contigs. For example, map errors may arise primarily from residual sizing error, unknown clone orientation, and a very low rate of missing or false cuts. Since the construction of contigs (overlapping clone maps) is more complex than the construction of an individual BAC map (consensus map derived from an ensemble of individual molecules), we developed a faster algorithm that uses an approximate maximum likelihood computation of the probability density for the hypothesis that each pair of BACs overlaps. Since the data error rate is low at this stage, the result is quite accurate. However, to eliminate any possible errors in the final contig, we also estimate the false-positive-match probability for each pair of BACs that appear to overlap and simply discard any maps that cannot be overlapped with high confidence. This may occasionally result in false negative errors in the form of gaps in the contigs, but allows high confidence in the actual contigs generated.
BAC Clone Isolation
BAC clones were isolated from total human genomic DNA libraries (Shizuya et al. 1992) made available through Research Genetics as human BAC library CITB. Clones were isolated by hybridization of STS markers derived from the DAZ region to high-density library filters. BAC clones were tested for Y-chromosome specificity by Southern blot analysis, PCR, and limited sequence analysis. Three clones—50D17, 132B16, and 148I14—were isolated from the first release of this library, made from DNA of a single male donor. The remaining BACs were isolated from the second library release and are derived from a different male donor.
DNA Purification and Preparation
DNA from BAC clones was purified using a modification of standard protocols, using QIAGEN −100 tips. Briefly, 150-ml cultures were used, and the amounts of buffers P1, P2, and P3 were increased to 7 ml. Incubation in lysis buffer P2 was decreased to 3 minutes. DNA was eluted from columns using warm (50°C) elution buffer. DNA was linearized by digestion with NotI for 1 hour at 37°C as per manufacturer's (New England Biolabs) protocol and was diluted with TE prior to mounting on optical mapping surfaces.
Surface Preparation
Optical mapping surfaces were prepared as described (Aston et al. 1999). In a closed vacuum system, glass cover slips were boiled first in concentrated nitric acid overnight, rinsed with water, boiled again overnight in concentrated hydrochloric acid, and then rinsed extensively with water. Clean cover slips were derivatized for two days at room temperature in ethanol containing 20–80 mg of 2% aminopropylmethyldiethoxysilane (GELEST, Inc.) or aminopropyltriethoxysilane (Sigma) solution which had been hydrolyzed at room temperature for 7 hours.
Alternatively, cover slips were heated at 80°C in piranha solution (H2SO4 : 30% H2O2, 9 : 1) for 50 minutes, rinsed with water, and then boiled in hydrochloric acid. Clean coverslips were derivatized in water containing a 60 : 3 ratio of trimethoxy : vinyltrimethoxy silanes (GELEST, Inc.) overnight at 65°C.
DNA Mounting and Digestion
NotI-digested BACs were applied to surfaces by capillary action, spotting, or peeling as described in Aston et al. (1999). The piranha/trimethyl/vinyl surfaces were covered with an acrylamide overlay before restriction digestion (Y. Skiadas and R. Qi, unpubl.). Digestions were performed by covering the DNA-containing surface with 100 μl of 1× digestion buffer containing 5–50 units of restriction enzyme and incubating at 37°C in a humid chamber. Digestion times generally ranged from 10 min to 2 hr. Digestions were stopped by aspirating off the enzyme solution, washing the surface with TE, and staining the DNA with a 0.1 μm solution of YOYO-I in 30% 2-mercaptoethanol in TE. Digested molecules were imaged as described in Aston et al. (1999). Each enzyme map was generated using between 50 and 150 individual molecules.
Acknowledgments
We thank Jonathan Vafai for assistance in data management and Leslie McGuinness for graphic design.
This work was supported by a grant from the National Institutes of Health (HG00225-08) to D.C.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dcschwartz@facstaff.wisc.edu; FAX (608) 265-6743.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.112100.
REFERENCES
- Affara NA, Lau Y-FC, Briggs H, Davey P, Jones MH, Khwaja O, Mitchell M, Sargent C. Report of the first international workshop on Y chromosome mapping. Cytogenet Cell Genet. 1994;67:360–387. doi: 10.1159/000133871. [DOI] [PubMed] [Google Scholar]
- Anantharaman TS, Mishra B, Schwartz DC. Genomics via optical mapping II: Restriction maps. J Comput Biol. 1997;4:91–118. doi: 10.1089/cmb.1997.4.91. [DOI] [PubMed] [Google Scholar]
- Anantharaman TS, Mishra B. “Genomics via optical mapping II(A): Restriction maps from partial molecules and variations.” Courant Technical Report #759. New York, New York, USA: Courant Institute; 1998. [Google Scholar]
- Anantharaman TS, Mishra B, Schwartz DC. “Genomics via optical mapping III: Contiging genomic DNA and variations.” Courant Technical Report #760. New York, New York, USA: Courant Institute; 1998. [Google Scholar]
- Aston C, Hiort C, Schwartz DC. Optical mapping: An approach for fine mapping. Methods Enzymol. 1999;303:55–73. doi: 10.1016/s0076-6879(99)03006-2. [DOI] [PubMed] [Google Scholar]
