Abstract
Francisella tularensis is one of the most virulent bacteria known and a Centers for Disease Control and Prevention Category A select agent. It is able to infect a variety of animals and insects and can persist in the environment, thus Francisella spp. must be able to survive in diverse environmental niches. However, F. tularensis has a surprising dearth of sensory and regulatory factors. Recent advancements in the field have identified new functions of encoded transcription factors and greatly expanded our understanding of virulence gene regulation. Here we review the current knowledge of environmental adaptation by F. tularensis, its transcriptional regulators and their relationship to animal virulence.
Keywords: Francisella, transcriptional regulators, Francisella pathogenicity island, two-component regulatory systems
Introduction
Francisella tularensis is a Gram-negative non-motile intracellular pathogen and the causative agent of the zoonotic human disease tularemia. Tularemia, also known as rabbit fever, is usually transmitted to humans by arthropod bites, oral consumption of contaminated food or water, or handling of infected animal carcasses (Evans et al., 1985; Thomas and Schaffner, 2010).
Francisella tularensis is divided into several subspecies (Staples et al., 2006; Kugeler et al., 2009). These subspecies are associated with important differences in geographic distribution. F. tularensis subsp. tularensis (Type A) is found exclusively in North America. It is highly virulent, with less than 10 bacteria causing disease in humans via the respiratory route, and if untreated, demonstrates a high mortality rate of 30–60% (Dienst, 1963). F. tularensis subsp. holarctica (Type B) is less virulent in humans and is predominant in Europe and Japan, but also found in North America. F. tularensis subsp. mediasiatica is found primarily in Asia and Russia, and is rarely associated with disease in humans. F. novicida is normally considered as the fourth subspecies of F. tularensis; however, recent genome-wide polymorphism analysis suggests that it is an independent species highly virulent in mice but not in humans (Johansson et al., 2010). An attenuated strain of F. tularensis subsp. holarctica, the live vaccine strain (LVS), was developed and is used as a live-attenuated vaccine in the former Soviet Union and in some US laboratory workers (Tigertt, 1962; Burke, 1977). Because F. tularensis is able to infect through the respiratory route and cause disease with a very small dose, can be easily disseminated, results in a high mortality rate, and has the potential to cause panic among the public, F. tularensis is given the highest priority classification by the Centers for Disease Control and Prevention as a Category A select agent and is a potential bioweapon.
Completed genome sequencing for different subspecies has revealed that despite high nucleotide identity, there are many DNA rearrangements, gene acquisitions, and gene losses between and even within different subspecies (Titball and Petrosino, 2007; Champion et al., 2009; Nalbantoglu et al., 2010; Sjodin et al., 2010). These differences are thought to reflect the environmental adaptation of these subspecies and are associated with their differences in virulence. Most of our current understanding about the genetics of F. tularensis pathogenesis have been acquired with F. novicida, as this subspecies can be genetically manipulated with relative ease and causes a disease in mice similar to that caused by the human virulent F. tularensis subsp. tularensis. However, due to the genetic, phenotypic, and host range variations between the species, care should be taken in extrapolating data from one subspecies to another. With the recently developed genetic tools for the human virulent subspecies tularensis (LoVullo et al., 2006, 2009), it is now possible and necessary to carry out studies in these strains, which is required gain a full understanding the molecular basis of Francisella pathogenesis. Nevertheless, the studies with F. novicida and LVS have provided and will continue to provide valuable information on the molecular pathogenesis of tularemia, and the results have been generally applied to F. tularensis.
Francisella tularensis exists naturally in a variety of environments. It can survive in water, wet soil, and animal carcasses for several weeks. F. tularensis has also been found in a broad range of hosts, including mammals, insects, arthropods, and fresh water protozoans (Ellis et al., 2002). Not only does F. tularensis infect different hosts, as an intracellular pathogen, F. tularensis also goes through various microenvironments within host cells, including the macrophage phagosome and the host cell cytosol after phagosomal escape (Santic et al., 2010). Essential to the success of F. tularensis as an intracellular pathogen is its ability to adapt to a wide variety of environments and host cell types. This is achieved via timely activation/repression of dedicated patterns of gene expression. Most of the virulence genes of F. tularensis identified so far localize to the Francisella pathogenicity island (FPI). This is a cluster of 17 genes that is critical for the intracellular survival and virulence of F. tularensis (Nano and Schmerk, 2007). FPI genes were the first identified Francisella virulence factors and are affected by essentially all environmental cues and transcriptional virulence regulators identified thus far, demonstrating their importance in F. tularensis virulence.
Environmental changes are often sensed and relayed into bacteria through two-component regulatory systems (TCS). These systems are conserved and ubiquitous in bacteria, typically composed of a histidine sensor kinase, regulated by environmental stimuli, and a response regulator that activates downstream responses (Stock et al., 2000). In contrast to the variety of environments F. tularensis naturally encounters and opposite to most Gram-negative pathogens, which have numerous TCS, F. novicida has very few TCS encoded in the genome, and other subspecies of F. tularensis, such as subsp. tularensis lack any classically arranged (adjacent and co-transcribed) TCS. Orphaned TCS members KdpD, QseC, and PmrA are the only TCS factors discovered so far that affect virulence/virulence factor regulation (Mohapatra et al., 2007; Bell et al., 2010). In addition, F. tularensis, unlike many other bacteria, encodes only one alternative sigma factor (Fuller et al., 2009). This makes F. tularensis a unique pathogen with relatively few easily identifiable regulatory factors to survive in diverse environments. It is possible that some regulators exist that do not show homology to know regulators in the database or that alternative means of regulation (e.g., sRNA, post-transcriptional) may predominate in the Francisella.
Environmental Stimuli
To survive in different environments, F. tularensis has to be able to sense and respond to signals from its surroundings to precisely regulate virulence gene expression. In fact, the protein profiles of F. tularensis (LVS) grown in broth versus inside macrophages revealed the altered expression of several proteins (Golovliov et al., 1997). In addition, global transcriptional profiling of F. tularensis Type A strain Schu S4 within infected macrophages confirmed that the expression of 658 genes was significantly changed, among which 298 were up-regulated and 360 were down-regulated, depending on the stage of the infection (Wehrly et al., 2009). These observations support the notion that F. tularensis is able to sense and respond to changing environments during the course of infection. Although the molecular mechanisms that underlie gene regulation are not fully understood, studies thus far have suggested that environmental stimuli including temperature, limited iron source, oxidative stress, and host intramacrophage components, lead to altered expression profiles.
