Abstract
Francisella tularensis is a Gram-negative, intracellular, zoonotic bacterium, and is the causative agent of tularemia with a broad host range. Arthropods such as ticks, mosquitoes, and flies maintain F. tularensis in nature by transmitting the bacteria among small mammals. While the tick is largely believed to be a biological vector of F. tularensis, transmission by mosquitoes and flies is largely believed to be mechanical on the mouthpart through interrupted feedings. However, the mechanism of infection of the vectors by F. tularensis is not well understood. Since F. tularensis has not been localized in the salivary gland of the primary human biting ticks, it is thought that bacterial transmission by ticks is through mechanical inoculation of tick feces containing F. tularensis into the skin wound. Drosophila melanogaster is an established good arthropod model for arthropod vectors of tularemia, where F. tularensis infects hemocytes, and is found in hemolymph, as seen in ticks. In addition, phagosome biogenesis and robust intracellular proliferation of F. tularensis in arthropod-derived cells are similar to that in mammalian macrophages. Furthermore, bacterial factors required for infectivity of mammals are often required for infectivity of the fly by F. tularensis. Several host factors that contribute to F. tularensis intracellular pathogenesis in D. melanogaster have been identified, and F. tularensis targets some of the evolutionarily conserved eukaryotic processes to enable intracellular survival and proliferation in evolutionarily distant hosts.
Keywords: arthropod, vector, tularemia, virulence factor, Drosophila, F. tularensis
Francisella tularensis – An Etiological Agent of the Arthropod-Borne Tularemia
Francisella tularensis is a Gram-negative, intracellular, zoonotic bacterium, and is the causative agent of tularemia (Ellis et al., 2002; Santic et al., 2010). The transmission of F. tularensis to humans is mediated by the bites of arthropods, such as ticks, flies, and mosquitoes, by inhalation, or by handling or ingesting contaminated meat or water (Figure 1; Ellis et al., 2002; Oyston et al., 2004; Santic et al., 2010). F. tularensis is among the most infectious pathogens known. The infective dose in humans is as low as 10 bacteria when injected subcutaneously and 25 bacteria when given as an aerosol (McCrumb, 1961; Saslaw and Carlisle, 1961). Since this bacterium is highly infectious, easily disseminated, and acquired via multiple routes, F. tularensis is one of the six pathogens classified by the CDC as a category A select agent (Dennis et al., 2001; Oyston et al., 2004; Santic et al., 2006).
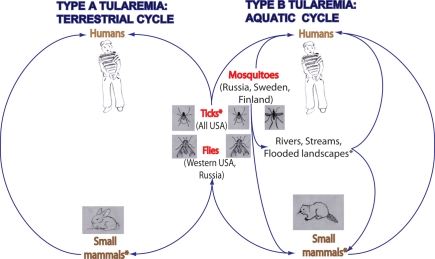
Figure 1.
The role that arthropods play in the transmission of F. tularensis type A and B. Arthropods maintain F. tularensis infections in nature by transmitting F. tularensis between small mammals, such as rabbits and beavers in order to maintain the reservoir. Type A strain of F. tularensis is commonly transmitted by ticks and by tabanid flies, whereas Type B strain is commonly transmitted by ticks, blood-feeding mosquitoes, and by tabanid flies. In USA, bites by ticks are the predominant mode of tularemia transmission. Transmission by biting flies is observed in western regions of the USA and in Russia. Transmission by mosquitoes is observed in the northern countries of Sweden, Finland, and Russia. Classification of hosts of F. tularensis based on the subspecies of F. tularensis associated with them indicates that there are two cycles of F. tularensis: terrestrial and aquatic. Type A has a terrestrial cycle with the main reservoirs being cottontail rabbits and ticks. Type B has mainly a water-borne cycle with semi-aquatic rodents as reservoirs of infection, such as muskrats and beaver in North America, and ground voles in the former Soviet Union. Type B infections have also been associated with rivers, streams, and temporarily flooded landscapes. Asterisk (*) indicates reservoirs of F. tularensis infections.
There are four recognized subspecies of F. tularensis: tularensis, holarctica, mediasiatica, and novicida (Forsman et al., 1994; Keim et al., 2007; Nigrovic and Wingerter, 2008). The four subspecies share about 97% genomic identity (Champion et al., 2009; Larsson et al., 2009). However, classification of novicida as a subspecies is still a matter of debate. Two subspecies of F. tularensis cause most human tularemia infections: subspecies tularensis, also known as type A, and subspecies holarctica, referred to as type B. The subspecies tularensis is the most virulent of F. tularensis ssp. for humans, whereas the subspecies holarctica causes milder infections and lower mortality rates in humans (Nigrovic and Wingerter, 2008). Both tularensis and the holarctica subspecies require level 3 bio-containment (Oyston et al., 2004; Keim et al., 2007; Nigrovic and Wingerter, 2008). The subspecies novicida and holarctica-derived LVS strain are often used to study the pathogenesis by F. tularensis, since they are attenuated in humans, but cause disease in animal models similar to the virulent subspecies (Santic et al., 2010). In addition, both of these attenuated species replicate intracellularly within human and mouse macrophages, an important step in the disease process in mammals (Oyston et al., 2004; Santic et al., 2010).
Arthropods carry disease causing agents and present a major problem worldwide as vectors of human diseases (Kay and Kemp, 1994). Ticks and flies are common arthropod vectors of F. tularensis transmission in the US (Keim et al., 2007). The Type A strain of F. tularensis is commonly transmitted by ticks and by tabanid flies, whereas the Type B strain is commonly transmitted by ticks, tabanid flies, and by blood-feeding mosquitoes (Figure 1; Keim et al., 2007; Nigrovic and Wingerter, 2008).
One major preventive measure to avoid tularemia, as any other arthropod-borne disease, is to use chemical repellants and pesticides in endemic regions (Nigrovic and Wingerter, 2008). However, potential resistance to pesticides and chemical repellant, contamination of food and the environment are major concerns associated with the usage of such pesticides (Kay and Kemp, 1994). Thus, it is desirable to develop alternative effective preventive measures (Kay and Kemp, 1994). One such measure would be to develop a tularemia vaccine (Nigrovic and Wingerter, 2008). Promising novel strategies are being developed to reduce microbial transmission by arthropod vectors. One example is illustrated in the study by McMeniman et al. (2009), who used Wolbachia infection to shorten the life span of the populations of mosquito Aedes aegypti. Shortening the lifespan of these mosquitoes results in fewer cases of mosquito-borne dengue fever illnesses in the human population.
Alternatively, reducing the transmission of vector-borne illness to humans can be achieved indirectly, by including in a vaccine formulation antigens that are important for the successful infection of the pathogen. For instance, the outer membrane lipoprotein A (OSPA) of Borrelia burgdorferi is up regulated and expressed in the tick but not in the mammalian host. However, an OSPA vaccine was shown to have 79% efficacy in a phase III human trial and was an FDA-approved vaccine from 1998 until 2002 (Earnhart et al., 2007). Recently a study has shown that antiserum against salp15, a tick salivary antigen, is protective, and enhances protection of OSPA and OSPC (another B. burgdorferi surface antigen) antiserum in a murine model of Lyme disease (Dai et al., 2009). Therefore, virulence determinants associated with F. tularensis–arthropod vectors might be important in developing vaccine antigens and/or therapeutic measures. Other studies have shown that vaccination against components of the saliva of arthropods or against antigens expressed in the gut of arthropods protected the host from infection and decreased the viability of the arthropod (Titus et al., 2006). In the context of vaccine development, a multi-subunit vaccine that targets F. tularensis itself, as well as components of the arthropod vector, might be worth exploring to control bacterial transmission to humans. This review focuses on Francisella–arthropod interactions, while other reviews related to other aspects of Francisella biology, genetics, physiology, and pathogenesis are included in this special topic issue (Broms et al., 2010; Chong and Celli, 2010; Meibom and Charbit, 2010; Asare and Abu Kwaik, 2011; Bosio, 2011; Cremer et al., 2011; Dai et al., 2011; Jones et al., 2011; Kilmury and Twine, 2011; Telford and Goethert, 2011; Zogaj and Klose, 2011).