- Cai W, Jing J, Irvin B, Ohler L, Rose E, Shizuya H, Kim U-J, Simon M, Anantharaman T, Mishra B, et al. High-resolution restriction maps of bacterial artificial chromosomes constructed by optical mapping. Proc Natl Acad Sci. 1998;95:3390–3395. doi: 10.1073/pnas.95.7.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Chumakov I, Weissenbach J. A first generation physical map of the human genome. Nature. 1993;366:698–701. doi: 10.1038/366698a0. [DOI] [PubMed] [Google Scholar]
- Collins F S, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L the Members of the DOE and NIH Planning Group. New Goals for the U.S. Human Genome Project: 1998–2003. Science. 1998;282:682–689. doi: 10.1126/science.282.5389.682. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tomé P, Hui L, Matise TC, McKusick KB, Beckmann JS, et al. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millassear P, Marc S, Hazan J, Seboun E, et al. A comprehensive genetic map of the human genome based on 5264 microsatellites. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- Dunham I, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, Ainscough R, Almeida JP, Babbage A, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Eichler E. Masquerading repeats: Paralogous pitfalls of the human genome. Genome Res. 1998;8:758–762. doi: 10.1101/gr.8.8.758. [DOI] [PubMed] [Google Scholar]
- Foote S, Vollrath D, Hilton A, Page DC. The human Y chromosome: Overlapping DNA clones spanning the euchromatic region. Science. 1992;258:60–66. doi: 10.1126/science.1359640. [DOI] [PubMed] [Google Scholar]
- Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, Park HS, Toyoda A, Ishii K, Totoki Y, Choi DK, et al. The DNA sequence of human chromosome 21. The chromosome 21 mapping and sequencing consortium. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- Jing J, Lai Z, Aston C, Carucci DJ, Gardner MJ, Mishra B, Anantharaman TS, Tettelin H, Cummings LM, Hoffman SL, et al. Optical mapping of Plasmodium falciparum chromosome 2. Genome Res. 1999;9:175–181. [PMC free article] [PubMed] [Google Scholar]
- Jing J, Reed J, Huang J, Hu X, Clarke V, Edington J, Housman D, Anantharaman TS, Huff EJ, Mishra B, et al. Automated high resolution optical mapping using arrayed, fluid-fixed DNA molecules. Proc Natl Acad Sci. 1998;95:8046–8051. doi: 10.1073/pnas.95.14.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U-J, Birren BW, Slepak T, Mancino V, Boysen C, Kang H-L, Simon MI, Shizuya H. Construction and characterization of a human bacterial artificial chromosome library. Genomics. 1996;34:213–218. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- Lin J, Qi R, Aston C, Jing J, Anantharaman TS, Mishra B, White O, Daly MJ, Minton KW, Venter JC, et al. Whole-genome shotgun optical mapping of Deinococcus radiodurans. Science. 1999;285:1558–1562. doi: 10.1126/science.285.5433.1558. [DOI] [PubMed] [Google Scholar]
- McPherson JD. Sequence ready—or not? Genome Res. 1997;7:1111–1113. doi: 10.1101/gr.7.12.1111. [DOI] [PubMed] [Google Scholar]
- Reijo R, Alagappan RK, Patrizio P, Page D. Severe oligospermia resulting from deletions of Azoospermia Factor gene on Y chromosome. Lancet. 1996;347:1290–1293. doi: 10.1016/s0140-6736(96)90938-1. [DOI] [PubMed] [Google Scholar]
- Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- Rowen L, Mahairas G, Hood L. Sequencing the human genome. Science. 1997;278:605–607. doi: 10.1126/science.278.5338.605. [DOI] [PubMed] [Google Scholar]
- The Sanger Centre and the Washington University Genome Sequencing Center. Toward a complete human genome sequence. Genome Res. 1998;8:1097–1108. doi: 10.1101/gr.8.11.1097. [DOI] [PubMed] [Google Scholar]
- Saxena R, Brown LG, Hawkins T, Alagappan RK, Skaletsky H, Reeve MP, Reijo R, Rozen S, Dinulos MB, Disteche CM, et al. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat Genet. 1996;14:292–299. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- Saxena, R., de Vries, J.W.A., Repping, S., Alagappan, R.K., Skaletsky, H., Brown, L.G., Ma, P., Chen, E., Hoovers, J.M.N., and Page, D.C. 2000. Four DAZ genes in two clusters found in AZFc region of human Y chromosome. Genomics, in press. [DOI] [PubMed]
- Seboun E, Barbaux S, Bourgeron T, Nishi S, Algonik A, Egashira M, Nikkawa N, Bishop C, Fellous M, McElreavey K, et al. Gene sequence, localization, and evolutionary conservation of DAZLA, a candidate male sterility gene. Genomics. 1997;41:227–235. doi: 10.1006/geno.1997.4635. [DOI] [PubMed] [Google Scholar]
- Shan Z, Hirschmann P, Seebacher T, Edelman A, Jauch A, Morell J, Urbitsch P, Vogt PH. A SPGY copy homologous to the mouse gene Dazla and the Drosophila gene boule is autosomal and expressed only in the human male gonad. Hum Mol Genet. 1996;5:2005–2011. doi: 10.1093/hmg/5.12.2005. [DOI] [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim U-J, Mancino V, Slepak T, Tachiiri T, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiadas J, Aston C, Samad A, Anantharaman TS, Mishra B, Schwartz DC. Optical PCR: Genomic analysis by long-range PCR and optical mapping. Mammalian Genome. 1999;10:1005–1009. doi: 10.1007/s003359901148. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Sutton GG, Kerlavage AR, Smith HO, Hunkapillar M. Shotgun sequencing of the human genome. Science. 1998;280:1540–1542. doi: 10.1126/science.280.5369.1540. [DOI] [PubMed] [Google Scholar]
- Weber JL, Myers EW. Human whole-genome shotgun sequencing. Genome Res. 1997;7:401–409. doi: 10.1101/gr.7.5.401. [DOI] [PubMed] [Google Scholar]