Temperature
A number of pathogenic bacteria that encounter a temperature shift during their life cycle have the ability to respond to this environmental cue with enhanced virulence. Shigella flexneri increases virB expression in response to elevated temperature (Tobe et al., 1991, 1995). Borrelia burgdorferi and Yersinia pestis have also adapted ways to change the expression profile of host-specific virulence genes according to an alteration in temperature (Konkel and Tilly, 2000). As a pathogen with different hosts, F. tularensis will go through a temperature shift upon entering a warm-blooded animal. When growing at 25 versus 37°C, F. novicida changes its outer surface by modifying the lipid A of lipopolysaccharide (Shaffer et al., 2007). Also, a recent transcriptomic study compared the global expression profiles of F. tularensis LVS cultivated in defined media at 26°C (environmental temperature) versus 37°C (mammalian body temperature; Horzempa et al., 2008). Approximately 11% of the genes in the entire LVS genome had significant changes in expression. Notably, around 40% of the genes that were up-regulated were previously identified or predicted to be important for intracellular growth and/or virulence (Horzempa et al., 2008). Another temperature change proteomic study with LVS and the Type A strain Schu S4 confirmed a significant increase in the level of FPI-encoded IglC, IglD, and PdpC, when the temperature shifted from 25 to 37°C (Lenco et al., 2009).
The regulatory mechanisms that underlie these temperature-dependent changes are not yet clear, but a few studies have suggested the involvement of an alternative sigma factor and its induction of heat-shock proteins, which is a mechanism used by many other bacteria (Ericsson et al., 1994; Meibom et al., 2008; Grall et al., 2009). However, unlike most other bacteria that have multiple sigma factors, F. tularensis encodes only one alternative sigma factor, RpoH, which is a sigma factor σ32 (Grall et al., 2009). Transcription profiling under heat-stress conditions in F. tularensis LVS showed that a number of heat-shock proteins and proteins involved in virulence were up-regulated. However, among all the genes that were regulated upon heat-stress, only 13 of them, which did not include FPI genes, had a putative σ32 binding site in their promoter region (Grall et al., 2009). This suggested that σ32 might not be acting directly on FPI genes.
Iron
Iron acquisition is a critical requirement for the survival of bacteria as it is involved in many metabolic processes. As one of the most common elements in the Earth's crust, iron is easily accessible for bacteria in the environment. However, for intracellular pathogens, their ability to acquire iron is critical because of the limited nature of freely available iron in mammalian host cells. Iron availability has a great impact on both virulence of bacteria pathogens and the host immune response (Wang and Cherayil, 2009; Skaar, 2010).
Pathogens respond to iron-restriction conditions by up-regulation of not only iron acquisition machinery but also virulence determinants (Ratledge and Dover, 2000; Skaar, 2010). A general mechanism for iron sensing is controlled by the ferric uptake regulator (Fur). Fur forms a complex with ferrous iron and represses gene expression by binding to the Fur box upstream of iron-regulated genes, which is then derepressed under iron-limiting conditions. In F. tularensis, an iron acquisition operon (fsl for Ft siderophore locus or fig for Ft iron genes) involved in siderophore production is located downstream of fur, and is induced under iron-restriction conditions in a Fur-dependent manner (Deng et al., 2006; Sullivan et al., 2006). A Fur box is also found upstream of iglC and pdpB, two FPI genes (Deng et al., 2006). Microarray analysis found that approximately 80 genes, including FPI genes, are differentially expressed in iron-starved F. tularensis LVS (Deng et al., 2006). This was confirmed by a proteomics analysis in F. tularensis LVS, which showed that the levels of a significant number of proteins change under iron-limiting conditions, which again included those in the FPI (Lenco et al., 2007). Although it did not appear in these two previously mentioned studies, the FPI-encoded PdpA is also up-regulated under iron-limiting conditions (Schmerk et al., 2009). Moreover, gallium, a transition metal that competes with iron for uptake and/or utilization, inhibits F. tularensis LVS and F. novicida growth in broth and in macrophages, and inhibits F. novicida virulence in a mouse model of pneumonic tularemia (Olakanmi et al., 2010), providing further evidence for an important role of iron in regulating F. tularensis virulence factors.
Oxidative Stress
One of the essential host cell innate immune defenses against invading pathogens is the generation of reactive oxygen species (ROS) and reactive nitrogen species (RNS; Fang, 2004). Many bacterial pathogens are able to sense the oxidative stress inside host cells and have ways to combat it. F. tularensis is in this group of pathogens as it is able to inactivate the phagocyte NADPH oxidase through a mechanism likely involving dephosphorylation of host kinases and phox components by bacterial enzymes/bacterial-mediated host signaling cascades (McCaffrey and Allen, 2006; Schulert et al., 2009; McCaffrey et al., 2010; Mohapatra et al., 2010). A screen for F. tularensis LVS proteins responding to heat and hydrogen peroxide identified chaperones DnaK, GroEL, and GroES (Ericsson et al., 1994). In addition to these proteins, Lenco et al. (2005) detected approximately 20 proteins that were up-regulated upon exposure to oxidative stress in F. tularensis LVS. In searching for the molecular mechanisms that underlie the response to oxidative stress, a proteomics assay was carried out comparing the protein profiles of wild-type F. novicida and an mglA (macrophage growth locus) mutant. In addition to virulence factors, homologs of oxidative, and general stress response proteins were identified in this study, including the previously identified chaperons, suggesting a role for MglA, a regulator of F. tularensis virulence, in the oxidative response (Guina et al., 2007).
Other Environmental Cues
In addition to reactive oxygen and nitrogen species, other components of the host environment may also be signals sensed by F. tularensis. A recent study identified spermine, a polyamine produced only in eukaryotic cells, as an intramacrophage component sensed by F. tularensis (Carlson et al., 2009). Infection with F. tularensis LVS or Schu S4 cultured in the presence or absence of spermine induced different patterns of cytokine production in macrophages. A genome-wide microarray assay indicated that the presence of spermine affected a significant portion of the F. tularensis transcriptome. This is in part mediated by the induction of the Francisella insertion sequence (IS) elements ISFtu1 and ISFtu2, which is sufficient to induce the expression of downstream genes in response to spermine (Carlson et al., 2009). This observation suggested a model of gene regulation in which intramacrophage compounds elicit substantial changes in Francisella gene expression, and result in low cytokine production and immune evasion.
As F. tularensis resides temporally in phagosomal vacuoles after entering the host cell, the decrease in pH inside the phagosome may play a role in the induction of virulence factors. In support of this, the expression of RipA, a cytoplasmic membrane protein involves in F. tularensis intracellular replication and host immune evasion, is regulated by pH with increased expression at neutral rather than acidic environment (Fuller et al., 2008, 2009; Huang et al., 2010). However, the early acidification of the phagosome is not singularly involved in FPI gene induction as virulence protein expression is increased even upon inhibition of phagosome acidification (Chong et al., 2008).