Epidemiology of Arthropod-Transmitted Tularemia
Tularemia outbreaks are usually rare and sporadic, and occur as an epidemic both in humans and in animals (Morner, 1992). Workers at increased risk for acquiring tularemia include laboratory workers, landscapers, farmers, veterinarians, hunters, trappers, cooks, and meat handlers (Nigrovic and Wingerter, 2008). There was a recent outbreak of pneumonic tularemia that occurred on Martha's Vineyard (MA, USA) during the summer of 2000, and 11 out of 15 confirmed cases of F. tularensis infection had pneumonia (Feldman et al., 2001). Although the cause of this outbreak was pronounced to be an incident of aerosolized F. tularensis caused by a lawnmower running over the carcass of an infected rabbit (Nigrovic and Wingerter, 2008), the origin of the infections was traced back to transmission by ticks (Keim et al., 2007). Genetic data indicates that the F. tularensis genotype from the landscape worker who contracted fatal Type A tularemia on Martha's Vineyard was a perfect MLVA genotype match for the F. tularensis genotypes obtained from ticks collected in the Squibnocket area on Martha's Vineyard, where he previously worked (Keim et al., 2007).
Arthropods, especially ticks, play a significant role in maintaining F. tularensis infections in nature, often by transmitting F. tularensis between small mammals, such as rabbits and other lagomorphs in order to maintain the reservoir (Figure 1; Francis, 1927; Morner, 1992; Keim et al., 2007; Petersen et al., 2009). Transmission of F. tularensis in nature has been documented in other less prevalent arthropod vectors, including fleas, lice, midges, and bedbugs (Hopla, 1974; Petersen et al., 2009). Geographic differences have been observed for the arthropod vectors transmitting F. tularensis (Keim et al., 2007; Petersen et al., 2009). These differences are linked to the geographic location and abundance of their host species (Petersen et al., 2009), usually small mammals. In the USA, Sweden, Finland, and Russia, arthropod bites, especially by ticks, are a common mode of tularemia transmission to humans (Petersen et al., 2009; Figure 1). Transmission, especially by the deer fly, Chrysops discalis, and by horse flies has been documented in western regions of the USA and Russia (Figure 1). In the Western USA, both deer flies and ticks are considered important vectors, whereas in the Eastern USA, only ticks are considered significant vectors (Petersen et al., 2009). In the USA, tick bites are the predominant mode of transmission (Petersen et al., 2009). The three tick species that are most important for human transmission include Dermacentor andersoni, D. variabilis, and A. americanum. D. variabilis and A. americanum are the two tick species found in regions of the USA reporting the highest incidence of tick-borne tularemia (Arkansas, Missouri, Oklahoma; Petersen et al., 2009). These two tick species have a high affinity for humans, which likely contributes to their success as vectors of tularemia (Parola and Raoult, 2001). In the northern countries of Sweden, Finland, and Russia, mosquitoes have been identified as the major vector transmitting tularemia to humans. In central Europe, contact with infected animals and ingestion of contaminated food or water are the more common modes of transmission in this region rather than arthropod transmission (Hubalek et al., 1996; Tarnvik et al., 2004; Petersen et al., 2009).
Common hosts associated with F. tularensis are rodents, ground squirrels, wild rabbits, semi-aquatic rodents, hares, ticks, tabanid flies, and mosquitoes (reviewed in Nigrovic and Wingerter, 2008). However, F. tularensis is found to be associated with numerous animals, including birds, fish, amphibians, arthropods, and protozoa (Morner, 1992). Hosts that are susceptible to F. tularensis infections include 190 mammals, 88 invertebrates, 23 birds, and 3 amphibians (Keim et al., 2007). Classification of hosts of F. tularensis, based on the subspecies of F. tularensis associated with them, indicates that there are two cycles of F. tularensis: terrestrial and aquatic (Figure 1; Morner, 1992). F. tularensis ssp. tularensis has a terrestrial cycle with the main reservoirs being cottontail rabbits and ticks. Arthropods such as ticks and flies are the most important vectors in this cycle (Morner, 1992; Nigrovic and Wingerter, 2008). F. tularensis ssp. holarctica or type B mainly has a water-borne cycle with semi-aquatic rodents as reservoirs of infection, such as muskrats and beaver in North America, and ground voles in the former Soviet Union (Morner, 1992). As part of this water-borne cycle, mosquitoes have been reported as significant vectors of tularemia in Sweden and Finland (Petersen and Schriefer, 2005; Nigrovic and Wingerter, 2008).
At a cellular level, F. tularensis has been reported to infect and replicate in macrophages of a broad range of mammals, as well as a plethora of other cell types, including fibroblasts, endothelial cells, hepatocytes, and muscle cells (Penn, 2005). Some studies with arthropods show restricted proliferation by F. tularensis in the natural arthropod hosts, the ticks, mosquitoes, and flies (reviewed in Petersen et al., 2009). In the fruit fly, a model of the fly arthropod vector for F. tularensis, the bacteria infect hemocytes (macrophage-like cells), other tissue, and are found in the hemolymph (Vonkavaara et al., 2008; Santic et al., 2009; Moule et al., 2010). A similar observation of infection that spread in diverse arthropod tissues was found in at least some species of ticks, such as D. andersoni, a natural host and vector of tularemia (Francis, 1927). Therefore, more studies are needed to decipher the infection process in the arthropod hosts. Overall, F. tularensis infects a plethora of host species, and arthropod-borne transmission plays an important role in the infectious life cycle of F. tularensis and subsequent pathogenesis in mammalian hosts. Therefore, understanding the interaction of F. tularensis with the arthropod vector at the molecular, cellular, and organismal level will advance our understanding of tularemia and transmission of F. tularensis.
Pathophysiology of Infection with F. tularensis
After infection of humans with F. tularensis, the incubation period is usually 3–6 days (Nigrovic and Wingerter, 2008), which is immediately followed by the onset of the disease (Oyston et al., 2004). Clinical manifestation of tularemia has been classified in two general groups. The ulceroglandular form is associated with systemic symptoms, and is often accompanied by a painful maculopapular lesion at the entry site. The typhoidal form is a severe form of tularemia without the skin or lymph node symptoms, but with gastrointestinal and pulmonary symptoms. The ulceroglandular form is more common and found in approximately 75% of patients, whereas the typhoidal form appears in approximately 25% of patients (Nigrovic and Wingerter, 2008). Although the mortality rate decreases significantly once an effective antibiotic is administered, the mortality rate for untreated pneumonia associated with tularemia can be as high as 60% (Nigrovic and Wingerter, 2008). Pneumonic tularemia occurs in approximately 30% of ulceroglandular tularemia and 80% of typhoidal tularemia (Nigrovic and Wingerter, 2008). Both ulceroglandular and typhoidal tularemia are associated with arthropod transmission of infection, but ulceroglandular tularemia is the most common form associated with an arthropod bite (Petersen et al., 2009). After successful infection, F. tularensis multiplies at the initial site of infection, and then spreads to the regional lymph nodes, liver, and spleen (Oyston et al., 2004; Santic et al., 2006). In small mammals such as guinea pigs, death is observed 3–5 days after infection due to F. tularensis-infected tick bites (Parker et al., 1924; Francis, 1927).
Ticks are established biological vectors of tularemia, as they are responsible for supporting F. tularensis infections in nature, facilitated by their lengthy lifecycle, which is about 2 years (Petersen et al., 2009). The study by Francis in 1927 established the tick D. andersoni as a biological host of F. tularensis. D. andersoni harbors F. tularensis in its feces, epithelial cells of the digestive tract and Malpighian tubules, as well as the coelomic fluid (Francis, 1927). Studies have been performed on ticks after taking a blood meal from F. tularensis-infected guinea pigs. The ticks were incubated for 30 days after the blood meal, dissected, and pathological analysis were conducted microscopically. Anatomical changes observed included the distention of the epithelial cells of the rectal sac, intestines, and Malpighian tubes. Invaded cells are swollen, and contain large numbers of F. tularensis, which are located in the protoplasm. Occasionally, F. tularensis multiplied in the gut wall, cells were swollen, and then ruptured, releasing their contents in a mass, which explains the recovery of F. tularensis from feces of ticks. Other studies have confirmed the localization of F. tularensis in the gut, in the hemolymph, and in excrements of ticks (Vyrostekova, 1993; Petersen et al., 2009). Surprisingly, F. tularensis was not localized in the salivary gland of the tick, suggesting that the transmission of F. tularensis by the tick D. andersoni was mechanically mediated through F. tularensis-containing feces directly into the skin wound (Francis, 1927). To date, F. tularensis infection has never been documented in the salivary glands of the primary human biting ticks (Petersen et al., 2009). In addition, in the bed bug as well, F. tularensis is not isolated from the salivary glands (Francis, 1927). However, one study reported that F. tularensis was localized in the salivary glands of the species D. marginatus, a non-primary biting tick (Hopla, 1974). Although, the transmission rate of F. tularensis to mammalian host by the adult tick is high and of a significant concern, the nymphal stage of this arthropod is not a significant vector of tularemia. A recent study compared the transmission rates among nymphal D. variabilis infected as larvae with wild-type strains of A1b, A2, and type B. As expected, D. variabilis larvae were able to acquire, maintain, and transstadially transmit F. tularensis. Significant replication of the bacteria also occurred in infected nymphs. However, transmission of F. tularensis to Swiss Webster mice was not observed with A1b, and low rates were observed with A2 (8.0%) and type B (13.5%) strains (Reese et al., 2010).