Another environmental change that might play an important role in the induction of F. tularensis virulence is stress/starvation. The host cell phagosome/cytosol is known to be a nutrient poor and stress-inducing environment for bacteria. Several of the F. tularensis virulence regulators have been implicated in the stringent response, i.e., a microbial reaction to starvation. For example, SspA is a stringent starvation transcriptional regulator that is highly related to and physically interacts with MglA in F. tularensis LVS and F. novicida (Charity et al., 2007). In addition, guanosine pentaphosphate (ppGpp), an alarmone that is the global regulator of gene expression during the stringent response, is also involved in F. tularensis virulence gene expression by promoting the interaction of FevR/PigR with the MglA–SspA–RNA polymerase (RNAP) complex (Charity et al., 2009). Moreover, RelA, the enzyme involved in ppGpp production, is also involved in F. tularensis LVS virulence and intracellular survival (Dean et al., 2009). These findings suggest that starvation plays a critical role in the induction of F. tularensis LVS virulence.
Transcriptional Regulators
Bacteria typically utilize specialized transcription factors that bind non-specifically or specifically to DNA recognition sequences. The location and nucleotide composition of transcription factor binding sites in part determines whether a transcription factor represses or activates the expression of a certain gene. An interesting feature of Francisella is that very few transcription factors have been identified by experimentation or homology searches (Table 1), but these few factors likely control the virulence cascades of this pathogen. While translational or post-translational regulation may play a role, the majority of evidence points to regulation at the level of transcription/mRNA transcripts. The FPI is the primary site of virulence-related genes; thus the remainder of this review will focus on transcription factors or regulators that have been identified to play a role in FPI gene regulation.
Table 1.
Transcription factors of Francisella tularensis identified by protein relatedness to known transcription factors/transcription factor motifs.
| Conservation in other Francisella | Experimentally studied in: | |||||||||
| FTT# (Type A; Schu S4) | Gene | Description | W96-3418 (Type A) | FSC198 (Type A) | OSU18 (Type B) | LVS (Type B) | U112 (F. novicida) | Schu S4 (Type A) | LVS (Type B) | U112 (F. novicida) |
| 30 | fur | Fe transcriptional regulator | + | + | + | + | + | Ramakrishnan et al. (2008) | Deng et al. (2006) | Kiss et al. (2008) |
| 94 | qseC | Sensor histidine kinase | + | + | + | + | + | Rasko et al. (2008) | Weiss et al. (2007), Durham-Colleran et al. (2010) | |
| 112 | Homolog of Bvg acc. factor | + | + | + | + | + | ||||
| 383 | fevR/pigR | Transcriptional regulator | + | + | + | + | + | Wehrly et al. (2009) | Charity et al. (2007, 2009) | Brotcke and Monack (2008) |
| 458 | sspA | Stringent starvation protein A regulator | + | + | + | + | + | Charity et al. (2007, 2009) | ||
| 492 | lysR | Homolog of LysR | + | + | + | + | + | |||
| 556 | oxyR | Oxidative stress regulator | + | + | + | + | + | Moule et al. (2010) | ||
| 748 | Homolog of TtgV | + | + | + | + | + | Mortensen et al. (2010) | Mortensen et al. (2010) | Weiss et al. (2007), Mortensen et al. (2010) | |
| 864 | LysR family regulator | + | + | − | − | + | ||||
| 932 | ROK family regulator | − | + | − | + | + | ||||
| 970 | Transcriptional repressor | + | + | + | + | + | ||||
| 977 | MarR-like regulator | + | − | − | − | − | ||||
| 981 | Sensor kinase | + | + | + | + | + | ||||
| 985 | Transcriptional regulator | + | + | + | + | + | ||||
| 1010 | Cro family regulator | − | − | − | − | − | ||||
| 1035 | rpoD | Sigma factor | + | + | + | + | + | Grall et al. (2009) | ||
| 1076 | hipA | Homolog of HipA | − | + | + | + | − | |||
| 1112 | rpoH | Sigma factor | + | + | + | + | + | Grall et al. (2009) | ||
| 1119 | LysR family regulator | + | + | + | + | + | ||||
| 1172 | Cold shock protein | + | + | − | + | − | ||||
| 1202 | LysR family regulator | + | + | − | + | + | ||||
| 1255 | AraC family regulator | + | + | + | + | + | ||||
| 1267 | LysR family regulator | + | + | + | + | + | ||||
| 1275 | mglA | Macrophage growth locus regulator | + | + | + | + | + | Bonquist et al. (2008), Charity et al. (2007, 2009) | Baron and Nano (1998), Brotcke et al. (2006), Guina et al. (2007) | |
| 1281 | rpoN | Sigma factor | − | − | − | − | − | |||
| 1285 | LysR family regulator | + | + | − | + | + | ||||
| 1392 | Homolog of Bvg acc. factor | + | + | + | + | + | ||||
| 1401 | LexA homolog | + | + | + | + | + | ||||
| 1543 | Response regulator | + | + | − | − | + | ||||
| 1557c | pmrA | Response regulator | + | + | + | + | + | Sammons-Jackson et al. (2008) | Bell et al. (2010), Mohapatra et al. (2007) | |
| 1594 | LysR family regulator | + | + | + | + | − | ||||
| 1652 | LysR family regulator | + | + | + | + | + | ||||
| 1613 | Homolog of BolA | − | − | − | − | − | ||||
| 1684 | Transcriptional regulator | + | + | − | + | + | ||||
| 1736 | kdpD | Sensor kinase KdpD | + | + | − | + | + | Bell et al. (2010), Weiss et al. (2007 | ||
RNA Polymerase
Bacteria react to various environmental conditions by employing different modes of regulation. F. tularensis can replicate in a wide range of hosts and cell types, which suggests an extensive ability to adapt to different niches through variable gene expression. Transcription is the process of transcribing DNA into RNA and is performed primarily by a complex of proteins that form the RNAP. RNAP binding involves the alpha subunit recognizing the upstream sequences (40 to −70 base pairs) of the gene, as well as the σ factor recognizing the −10 to −35 regions of the promoter. Francisella RNAP is unique, as it contains two distinct forms of the alpha subunit. Two separate genes encode these proteins and they are both incorporated into the enzyme (Charity et al., 2007). Bacterial σ factors recognize different promoter elements upstream of genes allowing the cell to respond to various environmental conditions by directing the RNAP to unique sets of genes. However, in contrast to Escherichia coli and Bacillus subtilis which have six to eight alternative sigma factors that are used under various circumstances, as mentioned previously, F. tularensis has only two (the housekeeping σ70 and the stress-related σ32).