Biting arthropods vectors insert their piercing mouthparts in the host skin, lacerate the skin, and then inject their anticoagulant-containing saliva to prevent blood clot (Atkins, 1978). Biological vectors allow the pathogen to multiply or develop before being transferred to another host, whereas mechanical vectors transmit pathogens to susceptible host without the development of the pathogen, by for instance transferring the pathogen on feet or mouth of the arthropod (Gray and Banerjee, 1999). F. tularensis transmission by mosquitoes and flies is not well understood, but it is believed to be mechanical, on the mouthpart through interrupted feedings. An infected biting fly in nature can transmit tularemia only up to 4 days following its initial infection. In a laboratory setting, F. tularensis is consistently recovered from deer flies for up to 5 days, but no longer than 14 days (Petersen et al., 2009). Similar to deer flies, the mosquito is not believed to support multiplication of F. tularensis (Triebenbach et al., 2010). A recent study indicated that Francisella DNA was detected in 30% of >2,500 mosquitoes captured in Alaska (Triebenbach et al., 2010). However, F. tularensis was not transstadially transmitted in mosquitoes tested. Furthermore, although adult female Anopheles gambiae and Ae. aegypti retained detectable levels of Francisella DNA for 3 days, F. tularensis was not transmitted to the mammalian host by these mosquitoes (Triebenbach et al., 2010). Thus, the absence of F. tularensis in the salivary glands of several arthropods makes a non-biting insect, such as Drosophila melanogaster, a more anatomically and physiologically relevant model of an arthropod vector of tularemia, which could be used to elucidate mechanisms of transmission by arthropod vectors of F. tularensis (Petersen et al., 2009). In D. melanogaster, after pricking (septic injury by needle) and introduction of F. tularensis in the hemolymph, bacteria were observed in the head, legs, and wings veins (Vonkavaara et al., 2008). Intracellular bacteria were localized in the cardia, at the invagination of the esophagus, and in hemocytes. Interestingly, when infection was attempted by oral route, F. tularensis survived in the gastric system for only 24 h after feeding, however the bacteria were cleared thereafter (Vonkavaara et al., 2008).
Drosophila melanogaster is a Tractable Arthropod Model for Tularemia
Drosophila melanogaster has been used as a model in almost every aspect of eukaryotic biology, and we understand more about the biology of this insect than almost any other multicellular organism (Boutros and Perrimon, 2000; Rubin and Lewis, 2000). This knowledge stems from Thomas Morgan's decision in early 1900 to use D. melanogaster as a model to study genetics (Rubin and Lewis, 2000). Interestingly, most biological processes are remarkably similar between flies and vertebrates, such as humans. For instance, sequence searches with 289 human cancer-related genes reveal that 61% of those genes have orthologs in D. melanogaster (Rubin et al., 2000). Conducting biological studies in Drosophila has allowed major scientific milestones in many fields, including microbial pathogenesis (Cherry and Silverman, 2006). Drosophila has been established as a useful model to dissect microbial pathogenesis of some important pathogens, such as Pseudomonas aeruginosa, Mycobacterium marinum, and Listeria monocytogenes, which successfully infect adult fruit flies (review in Cherry and Silverman, 2006). Thus, D. melanogaster is a general attractive model system for microbial pathogenesis. In addition, the signaling pathways regulating innate mammalian immune response are evolutionarily conserved and have similar function in insect immunity (Hoffmann et al., 1999). For instance, in D. melanogaster and in mammals, Toll family receptors (Hoffmann et al., 1999; Anderson, 2000) trigger host innate immune responses that are highly conserved. This conservation makes flies particularly useful for investigation of fundamental biological processes of great relevance to microbial pathogenesis. Furthermore, flies are inexpensive and grow quickly, and many studies have used forward and reverse genetics in Drosophila, which allowed the identification and characterization of many aspects of biological processes that are conserved through evolution.
Drosophila melanogaster is emerging as an attractive arthropod model of infection by F. tularensis and has facilitated the dissection of many processes of F. tularensis pathogenesis. Recent studies have used various arthropods as general models, as well as arthropod vector models of tularemia (Aperis et al., 2007; Read et al., 2008; Vonkavaara et al., 2008; Santic et al., 2009; Ahlund et al., 2010; Akimana et al., 2010; Asare et al., 2010; Moule et al., 2010) For example, the Drosophila-derived cell lines and the sualB cell line from An. gambiae have been used as models to study intracellular replication of F. tularensis (Read et al., 2008; Vonkavaara et al., 2008; Santic et al., 2009; Ahlund et al., 2010; Akimana et al., 2010; Asare et al., 2010). Recent studies have also shown that adult flies could be used as a model system to study Francisella pathogenesis (Vonkavaara et al., 2008; Santic et al., 2009). D. melanogaster is especially an attractive model system to study the pathogenesis of F. tularensis because arthropods are vectors for transmission of tularemia between mammals. This makes the Drosophila model system particularly useful for studying both general F. tularensis host–pathogen interactions and arthropod vector-specific factors.
Bacterial Virulence Factors in the Arthropod Model of Tularemia
To successfully establish a niche in a susceptible host, pathogens use virulence factors to invade, colonize, and survive within the host. After uptake by cells, F. tularensis escapes from the phagosome and propagates in the cytosol (Golovliov et al., 2003; Clemens et al., 2004; Santic et al., 2005b, 2008; McCaffrey and Allen, 2006; Chong et al., 2008). Multiplication results in cell death and release of bacteria (Lai et al., 2001), allowing them to spread to regional lymph nodes and to colonize the spleen, liver, and lung (Tempel et al., 2006). A substantial proportion of the bacterial burden can persist extracellularly in the bloodstream (Forestal et al., 2007; Yu et al., 2008). The virulence factors that are well studied and known to play a role in F. tularensis pathogenesis are involved in lipopolysaccharide biosynthesis or intracellular survival. Most research interest has been on a 30-kb genomic region called the Francisella pathogenicity island (FPI), which has been shown to be required for intracellular replication of F. tularensis within macrophages (Baron and Nano, 1998; Santic et al., 2005b; Bonquist et al., 2008; Schmerk et al., 2009), and which encodes a putative type VI-like secretion system (Nano and Schmerk, 2007; Filloux et al., 2008; Barker et al., 2009).
Studies with pathogenic bacteria in the fly have shown that virulence factors that function in the vertebrate hosts of these pathogens are often required for the pathogen to survive in the fly. Intra-macrophage proliferation is essential for F. tularensis pathogenesis. Similar to macrophages, replication of F. tularensis in S2 and SualB cells is dependent on MglA, MglB, IglA, IglC, IglD, PdpA, and PdpB, which are components or regulators of the FPI (Read et al., 2008; Vonkavaara et al., 2008; Santic et al., 2009). In addition, trafficking and robust intracellular proliferation of F. tularensis ssp. novicida in D. melanogaster-derived S2 cells are similar to trafficking and proliferation in mammalian macrophages (Santic et al., 2009). Within both host cells, F. tularensis transiently occupies a late endosome-like phagosome, followed by rapid bacterial escape into the cytosol, where the bacteria proliferate robustly (Golovliov et al., 2003; Clemens et al., 2004; Santic et al., 2005a,b, 2007, 2008; Checroun et al., 2006; Bonquist et al., 2008; Chong et al., 2008; Qin et al., 2009; Wehrly et al., 2009). This may suggest that some common mechanisms are utilized by F. tularensis to modulate phagosome biogenesis, escape into the cytosol, and to proliferate within mammalian and arthropod-derived cells.
All studies of proliferation of F. tularensis in adult flies indicate that this bacterium grows to high levels within flies and causes a lethal infection (Vonkavaara et al., 2008; Santic et al., 2009; Ahlund et al., 2010; Asare et al., 2010; Moule et al., 2010). F. tularensis kills the fly with a median time to death of 5–12.9 days post-infection, depending on the number of CFUs injected and the strain of F. tularensis used. Extremely high bacterial levels are observed within the fly due to bacteria growing extracellularly (Vonkavaara et al., 2008; Moule et al., 2010). Therefore, screening F. tularensis strains for lethality to D. melanogaster is likely to be an effective approach to identify important bacterial factors involved in arthropod–Francisella interaction. Consistent with this idea is an observation by Ahlund et al. (2010) that there is a significant correlation between fly survival and bacterial proliferation within mammalian cells. Genome-wide screens were conducted to identify factors required for intracellular proliferation within Drosophila-derived cells, and for in vivo growth and survival within the fly (Table 1). It has been shown that ~400 genes, representing 22% of the bacterial genome, are required for intracellular proliferation of F. tularensis within D. melanogaster-derived S2 cells (Asare and Abu Kwaik, 2010). Interestingly, many genes are required for intracellular proliferation in both Drosophila-derived S2 cells and human macrophages (Asare and Abu Kwaik, 2010; Moule et al., 2010). Among 149 F. tularensis ssp. novicida mutants attenuated in the fly, 41 of these mutants (28%) had previously been shown to be attenuated in the mouse model (Weiss et al., 2007a). Among ~250 F. tularensis ssp. novicida mutants that are attenuated in mice, 49 (20%) of them are attenuated in flies (Ahlund et al., 2010). Interestingly, among 168 mutants defective for intracellular growth in S2 cells, 80 are attenuated for lethality to D. melanogaster adult flies (Asare et al., 2010), indicating that >50% of genes required for intracellular proliferation in S2 derived cells play a crucial role in survival of the fly.
Table 1.
A combined list of genes essential for F. tularensis lethality to adult fruit flies.
| Gene loci (U112) | Gene product | Gene | Ahlund et al. (2010) | Asare et al. (2010) | Moule et al. (2010) |
|---|---|---|---|---|---|
| FTN_0019 | Aspartate carbamoyltransferase | pyrB | × | ||
| FTN_0020 | Carbamoyl-phosphate synthase large chain | carB | × | ||
| FTN_0021 | Carbamoyl-phosphate synthase small chain | carA | × | ||
| FTN_0024 | Dihydroorotase | pyrC | × | ||
| FTN_0030 | Hypothetical membrane protein | × | |||
| FTN_0035 | Orotidine-5-phosphate decarboxylase | pyrF | × | × | |
| FTN_0036 | Dihydroorotate dehydrogenase | pyrD | × | × | |
| FTN_0038 | Hypothetical protein | × | |||
| FTN_0051 | Conserved protein of unknown function | × | |||
| FTN_0052 | Protein of unknown function | × | |||
| FTN_0063 | Branched-chain amino acid aminotransferase protein (class IV) | ilvE | * | × | |
| FTN_0066 | Ferrous iron transport protein B | feoB | × | ||
| FTN_0068 | Oligoribonuclease | orn | × | ||
| FTN_0077 | Protein of unknown function | × | |||
| FTN_0078 | Shikimate 5-dehydrogenase | aroE1 | × | ||
| FTN_0090 | Acid phosphatase (precursor) | acpA | × | ||
| FTN_0096 | Conserved hypothetical membrane protein | × | |||
| FTN_0097 | Hydroxy/aromatic amino acid permease (HAAAP) family protein | × | |||
| FTN_0101 | Transcription regulator | × | |||
| FTN_0107 | GTP-binding protein LepA | lepA | × | ||
| FTN_0109 | Protein of unknown function | × | × | ||
| FTN_0111 | Riboflavin synthase beta-chain | ribH | × | ||
| FTN_0113 | Riboflavin synthase alpha chain | ribC | × | ||
| FTN_0115 | Overlaps Na+/H+ antiporter NHAP, fragment | × | × | ||
| FTN_0124 | Single-strand DNA binding protein | ssb | × | ||
| FTN_0141 | ABC transporter, ATP-binding protein | × | |||
| FTN_0162 | Cell division protein FtsQ | ftsQ | × | ||
| FTN_0190 | Major facilitator superfamily (MFS) transport protein, fragment | × | |||
| FTN_0192 | Cytochrome d terminal oxidase, polypeptide subunit II | cydB | × | ||
| FTN_0207 | Protein of unknown function containing a von Willebrand factor type A (vWA) domain | × | |||
| FTN_0214 | Valyl-tRNA synthetase | valS | × | ||
| FTN_0217 | l-lactate dehydrogenase | lldD | × | ||
| FTN_0266 | Chaperone Hsp90, heat shock protein HtpG | htpG | × | ||
| FTN_0275 | Hypothetical protein | × | |||
| FTN_0330 | Septum formation inhibitor-activating ATPase | minD | × | × | |
| FTN_0331 | Septum formation inhibitor | minC | × | ||
| FTN_0337 | Fumarate hydratase, class I | fumA | × | ||
| FTN_0338 | MutT/nudix family protein | × | |||
| FTN_0344 | Aspartate:alanine antiporter | × | |||
| FTN_0346 | OmpA family protein | × | |||
| FTN_0384 | Conserved hypothetical protein | × | |||
| FTN_0392 | Transcriptional regulator, LysR family | × | |||
| FTN_0404 | Peptide methionine sulfoxide reductase | msrB | × | ||
| FTN_0409 | Alcohol dehydrogenase class III, pseudogene | × | |||
| FTN_0412 | DNA repair protein recN | × | |||
| FTN_0416 | Lipid A 1-phosphatase | lpxE | × | ||
| FTN_0429 | Hypothetical protein | × | |||
| FTN_0439 | Protein of unknown function | × | |||
| FTN_0482 | Protein of unknown function | × | |||
| FTN_0493 | 5-Methylthioadenosine/S-adenosylhomocysteine nucleosidase | mtn | × | ||
| FTN_0494 | Hypothetical membrane protein | × | |||
| FTN_0495 | BNR/Asp-box repeat protein | × | |||
| FTN_0496 | Soluble lytic murein transglycosylase | slt | × | ||
| FTN_0504 | Lysine decarboxylase | × | |||
| FTN_0507 | Glycine cleavage system P protein, subunit 1 | gcvP1 | × | ||
| FTN_0513 | 1,4-α-Glucan branching enzyme | glgB | × | ||
| FTN_0516 | Glycogen synthase | glgA | × | ||
| FTN_0525 | Bifunctional aspartokinase/homoserine dehydrogenase I, pseudogene | thrA | × | ||
| FTN_0526 | Homoserine kinase (pseudogene) | thrB | × | ||
| FTN_0538 | Conserved hypothetical membrane protein | × | |||
| FTN_0545 | Glycosyl transferase, group 2 | × | |||
| FTN_0546 | Dolichyl-phosphate-mannose-protein mannosyltransferase family protein | × | |||
| FTN_0549 | Stringent starvation protein A | sspA | × | ||
| FTN_0554 | tRNA/rRNA methyltransferase | yibK | × | ||
| FTN_0567 | tRNA synthetase class II (D, K and N) | × | |||
| FTN_0577 | DNA mismatch repair enzyme with ATPase activity | mutL | × | ||
| FTN_0588 | Asparaginase | × | |||
| FTN_0593 | Succinyl-CoA synthetase, alpha subunit | sucD | × | ||
| FTN_0599 | Conserved hypothetical protein | × | |||
| FTN_0599 | Protein of unknown function | × | |||
| FTN_0600 | DNA gyrase subunit B | gyrB | × | ||
| FTN_0603 | Formamidopyrimidine-DNA glycosylase | mutM | × | ||
| FTN_0623 | 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase | ispD | × | ||
| FTN_0627 | Chitinase, glycosyl hydrolase family 18 | chiA | × | ||
| FTN_0649 | FAD-binding family protein, pseudogene | × | |||
| FTN_0651 | Cytidine deaminase | cdd | × | ||
| FTN_0652 | Uridine phosphorylase | udp | × | ||
| FTN_0653 | tRNA-(ms(2)io(6)a)-hydroxylase | miaE | × | ||
| FTN_0655 | Methylase | × | |||
| FTN_0660 | Cytosol aminopeptidase | pepA | × | ||
| FTN_0664 | Type IV pili fiber building block protein | × | |||
| FTN_0666 | Excinuclease ABC, subunit A | uvrA | × | ||
| FTN_0667 | Major facilitator superfamily (MFS) transport protein | yieO | × | ||
| FTN_0672 | Preprotein translocase, subunit A | secA | × | ||