MglA and SspA
Baron et al. was the first to identify genes that are required for the replication of F. novicida within macrophages, where they described mglA and mglB (Baron and Nano, 1998). As discussed previously, MglA is highly up-regulated upon infection of macrophages. An mglA mutant of F. novicida is unable to escape from macrophage phagosomes and is highly attenuated in the mouse model but does not provide protection against homolgous or heterologous Francisella subsp. challenge (Lauriano et al., 2004; Santic et al., 2005; Brotcke et al., 2006). MglA has been characterized as a transcription factor and is responsible for regulating ∼100 genes including those of the FPI (Lauriano et al., 2004; Brotcke et al., 2006; Guina et al., 2007), while proteomic analysis indicates that the abundance of ∼350 proteins is altered in the absence of MglA. In addition to the FPI, other virulence factors such as the metalloprotease PepO are regulated by MglA (Guina et al., 2007; West et al., 2008). MglA is similar to SspA (Baron and Nano, 1998; Brotcke et al., 2006), a transcription factor in E. coli that responds to nutrient limitation and is involved in regulation of genes contributing to the stringent starvation response. SspA of Francisella is important for the regulation of virulence genes, and the regulons of MglA and SspA are overlapping, including those genes within the FPI (Charity et al., 2007). MglA and SspA physically interact and both bind Francisella RNAP in a heterodimer, which is required for FPI gene activation (Charity et al., 2007).
FevR/PigR
An additional regulator of the FPI that is essential for intramacrophage replication and virulence in the mouse model has been named Francisella effector of virulence regulation or FevR (also called PigR; Buchan et al., 2008; Charity et al., 2009). Microarray analysis comparing fevR/pigR and mglA mutant strains of F. novicida to their parental strains revealed identical regulons for these two transcription factors (Brotcke and Monack, 2008), suggesting that FevR/PigR works in parallel with MglA and SspA to modulate gene expression. FevR/PigR has poor homology to DNA binding proteins, has not been demonstrated to bind to any specific DNA sequences, and appears to act upstream of MglA, SspA, and RNAP binding (Brotcke and Monack, 2008). Studies have varied on whether FevR/PigR, MglA, and SspA physically interact, but recently Charity et al. (2009) demonstrated that ectopic expression of fevR/pigR in LVS restored expression of the MglA/SspA-regulated genes to wild-type levels in a fevR/pigR mutant strain, failed to restore expression of the MglA/SspA-regulated genes in a mglA mutant strain, and partially restored expression of the MglA/SspA-regulated genes in a ΔrelA ΔspoT mutant strain, which suggested that PigR/FevR functions together with the MglA–SspA complex downstream of ppGpp (Charity et al., 2009). Detailed studies of ppGpp and RelA/SpoT gene regulation under stress conditions are described in a later part of this review.
MigR/CaiC
Another regulator, macrophage intracellular growth regulator (MigR, also called CaiC), is a large protein with a putative AMP-binding domain and homology with acyl-transferase/ligase proteins. This regulator was identified from a random pool of LVS mutagenized with the Tn5 transposon as one that regulates the iglABCD and fslABCD operons (Buchan et al., 2009). In F. tularensis LVS, the mutation of migR demonstrated a defect of growth and intracellular trafficking in macrophages (Buchan et al., 2009). However, mutation of migR in the Schu S4 strain retained its regulatory effect but growth inside macrophages was unaffected. MigR positively regulates fevR/pigR, and controls the expression of the FPI genes, iglABCD. The lack of a DNA binding domain in MigR calls into question its role as a transcriptional regulator, instead it seems likely that the regulatory effect of MigR on fevR/pigR is indirect. MigR activity could affect RelA or SpoT (stringent response regulators) to induce the synthesis of (p)ppGpp, which has been demonstrated to contribute to the expression of fevR, and iglABCD, and thus is involved in the pathogenesis of F. tularensis (Charity et al., 2009).
(p)ppGpp
Under nutrient starvation conditions, certain bacterial species resist environmental stress by the production of hyperphosphorylated guanosine diphosphate and triphosphate analogs, collectively termed (p)ppGpp. During this process, uncharged tRNA molecules bind to the ribosome, which results in ribosome stalling, activation of ribosome-associated RelA, and production of (p)ppGpp, which is subsequently converted to ppGpp. These ppGpp small molecules bind to RNAP to affect expression of a wide range of physiological systems. In some bacteria, inactivation of both relA and spoT is required to completely abolish (p)ppGpp synthesis; SpoT is a bifunctional enzyme capable of both synthesis and degradation of the signal molecule, thereby preventing uncontrolled accumulation of (p)ppGpp (Jain et al., 2006). It has been proposed that the RelA-dependent response is linked to amino acid starvation and that the SpoT-dependent response is linked to fatty acid metabolism in bacteria (Battesti and Bouveret, 2009). In some pathogens, regulation of (p)ppGpp synthesis is dependent on SpoT, and virulence in Legionella pneumophila is critically dependent on the degradation of (p)ppGpp by SpoT (Dalebroux et al., 2009). Both relA and spoT genes have been annotated in the F. novicida U112 (Rohmer et al., 2006) and F. tularensis Schu S4 (Larsson et al., 2005; Dean et al., 2009) genome sequences. Inactivation of the relA gene in F. novicida rendered it defective for (p)ppGpp production, and showed increased biofilm formation and reduced resistance to in vitro stress (Dean et al., 2009). The mutant was attenuated in the J774.1 macrophage cell line and in the mouse model, and induced a protective immune response against homologous challenge. The double deletion of relA and spoT in F. tularensis LVS resulted in complete disappearance of (p)ppGpp production and increased the robustness of the relA mutant phenotypes. ppGpp promotes interaction of FevR/PigR with the MglA–SspA complex and RNAP by an unknown mechanism (Charity et al., 2009).
PmrA and KdpD
Bacteria have evolved sensory systems including two-component systems to sense and respond to their immediate surroundings. TCS typically consist of a membrane-bound sensor kinase and a cytoplasmic response regulator whose genes are usually adjacent in a single transcription unit. The communication between the sensor kinase and response regulator involve phosphotransfer at conserved residues.
The F. tularensis genome encodes only two such systems, neither of which is chromosomally paired (Mohapatra et al., 2007). One of these unpaired response regulators is homologous to PmrA in Salmonella enterica, where it is primarily involved in the induction of genes that mediate LPS modification (Gunn, 2008). Inactivation of this gene in F. novicida led to increased bacterial susceptibility to killing by antimicrobial peptides, decreased intramacrophage growth, lack of phagosomal escape in macrophages and attenuation in the mouse model while providing protection against homologous wild-type challenge (Mohapatra et al., 2007). Microarray analysis of this mutant revealed a change in expression of 65 genes when compared to wild-type bacteria grown under the same conditions (Mohapatra et al., 2007). PmrA is a DNA binding protein that is phosphorylated at Asp51 primarily by the unlinked putative histidine kinase KdpD, forming the first TCS described for F. tularensis (Bell et al., 2010). Phosphorylation of PmrA is complicated in F. novicida by the presence of a secondary KdpD target, KdpE, and an additional kinase, FTN1453, both of which negatively affect the phosphorylation state of PmrA. However, neither FTN1453 or KdpE is present/functional in the F. tularensis Schu4 strain, resulting in increased net phosphorylation of PmrA (Bell et al., 2010).