| FTN_0673 | DNA-3-methyladenine glycosylase I (pseudogene) | tag | × | ||
| FTN_0692 | Quinolinate sythetase A | nadA | × | ||
| FTN_0696 | Hypothetical membrane protein | × | |||
| FTN_0728 | Predicted Co/Zn/Cd cation transporter | × | |||
| FTN_0732 | Hypothetical protein | × | |||
| FTN_0741 | Proton-dependent oligopeptide transporter (POT) family protein, di- or tripeptide:H+ symporter | × | |||
| FTN_0750 | l-Serine dehydratase 1 | sdaA | × | ||
| FTN_0759 | Conserved hypothetical protein | × | |||
| FTN_0760 | Conserved hypothetical protein | × | |||
| FTN_0768 | Tryptophan-rich sensory protein | tspO | × | ||
| FTN_0770 | Major facilitator superfamily (MFS) transport protein, pseudogene | bcr1 | × | ||
| FTN_0772 | Conserved protein of unknown function | × | |||
| FTN_0773 | 4Fe–4S ferredoxin (electron transport) family protein, pseudogene | yjeS | × | ||
| FTN_0774 | Conserved hypothetical protein | × | |||
| FTN_0781 | Transaldolase | talA | × | ||
| FTN_0783 | Isochorismatase hydrolase family protein | × | |||
| FTN_0790 | Recombination associated protein | rdgC | × | ||
| FTN_0791 | Protein of unknown function | × | |||
| FTN_0806 | Glycosyl hydrolase family 3 | × | |||
| FTN_0810 | ROK family protein | × | |||
| FTN_0824 | Major facilitator superfamily (MFS) transport protein, pseudogene | × | |||
| FTN_0826 | Aldo/keto reductase | × | |||
| FTN_0838 | Exodeoxyribonuclease III | xthA | × | ||
| FTN_0840 | Modulator of drug activity B | mdaB | × | × | |
| FTN_0848 | Amino acid antiporter | × | × | ||
| FTN_0855 | Protein of unknown function | × | |||
| FTN_0861 | Type IV pili fiber building block protein | pilA | × | × | |
| FTN_0875 | Major facilitator superfamily (MFS) transport protein | × | |||
| FTN_0877 | Cardiolipin synthetase | cls | × | ||
| FTN_0885 | Proton-dependent oligopeptide transporter (POT) family protein | yhiP | × | ||
| FTN_0886 | Hypothetical membrane protein | × | |||
| FTN_0887 | Hypothetical protein | × | |||
| FTN_0888 | Hypothetical membrane protein | × | |||
| FTN_0889 | Helix-turn-helix family protein | × | |||
| FTN_0891 | Holliday junction DNA helicase, subunit B | ruvB | × | ||
| FTN_0898 | Amino acid permease | × | |||
| FTN_0900 | Protein of unknown function with predicted hydrolase and phosphorylase activity | × | |||
| FTN_0921 | FKBP-type peptidyl-prolyl cis-trans isomerase | × | |||
| FTN_0928 | Sulfate adenylyltransferase subunit 2 | cysD | × | ||
| FTN_0949 | 50S ribosomal protein L9 | rplI | × | ||
| FTN_0954 | Histidine acid phosphatase | × | |||
| FTN_0959 | Oxidative stress transcriptional regulator | oxyR | × | ||
| FTN_0972 | Hypothetical protein | × | |||
| FTN_0975 | Hypothetical protein | × | |||
| FTN_0976 | ThiF family protein, pseudogene | × | |||
| FTN_0978 | Ubiquinone biosynthesis protein | × | |||
| FTN_0982 | Glutaredoxin 1 | grxA | × | ||
| FTN_0984 | ABC transporter, ATP-binding protein | × | |||
| FTN_0997 | Proton-dependent oligopeptide transporter (POT) family protein, di- or tripeptide:H+ symporter | × | |||
| FTN_1006 | Transporter-associated protein, HlyC/CorC family | × | |||
| FTN_1014 | Nicotinamide mononucleotide transport (NMT) family protein | × | |||
| FTN_1016 | Hypothetical protein | × | |||
| FTN_1026 | Major facilitator superfamily (MFS) transport protein, pseudogene | × | |||
| FTN_1027 | Holliday junction endodeoxyribonuclease | ruvC | × | × | |
| FTN_1034 | Iron-sulfur cluster-binding protein | rnfB | × | ||
| FTN_1038 | Conserved hypothetical membrane protein | × | |||
| FTN_1058 | Trigger factor (TF) protein | tig | × | ||
| FTN_1066 | Metal ion transporter protein | × | |||
| FTN_1073 | DNA/RNA endonuclease G | × | |||
| FTN_1091 | 3-Phosphoshikimate 1-carboxyvinyltransferase | aroA | × | ||
| FTN_1099 | Transcriptional regulator, LysR family | × | |||
| FTN_1115 | Type IV pili nucleotide binding protein, ABC transporter, ATP-binding protein | pilB | × | ||
| FTN_1116 | Type IV pili polytopic inner membrane protein | pilC | × | ||
| FTN_1135 | 3-Dehydroquinate synthetase | aroB | × | ||
| FTN_1137 | Type IV pilin multimeric outer membrane protein | pilQ | × | × | |
| FTN_1168 | Exodeoxyribonuclease VII large subunit | xseA | × | ||
| FTN_1170 | Conserved protein of unknown function | × | |||
| FTN_1171 | Conserved hypothetical lipoprotein | × | |||
| FTN_1174 | Glutamate racemase | murI | × | ||
| FTN_1176 | Excinuclease ABC, subunit B | uvrB | × | ||
| FTN_1179 | Transcriptional regulator, LysR family | × | |||
| FTN_1186 | Metallopeptidase family M13 protein, pseudogene | × | × | ||
| FTN_1196 | Conserved hypothetical UPF0133 protein | ybaB | × | ||
| FTN_1198 | Guanosine-3,5-bis(diphosphate) 3-pyrophosphohydrolase/(p)ppGpp synthase | spoT | × | ||
| FTN_1214 | Glycosyl transferase, family 2 | × | |||
| FTN_1215 | Capsule polysaccharide export protein KpsC | kpsC | × | ||
| FTN_1220 | Bacterial sugar transferase family protein | × | |||
| FTN_1223 | Conserved hypothetical membrane protein | × | |||
| FTN_1240 | BolA family protein | × | |||
| FTN_1241 | DedA family protein | × | |||
| FTN_1243 | DNA repair protein recO | recO | × | ||
| FTN_1257 | Membrane protein of unknown function | × | |||
| FTN_1261 | Protein of unknown function | × | |||
| FTN_1268 | Mycobacterial cell entry (mce) related family protein | × | |||
| FTN_1275 | Drug:H+ antiporter-1 (DHA2) family protein | × | |||
| FTN_1276 | Membrane fusion protein | × | |||
| FTN_1278 | NH(3)-dependent NAD(+) synthetase | nadE | × | ||
| FTN_1282 | LysR transcriptional regulator family protein | × | |||
| FTN_1290 | Macrophage growth locus, subunit A | mglA | × | ||
| FTN_1291 | Macrophage growth locus, subunit B | mglB | × | ||
| FTN_1300 | LysR transcriptional regulator family protein | × | |||
| FTN_1309 | Protein of unknown function | pdpA | × | ||
| FTN_1310 | Conserved hypothetical protein; conserved hypothetical protein | pdpB; pdpB1 | × | × | |
| FTN_1311 | Protein of unknown function | iglEa | × | ||
| FTN_1312 | Conserved hypothetical protein | vgrGa | × | ||
| FTN_1313 | Hypothetical protein | iglFa | × | ||
| FTN_1314 | Conserved hypothetical protein | iglGa | × | ||
| FTN_1315 | Protein of unknown function | iglHa | × | ||
| FTN_1316 | Conserved protein of unknown function | dotUa | × | ||
| FTN_1317 | Protein of unknown function | iglIa | × | ||
| FTN_1318 | Hypothetical protein | iglJa | × | ||
| FTN_1319 | Conserved hypothetical protein; conserved hypothetical protein | pdpC | × | × | |
| FTN_1321 | Intracellular growth locus, subunit D; subunit D | iglD; iglD1 | × | × | × |
| FTN_1322 | Intracellular growth locus, subunit C; subunit C1 | iglC; iglC1 | × | * | × |
| FTN_1323 | Conserved protein of unknown function | iglB | × | ||
| FTN_1324 | Conserved protein of unknown function | iglA | × | ||
| FTN_1343 | Conserved protein of unknown function | × | |||
| FTN_1357 | ATP-dependent exoDNAse (exonuclease V) beta subunit | recB | × | × | |
| FTN_1359 | Exodeoxyribonuclease V gamma chain | recC | × | ||
| FTN_1368 | Fe2+ transport system protein A | feoA | × | ||
| FTN_1372 | Protein of unknown function | × | |||
| FTN_1376 | Disulfide bond formation protein, DsbB family | × | |||
| FTN_1386 | Protein of unknown function | × | |||
| FTN_1406 | Conserved hypothetical membrane protein | x | |||
| FTN_1409 | Major facilitator superfamily (MFS) transport protein, pseudogene | × | × | ||
| FTN_1412 | DNA-directed RNA polymerase e subunit | x | |||