Coimmunoprecipitation assays suggested that PmrA, MglA, and SspA are a part of the same protein complex and likely physically interact (Bell et al., 2010). The pdpD gene is regulated by PmrA, MglA, and SspA; however, pmrA is positively autoregulated by PmrA but not affected by MglA or SspA (Bell et al., 2010). Phosphorylation aids PmrA binding to regulated promoters (Bell et al., 2010). A strain expressing PmrA D51A retains some DNA binding, but exhibits reduced expression of the PmrA-regulon, a deficiency in intramacrophage replication and attenuation in the mouse model. Thus, Francisella gene regulation includes a TCS consisting of KdpD and PmrA. Once phosphorylated, PmrA binds to regulated gene promoters recruiting free or RNAP-bound MglA and SspA to initiate FPI gene transcription. Interestingly, recent published and unpublished data suggest that the gene regulation by PmrA varies between Francisella subsp. and even within different isolates of the same subspecies (Sammons-Jackson et al., 2008). The external signals that are required for KdpD and PmrA two-component system activation remain to be identified but are a focus of current investigation.
Hfq and Small RNA
It is increasingly apparent that in most living organisms, small non-coding RNAs (sRNA) are responsible for wide range of changes in gene expression (Gottesman and Storz, 2010). The regulatory mechanism of these sRNAs is based on pairing with complementary sequences within mRNA messages (Papenfort and Vogel, 2010), though in some cases the regulatory RNA directly affects protein function (Yang et al., 1996). The sRNA–mRNA interaction impacts mRNA stability and translational efficiency. Sometimes sRNA activity is dependent upon interaction with a specific protein, which can aid in stabilization of the regulatory RNA or in targeting to a specific mRNA (Thomas and Schaffner, 2010). One such protein, host factor for bacteriophage Q β replication (Hfq), has been identified in many bacterial species, including F. tularensis, and often is involved in the regulation of a wide range of genes (Meibom et al., 2009). Not surprisingly, mutation of hfq in F. tularensis LVS resulted in pleiotropic effects including an increased sensitivity to elevated salt concentrations, detergents, and high temperatures (Meibom et al., 2009). In addition to these in vitro phenotypes, the hfq mutant was attenuated for growth in several types of macrophages and demonstrated reduced virulence in mice. Microarray and qRT-PCR analysis of the mutant revealed an effect on the expression of 104 genes, including 10 genes within the FPI. Strikingly, Hfq negatively regulates only one of two divergently expressed putative operons in the FPI, in contrast to the other known regulators that appear to affect all FPI-encoded genes. Because of the large size of the Hfq regulon, it is difficult to attribute specific phenotypes of the mutant to specific genes or to assess the impact of reduced FPI gene expression on virulence in this mutant.
A recent study identified several sRNAs in the F. tularensis (LVS) genome (Postic et al., 2010) by in silico predictions and experimental assays. These non-coding regulatory RNAs, include members of well conserved families of structural and housekeeping RNAs as well as less conserved RNAs that may have novel functions/regulate novel targets in Francisella. Thus, the combined work on Hfq and the discovery of these sRNAs suggest a non-protein based means of gene regulation in the Francisella that may somewhat explain the paucity of protein regulatory factors.
Conclusion
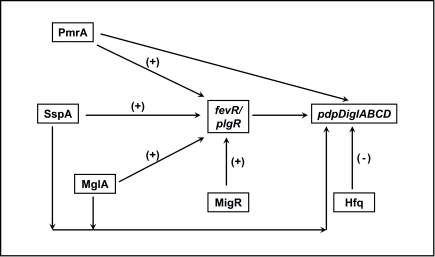
Transcription factors are important regulators of virulence in virtually all known bacterial pathogens. However, virulence gene regulation in Francisella spp. is unique with regard to the fact that it has only one alternative sigma factor, no classically arranged TCS, and the involvement of MglA, SspA, PmrA, MigR, Hfq, sRNA, and FevR/PigR in regulatory control of the FPI. The published literature suggests that MglA, SspA, PmrA, MigR, Hfq, and FevR/PigR co-operate with each other to regulate the Francisella virulence network. Only one identified two-component system exists in virulent Type A F. tularensis, where the sensor kinase KdpD becomes phosphorylated by an unidentified environmental signal and relays that signal to the response regulator PmrA through phosphorylation of D51. PmrA binds to regulated promoters (including those within the FPI) and interacts with the MglA–SspA–RNAP complex to regulate virulence gene transcription (Bell et al., 2010). Microarray data have demonstrated that fevR/pigR is transcriptionally regulated by pmrA, mglA, and sspA (Figure 1). Thus, several environmental cues are likely integrated to activate fevR/pigR, which then plays a key role in FPI-mediated transcription. Additionally, several nutrient starvation and stress response proteins, such as RelA and SpoT (ppGpp), and sRNA species/sRNA carrier proteins are involved in virulence gene regulation in Francisella (see Figure 2 for model). Present and future studies will investigate the molecular interactions between the known FPI regulators and identify new regulators/DNA binding proteins that aid Francisella pathogenesis in various host and non-host niches.
Figure 1.
Interactions between transcriptional regulators of Francisella that control FPI gene expression. PmrA, SspA, MglA, and MigR all positively regulate expression of fevR/pigR. The fevR/pigR product then plays a role, in conjunction with all of the other illustrated factors, in FPI gene transcription. PmrA, MglA, and SspA also directly regulate the genes of the FPI (see Figure 2). (+), Positive transcriptional regulation. (−), Negative transcriptional regulation.
Figure 2.
Model for regulation of Francisella pathogenicity island gene expression. Unknown environmental signals are sensed by KdpD and relayed to the DNA binding protein PmrA. MglA and SspA heterodimerize and associate with RNA polymerase (RNAP). The regulatory activity of the MglA/SspA/RNAP complex is dependent on association of PmrA to the complex as well as the production of ppGpp, which promotes interaction of FevR/PigR with the MglA/SspA/RNAP complex.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by funding from The Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (NIH award 1-U54-AI-057153). We wish to thank Tom Zhart for his contributions to Table 1.
References
- Baron G. S., Nano F. E. (1998). MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol. Microbiol. 29, 247–259 [DOI] [PubMed] [Google Scholar]
- Battesti A., Bouveret E. (2009). Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein–SpoT interaction. J. Bacteriol. 191, 616–624 10.1128/JB.01195-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell B. L., Mohapatra N. P., Gunn J. S. (2010). Regulation of virulence gene transcripts by the Francisella novicida orphan response regulator PmrA: role of phosphorylation and evidence of MglA/SspA interaction. Infect. Immun. 78, 2189–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonquist L., Lindgren H., Golovliov I., Guina T., Sjostedt A. (2008). MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect. Immun. 76, 3502–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotcke A., Monack D. M. (2008). Identification of fevR, a novel regulator of virulence gene expression in Francisella novicida. Infect. Immun. 76, 3473–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotcke A., Weiss D. S., Kim C. C., Chain P., Malfatti S., Garcia E., Monack D. M. (2006). Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect. Immun. 74, 6642–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan B. W., McCaffrey R. L., Lindemann S. R., Allen L. A., Jones B. D. (2009). Identification of migR, a regulatory element of the Francisella tularensis live vaccine strain iglABCD virulence operon required for normal replication and trafficking in macrophages. Infect. Immun. 77, 2517–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan B. W., McLendon M. K., Jones B. D. (2008). Identification of differentially regulated Francisella tularensis genes by use of a newly developed Tn5-based transposon delivery system. Appl. Environ. Microbiol. 74, 2637–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D. S. (1977). Immunization against tularemia: analysis of the effectiveness of live Francisella tularensis vaccine in prevention of laboratory-acquired tularemia. J. Infect. Dis. 135, 55–60 [DOI] [PubMed] [Google Scholar]
- Carlson P. E., Jr., Horzempa J., O'Dee D. M., Robinson C. M., Neophytou P., Labrinidis A., Nau G. J. (2009). Global transcriptional response to spermine, a component of the intramacrophage environment, reveals regulation of Francisella gene expression through insertion sequence elements. J. Bacteriol. 191, 6855–6864 10.1128/JB.00995-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion M. D., Zeng Q., Nix E. B., Nano F. E., Keim P., Kodira C. D., Borowsky M., Young S., Koehrsen M., Engels R., Pearson M., Howarth C., Larson L., White J., Alvarado L., Forsman M., Bearden S. W., Sjostedt A., Titball R., Michell S. L., Birren B., Galagan J. (2009). Comparative genomic characterization of Francisella tularensis strains belonging to low and high virulence subspecies. PLoS Pathog. 5, e1000459. 10.1371/journal.ppat.1000459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charity J. C., Blalock L. T., Costante-Hamm M. M., Kasper D. L., Dove S. L. (2009). Small molecule control of virulence gene expression in Francisella tularensis. PLoS Pathog. 5, e1000641. 10.1371/journal.ppat.1000641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charity J. C., Costante-Hamm M. M., Balon E. L., Boyd D. H., Rubin E. J., Dove S. L. (2007). Twin RNA polymerase-associated proteins control virulence gene expression in Francisella tularensis. PLoS Pathog. 3, e84. 10.1371/journal.ppat.0030084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A., Wehrly T. D., Nair V., Fischer E. R., Barker J. R., Klose K. E., Celli J. (2008). The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect. Immun. 76, 5488–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux Z. D., Edwards R. L., Swanson M. S. (2009). SpoT governs Legionella pneumophila differentiation in host macrophages. Mol. Microbiol. 71, 640–658 [DOI] [PubMed] [Google Scholar]
- Dean R. E., Ireland P. M., Jordan J. E., Titball R. W., Oyston P. C. (2009). RelA regulates virulence and intracellular survival of Francisella novicida. Microbiology 155, 4104–4113 [DOI] [PubMed] [Google Scholar]
- Deng K., Blick R. J., Liu W., Hansen E. J. (2006). Identification of Francisella tularensis genes affected by iron limitation. Infect. Immun. 74, 4224–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienst F. T., Jr. (1963). Tularemia: a perusal of three hundred thirty-nine cases. J. La. State. Med. Soc. 115, 114–127 [PubMed] [Google Scholar]
- Durham-Colleran M. W., Verhoeven A. B., van Hoek M. L. (2010). Francisella novicida forms in vitro biofilms mediated by an orphan response regulator. Microb. Ecol. 59, 457–465 [DOI] [PubMed] [Google Scholar]
- Ellis J., Oyston P. C., Green M., Titball R. W. (2002). Tularemia. Clin. Microbiol. Rev. 15, 631–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson M., Tarnvik A., Kuoppa K., Sandstrom G., Sjostedt A. (1994). Increased synthesis of DnaK, GroEL, and GroES homologs by Francisella tularensis LVS in response to heat and hydrogen peroxide. Infect. Immun. 62, 178–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. E., Gregory D. W., Schaffner W., McGee Z. A. (1985). Tularemia: a 30-year experience with 88 cases. Medicine (Baltimore) 64, 251–269 [PubMed] [Google Scholar]
- Fang F. C. (2004). Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2, 820–832 [DOI] [PubMed] [Google Scholar]
- Fuller J. R., Craven R. R., Hall J. D., Kijek T. M., Taft-Benz S., Kawula T. H. (2008). RipA, a cytoplasmic membrane protein conserved among Francisella species, is required for intracellular survival. Infect. Immun. 76, 4934–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller J. R., Kijek T. M., Taft-Benz S., Kawula T. H. (2009). Environmental and intracellular regulation of Francisella tularensis ripA. BMC Microbiol. 9, 216. 10.1186/1471-2180-9-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovliov I., Ericsson M., Sandstrom G., Tarnvik A., Sjostedt A. (1997). Identification of proteins of Francisella tularensis induced during growth in macrophages and cloning of the gene encoding a prominently induced 23-kilodalton protein. Infect. Immun. 65, 2183–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Storz G. (2010). Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. [Epub ahead of print]. 10.1101/cshperspect.a003798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grall N., Livny J., Waldor M., Barel M., Charbit A., Meibom K. L. (2009). Pivotal role of the Francisella tularensis heat-shock sigma factor RpoH. Microbiology 155, 2560–2572 10.1099/mic.0.029058-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guina T., Radulovic D., Bahrami A. J., Bolton D. L., Rohmer L., Jones-Isaac K. A., Chen J., Gallagher L. A., Gallis B., Ryu S., Taylor G. K., Brittnacher M. J., Manoil C., Goodlett D. R. (2007). MglA regulates Francisella tularensis subsp. novicida (Francisella novicida) response to starvation and oxidative stress. J. Bacteriol. 189, 6580–6586 10.1128/JB.00809-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn J. S. (2008). The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 16, 284–290 10.1016/j.tim.2008.03.007 [DOI] [PubMed] [Google Scholar]