| FTN_1417 | Phosphomannomutase | manB | × | × | |
| FTN_1428 | Transferase | wbtO | × | ||
| FTN_1439 | 3-Ketoacyl-CoA thiolase | fadA | × | ||
| FTN_1441 | Sugar transport protein, pseudogene | × | × | ||
| FTN_1448 | Protein of unknown function | × | |||
| FTN_1452 | Two-component response regulator | × | |||
| FTN_1457 | Protein of unknown function | × | |||
| FTN_1465 | Two-component response regulator | pmrA | × | ||
| FTN_1488 | Prophage maintenance system killer protein (DOC) | × | |||
| FTN_1491 | Adenine specific DNA methylase | × | |||
| FTN_1494 | Pyruvate dehydrogenase complex, E1 component, pyruvate dehydrogenase | aceE | × | ||
| FTN_1501 | Monovalent cation:proton antiporter-1 | × | |||
| FTN_1513 | Site-specific recombinase | xerC | × | ||
| FTN_1518 | GDP pyrophosphokinase/GTP pyrophosphokinase | relA | × | ||
| FTN_1530 | Diaminopimelate decarboxylase | lysA | × | ||
| FTN_1534 | Conserved protein of unknown function | × | |||
| FTN_1542 | Conserved protein of unknown function | × | |||
| FTN_1549 | Drug:H+ antiporter-1 (DHA1) family protein | × | |||
| FTN_1552 | Acid phosphatase, PAP2 family | × | |||
| FTN_1557 | Oxidoreductase iron/ascorbate family protein | × | |||
| FTN_1580 | Helicase | × | |||
| FTN_1582 | Hypothetical membrane protein | × | |||
| FTN_1584 | Glycerol-3-phosphate dehydrogenase | glpD | × | ||
| FTN_1593 | ABC-type oligopeptide transport system, periplasmic component | oppA | × | ||
| FTN_1595 | Signal recognition particle receptor FtsY | ftsY | × | ||
| FTN_1599 | Nucleoside permease NUP family protein | nupC | × | ||
| FTN_1600 | Nucleoside permease NUP family protein | nupC1 | × | ||
| FTN_1608 | Disulfide bond formation protein | dsbB | × | ||
| FTN_1611 | Major facilitator superfamily (MFS) transport protein | – | × | ||
| FTN_1612 | Hypothetical protein | × | |||
| FTN_1617 | Sensor histidine kinase | qsec | × | ||
| FTN_1618 | Conserved hypothetical protein | × | |||
| FTN_1621 | Predicted NAD/FAD-dependent oxidoreductase | × | |||
| FTN_1630 | Preprotein translocase, subunit G, membrane protein | secG | × | ||
| FTN_1654 | Major facilitator superfamily (MFS) transport protein | × | |||
| FTN_1657 | Major facilitator superfamily (MFS) transport protein | × | |||
| FTN_1658 | Histidyl-tRNA synthetase | hisS | × | ||
| FTN_1678 | NADH dehydrogenase I, C subunit | nuoC | × | ||
| FTN_1682 | Conserved hypothetical protein | × | |||
| FTN_1683 | Drug:H+ antiporter-1 (DHA1) family protein | × | |||
| FTN_1685 | Drug:H+ antiporter-1 (DHA1) family protein | × | |||
| FTN_1692 | Secretion protein | × | |||
| FTN_1704 | Protein-l-isoaspartate O-methyltransferase | pcm | × | ||
| FTN_1714 | Transcriptional regulatory protein, pseudogene | kdpE | × | ||
| FTN_1715 | Two-component sensor protein | kdpD | × | ||
| FTN_1716 | Potassium-transporting ATPase C chain | kdpC | × | ||
| FTN_1718 | Potassium-transporting ATPase, A chain, pseudogene | kdpA | × | ||
| FTN_1719 | NAD-dependent formate dehydrogenase, fragment | × | |||
| FTN_1733 | Conserved hypothetical membrane, pseudogene | × | |||
| FTN_1743 | Chaperone ClpB | clpB | × | ||
| FTN_1745 | Phosphoribosylglycinamide formyltransferase 2 | purT | × | ||
| FTN_1750 | Acyltransferase | × | |||
| FTN_1753 | Oxidase-like protein, pseudogene | × | |||
| FTN_1762 | ABC transporter, ATP-binding protein | yjjK | × | ||
| FTN_1763 | Major facilitator superfamily (MFS) transport protein | × | |||
| FTN_1776 | Anthranilate synthase component II, pseudogene | trpG1 | × |
An × marks a screen, where the gene was identified to be essential for lethality to flies, whereas * marks a gene not essential for lethality to flies, but the gene is important for reduction of bacterial load in the indicated screen (mentioned because it was found in at least one other screen). Genes found in multiple screens are shown in a bold font.
Overall, F. tularensis grows in large numbers in D. melanogaster resulting in lethality, similar to mammals (Vonkavaara et al., 2008; Santic et al., 2009; Ahlund et al., 2010; Asare et al., 2010; Moule et al., 2010). In addition, contrasting studies in flies to those in mammalian models (Ahlund et al., 2010; Asare et al., 2010; Moule et al., 2010) indicates that F. tularensis might have acquired some of the mechanisms to proliferate within mammalian cells through patho-adaptation to the arthropod host. Some of the virulence factors that have been possibly acquired through patho-adaptation in insect hosts include most genes of the FPI. However, additional distinct molecular mechanisms are also required for proliferation within both evolutionarily distant hosts, as numerous factors important for infectivity of D. melanogaster are not required for infectivity of mammalian hosts and vice versa (Ahlund et al., 2010; Asare et al., 2010).
Host Virulence Determinants for Intracellular Proliferation of F. tularensis in the Arthropod-Derived Cells
To reduce transmission and morbidity associated with arthropod-borne tularemia, not only bacterial factors are important, but also arthropod host factors can be used to develop therapeutic measures against F. tularensis. For example, it has been shown in Lyme disease that a tick antigen Salp15, a salivary gland protein, can be a protective immunogen to some degree, and can be used to enhance the potency of a bacterial vaccine antigen OSPA (Dai et al., 2009).
Like other intracellular bacterial pathogens, F. tularensis has evolved varying strategies to avoid being attacked by the host macrophages (Aderem and Underhill, 1999). Within mammalian and arthropod-derived cells, F. tularensis escapes the acidified late endosome-like phagosome to reach the host cell cytosol, where replication occurs (Santic et al., 2009). Therefore, it is reasonable to assume that F. tularensis targets evolutionarily conserved eukaryotic factors for intracellular survival and growth. Some of the strategies to evade the host defense efforts by F. tularensis involve its ability to modulate the host cellular and molecular machinery. While several bacterial determinants that facilitate intracellular infection by F. tularensis have been characterized (Asare et al., 2010; Moule et al., 2010), such as genes of the FPI, less is known about the host factors that are exploited or subverted by F. tularensis.
Some of the immune system processes are known to be manipulated by F. tularensis to avoid being attacked by the host. For instance, F. tularensis ssp. novicida delays inflammasome activation (reviewed in Weiss et al., 2007b). However, until recently there has been no comprehensive genome-wide analysis that has been conducted to identify all host genes that are important for F. tularensis infection. Since until recently it has been difficult to conduct extensive genetic manipulation in the mammalian hosts, many investigators have used D. melanogaster to model microbial diseases (Cherry, 2008). The genetic tractability of Drosophila has enabled the identification of host-encoded factors that affect the pathogen–host interaction at both the cellular and molecular levels in many pathogens, such as L. monocytogenes, M. marinum, and Legionella pneumophila (Dionne et al., 2003; Cheng et al., 2005; Dorer et al., 2006). It has also been shown that infection of D. melanogaster cells by intracellular bacterial pathogens is similar to infection of mammalian host cells. Thus, it is likely that the intracellular infection requires conserved host factors in mammals and arthropods.
In contrast to many other pathogens for which D. melanogaster has been used to identify host factors required for the pathogen–host interaction (Cherry, 2008), F. tularensis is naturally transmitted to mammalian hosts by arthropod vectors. While many pathogens can only be transmitted by a single specific arthropod vector species, F. tularensis is associated with various arthropods ranging from ticks to multiple species of mosquitoes to biting flies such as deer flies. This makes the D. melanogaster model system particularly useful for studying both general F. tularensis host–pathogen interaction and insect-specific factors. Thus, we can expect that F. tularensis targets many insect specific factors that D. melanogaster is likely to harbor.
A recent study has used a genome-wide RNAi screen to identify host factors that contribute to intracellular proliferation of F. tularensis within D. melanogaster-derived cells. In this screen at least 186 host factors have been shown to be required for intracellular bacterial proliferation (Akimana et al., 2010). The discovery of these genes initiated studies to uncover host processes that are likely important in the arthropod vector. The predominant functional category of the host factors identified in the screen are involved in signal transduction, indicating that F. tularensis modulate many host signaling molecules for its own advantage (Hrstka et al., 2005; Al-Khodor and Abu Kwaik, 2010). Silencing mammalian homolog of the factors identified in the D. melanogaster RNAi screen shows that four conserved factors are also required for replication of F. tularensis in human cells (Table 2): the Ras/Rho guanyl-nucleotide exchange factor activity SOS2, the PI4 kinase PI4KCA, the ubiquitin hydrolase USP22, and the ubiquitin ligase CDC27 (Akimana et al., 2010; Al-Khodor and Abu Kwaik, 2010). Furthermore, one of these evolutionally conserved factors, the CDC27 ubiquitin ligase, is required for evading lysosomal fusion and for bacterial escape into the cytosol (Akimana et al., 2010).
Table 2.
List of evolutionally conserved host factors involved in intracellular trafficking of F. tularensis in both D. melanogaster and human cells.
| Category | Description | Drosophila Melanogaster gene | Human homolog gene |
|---|---|---|---|
| Cell cycle | Mitosis | cdc27 | CDC27*a |
| Proteolysis | Ubiquitin thiolesterase activity | not | USP22a |
| Signal transduction | Ras/Rho guanyl-nucleotide exchange factor activity | sos | SOS2b |
| Signal transduction | 1-Phosphatidylinositol 4-kinase activity | CG10260 | PI4KCAa |
The SOS2 mammalian host factor and its arthropod homolog sos has been shown to be important for proliferation of F. tularensis in S2 cells and human cells. Intracellular F. tularensis ssp. novicida triggers temporal and early activation of Ras through the SOS2/GrB2/ PKCα/PKCβI, and that this signaling cascade is essential for intracellular bacterial proliferation within the cytosol, and associated with down-regulation of early caspase-3 activation, which promotes survival of the infected cells (Al-Khodor and Abu Kwaik, 2010). Thus, using D. melanogaster as a model, host factors important for F. tularensis intracellular proliferation in the arthropod host have been identified, and some are conserved in mammalian cells (Table 2).
Environmental Factors Relevant to Arthropod-Mammalian Adaptation
Although this review focuses largely on the genes required for arthropod and mammalian infection, other important studies identified some environmental factors that are relevant to arthropod–mammalian transition of F. tularensis. Horzempa et al. (2008) has examined the impact of arthropod-like versus mammalian-like temperatures, 26°C versus 37°C, respectively on gene regulation of F. tularensis. Interestingly, they found that the FPI genes pdpC, iglC, and iglD were down-regulated at 26°C (Horzempa et al., 2008), yet these genes are required for F. tularensis survival in D. melanogaster as shown in Table 1 (Asare et al., 2010; Moule et al., 2010). However, pdpC, which is significantly down-regulated in arthropod-like temperature, is dispensable for infection of Sua1B mosquito-derived cells (Read et al., 2008; Vonkavaara et al., 2008; Santic et al., 2009). It will be interesting to test whether pdpC has a similar role in F. tularensis as the role of OspC in B. burgdorferi; i.e., requirement for initial mammalian infection (Schwan et al., 1995; Tilly et al., 2006). Alternatively, FTL_1581, a hypothetical lipoprotein induced by mammalian temperature (Horzempa et al., 2008) could have a similar role as OspC. In addition, F. tularensis ssp. novicida has been shown to alter its outer membrane at 25 versus 37°C by differentially modifying the lipid A component of the lipopolysaccharide, but this modification does not alter the virulence of F. tularensis (Shaffer et al., 2007). Another interesting environment factor is that spermine and spermidine are novel triggers to alert F. tularensis of its eukaryotic host environment (Carlson et al., 2009). All these differences in mammalian-like versus arthropod-like conditions observed reveal bits of patho-adaptation by F. tularensis in arthropods and human that still needs to be elucidated. However, it is important to note that the temperature is only one variable between the environments of the two hosts and that the actual composition of the environments and the host–microbe interaction within these distinct hosts are much more complex than just the temperature variable.
Concluding Remarks and Future Directions
Arthropod-borne transmission of F. tularensis is responsible for maintaining tularemia in nature and is of significant concern worldwide. So far, there are many unanswered questions pertaining to F. tularensis–arthropod vector interaction and its role in patho-adaptation to infect mammals. The study of arthropod vectors–F. tularensis interaction or comparing these studies to mammalian studies helps us understand patho-adaption aspect of this bacterium in its diverse hosts.
Outbreaks of F. tularensis are connected to the arthropod transmission. Thus, it is desirable to develop strategies to reduce arthropod vector transmission of tularemia. Francis showed that the transmission of F. tularensis occurs through the tick feces rather than through the salivary gland, unlike other blood-feeding arthropods, such as Lyme disease transmitting ticks. Although ticks transmit F. tularensis transovarial, one other possibility is transmission of this pathogen from one tick developmental stage to the other through feces, which is a frequent method of transmission in small mammals and birds.
While mammals such as guinea pigs, mice, and humans are very susceptible to F. tularensis infections, arthropod vectors that are natural host of F. tularensis are able to limit the severity of infection by F. tularensis. It will be interesting to identify factors underlining the difference in theses two evolutionary distant hosts. Many bacterial factors are required for intracellular proliferation within both arthropod-derived and human-derived cells. In addition, many eukaryotic host factors conserved in arthropods and mammals are required for intracellular proliferation of F. tularensis within the two evolutionarily distant hosts. Therefore, it is likely that patho-adaptation of F. tularensis in the arthropod vector has allowed this bacterium to successfully infect the human host.
Many studies to date have utilized D. melanogaster as a general model and have shown that it is a tractable genetic arthropod vector model of tularemia. A unique advantage of using D. melanogaster as a model of F. tularensis is that F. tularensis infections are transmitted to mammalian hosts by at least three established arthropod vectors: ticks, biting flies, and mosquitoes, whereas in almost all other arthropod-borne diseases, only one arthropod vector is solely responsible for transmitting the disease. Studies utilizing the well studied and genetically tractable model D. melanogaster, are likely to help us understand the arthropod host, since F. tularensis likely uses similar virulence strategies to infect its diverse arthropods hosts. However, additional studies are needed to fully establish D. melanogaster as a vector model to decipher F. tularensis–arthropod vector interaction.
As shown in Table 1, three large-scale screens using F. tularensis transposon insertion mutants have led to the rapid identification of 250 different genes required for F. tularensis in vivo infection of D. melanogaster. Overall, there is a poor overlap between hits identified in these studies. The FPI genes iglB, iglC, iglD, and mglA have been previously identified to be required for F. tularensis infection of the D. melanogaster and were expected to become hits in all theses screens, but only iglC and iglD were identifies by all the screens. These results are not surprising since an inherent problem of large-scale screens is the presence of false positive and false negative hits. In addition, transposon mutants might not exhibit a loss of function phenotype. These results suggest that this overwhelming amount of data need to be analyzed by first looking at overlapping information from these studies; and also suggest that even non-overlapping data is essential and should be further analyzed as well.
Interestingly, studies with the insect model D. melanogaster have shown that the FPI genes are equally important in the arthropod models. Furthermore, a large number of bacterial factors are required for proliferation within both D. melanogaster and mammalian cells. Since the tick is a major vector of F. tularensis infections, future studies should determine the role of these factors in the tick.