- Horzempa J., Carlson P. E., Jr., O'Dee D. M., Shanks R. M., Nau G. J. (2008). Global transcriptional response to mammalian temperature provides new insight into Francisella tularensis pathogenesis. BMC Microbiol. 8, 172. 10.1186/1471-2180-8-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. T., Mortensen B. L., Taxman D. J., Craven R. R., Taft-Benz S., Kijek T. M., Fuller J. R., Davis B. K., Allen I. C., Brickey W. J., Gris D., Wen H., Kawula T. H., Ting J. P. (2010). Deletion of ripA alleviates suppression of the inflammasome and MAPK by Francisella tularensis. J. Immunol. 185, 5476–5485 10.4049/jimmunol.1002154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V., Kumar M., Chatterji D. (2006). ppGpp: stringent response and survival. J. Microbiol. 44, 1–10 [PubMed] [Google Scholar]
- Johansson A., Celli J., Conlan W., Elkins K. L., Forsman M., Keim P. S., Larsson P., Manoil C., Nano F. E., Petersen J. M., Sjostedt A. (2010). Objections to the transfer of Francisella novicida to the subspecies rank of Francisella tularensis. Int. J. Syst. Evol. Microbiol. 60, 1717–1718; author reply 1718–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss K., Liu W., Huntley J. F., Norgard M. V., Hansen E. J. (2008). Characterization of fig operon mutants of Francisella novicida U112. FEMS Microbiol. Lett. 285, 270–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel M. E., Tilly K. (2000). Temperature-regulated expression of bacterial virulence genes. Microbes Infect. 2, 157–166 10.1016/S1286-4579(00)00272-0 [DOI] [PubMed] [Google Scholar]
- Kugeler K. J., Mead P. S., Janusz A. M., Staples J. E., Kubota K. A., Chalcraft L. G., Petersen J. M. (2009). Molecular epidemiology of Francisella tularensis in the United States. Clin. Infect. Dis. 48, 863–870 [DOI] [PubMed] [Google Scholar]
- Larsson P., Oyston P. C., Chain P., Chu M. C., Duffield M., Fuxelius H. H., Garcia E., Halltorp G., Johansson D., Isherwood K. E., Karp P. D., Larsson E., Liu Y., Michell S., Prior J., Prior R., Malfatti S., Sjostedt A., Svensson K., Thompson N., Vergez L., Wagg J. K., Wren B. W., Lindler L. E., Andersson S. G., Forsman M., Titball R. W. (2005). The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37, 153–159 [DOI] [PubMed] [Google Scholar]
- Lauriano C. M., Barker J. R., Yoon S. S., Nano F. E., Arulanandam B. P., Hassett D. J., Klose K. E. (2004). MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. U.S.A. 101, 4246–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenco J., Hubalek M., Larsson P., Fucikova A., Brychta M., Macela A., Stulik J. (2007). Proteomics analysis of the Francisella tularensis LVS response to iron restriction: induction of the F. tularensis pathogenicity island proteins IglABC. FEMS Microbiol. Lett. 269, 11–21 [DOI] [PubMed] [Google Scholar]
- Lenco J., Link M., Tambor V., Zakova J., Cerveny L., Stulik A. J. (2009). iTRAQ quantitative analysis of Francisella tularensis ssp. holarctica live vaccine strain and Francisella tularensis ssp. tularensis Schu S4 response to different temperatures and stationary phases of growth. Proteomics 9, 2875–2882 10.1002/pmic.200700820 [DOI] [PubMed] [Google Scholar]
- Lenco J., Pavkova I., Hubalek M., Stulik J. (2005). Insights into the oxidative stress response in Francisella tularensis LVS and its mutant DeltaiglC1+2 by proteomics analysis. FEMS Microbiol. Lett. 246, 47–54 [DOI] [PubMed] [Google Scholar]
- LoVullo E. D., Molins-Schneekloth C. R., Schweizer H. P., Pavelka M. S., Jr. (2009). Single-copy chromosomal integration systems for Francisella tularensis. Microbiology 155, 1152–1163 10.1099/mic.0.022491-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoVullo E. D., Sherrill L. A., Perez L. L., Pavelka M. S., Jr. (2006). Genetic tools for highly pathogenic Francisella tularensis subsp. tularensis. Microbiology 152, 3425–3435 10.1099/mic.0.29121-0 [DOI] [PubMed] [Google Scholar]
- McCaffrey R. L., Allen L. A. (2006). Francisella tularensis LVS evades killing by human neutrophils via inhibition of the respiratory burst and phagosome escape. J. Leukoc. Biol. 80, 1224–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey R. L., Schwartz J. T., Lindemann S. R., Moreland J. G., Buchan B. W., Jones B. D., Allen L. A. (2010). Multiple mechanisms of NADPH oxidase inhibition by type A and type B Francisella tularensis. J. Leukoc. Biol. 88, 791–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom K. L., Dubail I., Dupuis M., Barel M., Lenco J., Stulik J., Golovliov I., Sjostedt A., Charbit A. (2008). The heat-shock protein ClpB of Francisella tularensis is involved in stress tolerance and is required for multiplication in target organs of infected mice. Mol. Microbiol. 67, 1384–1401 [DOI] [PubMed] [Google Scholar]
- Meibom K. L., Forslund A. L., Kuoppa K., Alkhuder K., Dubail I., Dupuis M., Forsberg A., Charbit A. (2009). Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infect. Immun. 77, 1866–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra N. P., Soni S., Bell B. L., Warren R., Ernst R. K., Muszynski A., Carlson R. W., Gunn J. S. (2007). Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infect. Immun. 75, 3305–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra N. P., Soni S., Rajaram M. V., Dang P. M., Reilly T. J., El-Benna J., Clay C. D., Schlesinger L. S., Gunn J. S. (2010). Francisella acid phosphatases inactivate the NADPH oxidase in human phagocytes. J. Immunol. 184, 5141–5150 10.4049/jimmunol.0903413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen B. L., Fuller J. R., Taft-Benz S., Kijek T. M., Miller C. N., Huang M. T., Kawula T. H. (2010). Effects of the putative transcriptional regulator IclR on Francisella tularensis pathogenesis. Infect. Immun. 78, 5022–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moule M. G., Monack D. M., Schneider D. S. (2010). Reciprocal analysis of Francisella novicida infections of a Drosophila melanogaster model reveal host–pathogen conflicts mediated by reactive oxygen and imd-regulated innate immune response. PLoS Pathog. 6, e1001065. 10.1371/journal.ppat.1001065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbantoglu U., Sayood K., Dempsey M. P., Iwen P. C., Francesconi S. C., Barabote R. D., Xie G., Brettin T. S., Hinrichs S. H., Fey P. D. (2010). Large direct repeats flank genomic rearrangements between a new clinical isolate of Francisella tularensis subsp. tularensis A1 and Schu S4. PLoS ONE 5, e9007. 10.1371/journal.pone.0009007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano F. E., Schmerk C. (2007). The Francisella pathogenicity island. Ann. N. Y. Acad. Sci. 1105, 122–137 [DOI] [PubMed] [Google Scholar]
- Olakanmi O., Gunn J. S., Su S., Soni S., Hassett D. J., Britigan B. E. (2010). Gallium disrupts iron uptake by intracellular and extracellular Francisella strains and exhibits therapeutic efficacy in a murine pulmonary infection model. Antimicrob. Agents Chemother. 54, 244–253 10.1128/AAC.00655-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K., Vogel J. (2010). Regulatory RNA in bacterial pathogens. Cell Host Microbe 8, 116–127 10.1016/j.chom.2010.06.008 [DOI] [PubMed] [Google Scholar]
- Postic G., Frapy E., Dupuis M., Dubail I., Livny J., Charbit A., Meibom K. L. (2010). Identification of small RNAs in Francisella tularensis. BMC Genomics 11, 625. 10.1186/1471-2164-11-625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan G., Meeker A., Dragulev B. (2008). fslE is necessary for siderophore-mediated iron acquisition in Francisella tularensis Schu S4. J. Bacteriol. 190, 5353–5361 10.1128/JB.00181-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko D. A., Moreira C. G., Li de R., Reading N. C., Ritchie J. M., Waldor M. K., Williams N., Taussig R., Wei S., Roth M., Hughes D. T., Huntley J. F., Fina M. W., Falck J. R., Sperandio V. (2008). Targeting QseC signaling and virulence for antibiotic development. Science 321, 1078–1080 10.1126/science.1160354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratledge C., Dover L. G. (2000). Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54, 881–941 10.1146/annurev.micro.54.1.881 [DOI] [PubMed] [Google Scholar]
- Rohmer L., Brittnacher M., Svensson K., Buckley D., Haugen E., Zhou Y., Chang J., Levy R., Hayden H., Forsman M., Olson M., Johansson A., Kaul R., Miller S. I. (2006). Potential source of Francisella tularensis live vaccine strain attenuation determined by genome comparison. Infect. Immun. 74, 6895–6906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammons-Jackson W. L., McClelland K., Manch-Citron J. N., Metzger D. W., Bakshi C. S., Garcia E., Rasley A., Anderson B. E. (2008). Generation and characterization of an attenuated mutant in a response regulator gene of Francisella tularensis live vaccine strain (LVS). DNA Cell Biol. 27, 387–403 10.1089/dna.2007.0687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M., Al-Khodor S., Abu Kwaik Y. (2010). Cell biology and molecular ecology of Francisella tularensis. Cell. Microbiol. 12, 129–139 [DOI] [PubMed] [Google Scholar]
- Santic M., Molmeret M., Klose K. E., Jones S., Kwaik Y. A. (2005). The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7, 969–979 [DOI] [PubMed] [Google Scholar]
- Schmerk C. L., Duplantis B. N., Wang D., Burke R. D., Chou A. Y., Elkins K. L., Ludu J. S., Nano F. E. (2009). Characterization of the pathogenicity island protein PdpA and its role in the virulence of Francisella novicida. Microbiology 155, 1489–1497 10.1099/mic.0.025379-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulert G. S., McCaffrey R. L., Buchan B. W., Lindemann S. R., Hollenback C., Jones B. D., Allen L. A. (2009). Francisella tularensis genes required for inhibition of the neutrophil respiratory burst and intramacrophage growth identified by random transposon mutagenesis of strain LVS. Infect. Immun. 77, 1324–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer S. A., Harvey M. D., Goodlett D. R., Ernst R. K. (2007). Structural heterogeneity and environmentally regulated remodeling of Francisella tularensis subspecies novicida lipid A characterized by tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 18, 1080–1092 10.1016/j.jasms.2007.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjodin A., Svensson K., Lindgren M., Forsman M., Larsson P. (2010). Whole-genome sequencing reveals distinct mutational patterns in closely related laboratory and naturally propagated Francisella tularensis strains. PLoS ONE 5, e11556. 10.1371/journal.pone.0011556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar E. P. (2010). The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 6, e1000949. 10.1371/journal.ppat.1000949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples J. E., Kubota K. A., Chalcraft L. G., Mead P. S., Petersen J. M. (2006). Epidemiologic and molecular analysis of human tularemia, United States, 1964-2004. Emerg. Infect. Dis. 12, 1113–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A. M., Robinson V. L., Goudreau P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215 10.1146/annurev.biochem.69.1.183 [DOI] [PubMed] [Google Scholar]
- Sullivan J. T., Jeffery E. F., Shannon J. D., Ramakrishnan G. (2006). Characterization of the siderophore of Francisella tularensis and role of fslA in siderophore production. J. Bacteriol. 188, 3785–3795 10.1128/JB.00027-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L. D., Schaffner W. (2010). Tularemia pneumonia. Infect. Dis. Clin. North Am. 24, 43–55 [DOI] [PubMed] [Google Scholar]
- Tigertt W. D. (1962). Soviet viable Pasteurella tularensis vaccines. A review of selected articles. Bacteriol. Rev. 26, 354–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titball R. W., Petrosino J. F. (2007). Francisella tularensis genomics and proteomics. Ann. N. Y. Acad. Sci. 1105, 98–121 [DOI] [PubMed] [Google Scholar]
- Tobe T., Nagai S., Okada N., Adler B., Yoshikawa M., Sasakawa C. (1991). Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol. Microbiol. 5, 887–893 [DOI] [PubMed] [Google Scholar]
- Tobe T., Yoshikawa M., Sasakawa C. (1995). Thermoregulation of virB transcription in Shigella flexneri by sensing of changes in local DNA superhelicity. J. Bacteriol. 177, 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Cherayil B. J. (2009). Ironing out the wrinkles in host defense: interactions between iron homeostasis and innate immunity. J. Innate Immun. 1, 455–464 10.1159/000210016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrly T. D., Chong A., Virtaneva K., Sturdevant D. E., Child R., Edwards J. A., Brouwer D., Nair V., Fischer E. R., Wicke L., Curda A. J., Kupko J. J., 3rd, Martens C., Crane D. D., Bosio C. M., Porcella S. F., Celli J. (2009). Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell. Microbiol. 11, 1128–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D. S., Brotcke A., Henry T., Margolis J. J., Chan K., Monack D. M. (2007). In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U.S.A. 104, 6037–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West T. E., Pelletier M. R., Majure M. C., Lembo A., Hajjar A. M., Skerrett S. J. (2008). Inhalation of Francisella novicida Delta mglA causes replicative infection that elicits innate and adaptive responses but is not protective against invasive pneumonic tularemia. Microbes Infect. 10, 773–780 10.1016/j.micinf.2008.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Liu M. Y., Romeo T. (1996). Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J. Bacteriol. 178, 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]