To reduce transmission and morbidity associated with arthropod-borne tularemia, not only bacterial factors are important, but also arthropod host factors may be used to develop therapeutic measures against F. tularensis. Recent microbial pathogenesis studies are uncovering more about bacterial effectors that modulate important host processes. Numerous Drosophila genes that are essential for F. tularensis infection have been identified. To further confirm the role of arthropod host genes play in the pathogenesis by F. tularensis, one could study and screen for host genes important for F. tularensis virulence in vivo in the arthropod hosts by using D. melanogaster mutants defective in host genes essential for F. tularensis virulence. It has also been shown that F. tularensis targets some evolutionarily conserved host factors for intracellular survival and growth. Determining whether other D. melanogaster genes have mammalian homologs, and whether these homologs are also involved in intracellular infection or other biological function, will be at the crux of our understanding of bacteria–arthropod interaction and its role in patho-adaptation to infect mammals. In the future, bioinformatics studies should facilitate the dissection of biochemical pathways that are important for F. tularensis infection by using both bacterial and host genes shown to date to be essential for F. tularensis in the arthropod host. The accumulated knowledge of vector–F. tularensis interactions will ultimately allow the development of strategies to prevent and treat tularemia.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Yousef Abu Kwaik is supported by Public Health Service Awards R01AI43965 and R01AI069321 from National Institute of Allergy and Infectious Diseases (NIAID) and by the Commonwealth of Kentucky Research Challenge Trust Fund. We thank Tasneem Al-Quadan for assistance. We also thank Snake Jones and Rexford Asare for proofreading the manuscript.
References
- Aderem A., Underhill D. M. (1999). Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 17, 593–623 10.1146/annurev.immunol.17.1.593 [DOI] [PubMed] [Google Scholar]
- Ahlund M. K., Ryden P., Sjostedt A., Stoven S. (2010). Directed screen of Francisella novicida virulence determinants using Drosophila melanogaster. Infect. Immun. 78, 3118–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimana C., Al-Khodor S., Abu Kwaik Y. (2010). Host factors required for modulation of phagosome biogenesis and proliferation of Francisella tularensis within the cytosol. PLoS ONE 5, e11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khodor S., Abu Kwaik Y. (2010). Triggering Ras signalling by intracellular Francisella tularensis through recruitment of PKCalpha and betaI to the SOS2/GrB2 complex is essential for bacterial proliferation in the cytosol. Cell. Microbiol. 12, 1604–1621 [DOI] [PubMed] [Google Scholar]
- Anderson K. V. (2000). Toll signaling pathways in the innate immune response. Curr. Opin. Immunol. 12, 13–19 [DOI] [PubMed] [Google Scholar]
- Aperis G., Fuchs B. B., Anderson C. A., Warner J. E., Calderwood S. B., Mylonakis E. (2007). Galleria mellonella as a model host to study infection by the Francisella tularensis live vaccine strain. Microbes Infect. 9, 729–734 10.1016/j.micinf.2007.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asare R., Abu Kwaik Y. (2010). Molecular complexity orchestrates modulation of phagosome biogenesis and escape to the cytosol of macrophages by Francisella tularensis. Environ. Microbiol. 12, 2559–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asare R., Abu Kwaik Y. (2011). Exploitation of host cell biology and evasion of immunity by Francisella tularensis. Front. Microbio. 1:145. 10.3389/fmicb.2010.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asare R., Akimana C., Jones S., Abu Kwaik Y. (2010). Molecular bases of proliferation of Francisella tularensis in arthropod vectors. Environ. Microbiol. 12, 2587–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins M. D. (1978). Insects in Perspective. New York: Prentice Hall, 359 [Google Scholar]
- Barker J. R., Chong A., Wehrly T. D., Yu J. J., Rodriguez S. A., Liu J., Celli J., Arulanandam B. P., Klose K. E. (2009). The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol. Microbiol. 74, 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron G. S., Nano F. E. (1998). MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol. Microbiol. 29, 247–259 [DOI] [PubMed] [Google Scholar]
- Bonquist L., Lindgren H., Golovliov I., Guina T., Sjostedt A. (2008). MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect. Immun. 76, 3502–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio C. M. (2011). The subversion of the immune system by Francisella tularensis. Front. Microbio. 2:9. 10.3389/fmicb.2011.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M., Perrimon N. (2000). Drosophila genome takes flight. Nat. Cell Biol. 2, E53–E54 [DOI] [PubMed] [Google Scholar]
- Broms J. E., Sjostedt A., Lavander M. (2010). The role of the Francisella tularensis pathogenicity island in type VI secretion, intracellular survival, and modulation of host cell signalling. Front. Microbio. 1:136. 10.3389/fmicb.2010.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson P. E., Jr., Horzempa J., O'Dee D. M., Robinson C. M., Neophytou P., Labrinidis A., Nau G. J. (2009). Global transcriptional response to spermine, a component of the intramacrophage environment, reveals regulation of Francisella gene expression through insertion sequence elements. J. Bacteriol. 191, 6855–6864 10.1128/JB.00995-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion M. D., Zeng Q., Nix E. B., Nano F. E., Keim P., Kodira C. D., Borowsky M., Young S., Koehrsen M., Engels R., Pearson M., Howarth C., Larson L., White J., Alvarado L., Forsman M., Bearden S. W., Sjostedt A., Titball R., Michell S. L., Birren B., Galagan J. (2009). Comparative genomic characterization of Francisella tularensis strains belonging to low and high virulence subspecies. PLoS Pathog. 5, e1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checroun C., Wehrly T. D., Fischer E. R., Hayes S. F., Celli J. (2006). Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. U.S.A. 103, 14578–14583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L. W., Viala J. P., Stuurman N., Wiedemann U., Vale R. D., Portnoy D. A. (2005). Use of RNA interference in Drosophila S2 cells to identify host pathways controlling compartmentalization of an intracellular pathogen. Proc. Natl. Acad. Sci. U.S.A. 102, 13646–13651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S. (2008). Genomic RNAi screening in Drosophila S2 cells: what have we learned about host-pathogen interactions? Curr. Opin. Microbiol. 11, 262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S., Silverman N. (2006). Host-pathogen interactions in Drosophila: new tricks from an old friend. Nat. Immunol. 7, 911–917 [DOI] [PubMed] [Google Scholar]
- Chong A., Celli J. (2010). The Francisella intracellular life cycle: towards molecular mechanisms of intracellular survival and proliferation. Front. Microbio. 1:138. 10.3389/fmicb.2010.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A., Wehrly T. D., Nair V., Fischer E. R., Barker J. R., Klose K. E., Celli J. (2008). The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect. Immun. 76, 5488–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens D. L., Lee B. Y., Horwitz M. A. (2004). Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 72, 3204–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T. J., Butchar J., Tridandapani S. (2011). Francisella subverts innate immune signaling: focus on PI3K/Akt. Front. Microbio. 2:13. 10.3389/fmicb.2011.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Wang P., Adusumilli S., Booth C. J., Narasimhan S., Anguita J., Fikrig E. (2009). Antibodies against a tick protein, Salp15, protect mice from the Lyme disease agent. Cell Host Microbe 6, 482–492 10.1016/j.chom.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S., Mohapatra N. P., Schlesinger L. S., Gunn J. S. (2011). Regulation of Francisella tularensis virulence. Front. Microbio. 1:144. 10.3389/fmicb.2010.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis D. T., Inglesby T. V., Henderson D. A., Bartlett J. G., Ascher M. S., Eitzen E., Fine A. D., Friedlander A. M., Hauer J., Layton M., Lillibridge S. R., McDade J. E., Osterholm M. T., O'Toole T., Parker G., Perl T. M., Russell P. K., Tonat K. (2001). Tularemia as a biological weapon: medical and public health management. JAMA 285, 2763–2773 10.1001/jama.285.21.2763 [DOI] [PubMed] [Google Scholar]
- Dionne M. S., Ghori N., Schneider D. S. (2003). Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum. Infect. Immun. 71, 3540–3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer M. S., Kirton D., Bader J. S., Isberg R. R. (2006). RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog. 2, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnhart C. G., Buckles E. L., Marconi R. T. (2007). Development of an OspC-based tetravalent, recombinant, chimeric vaccinogen that elicits bactericidal antibody against diverse Lyme disease spirochete strains. Vaccine 25, 466–480 10.1016/j.vaccine.2006.07.052 [DOI] [PubMed] [Google Scholar]
- Ellis J., Oyston P. C., Green M., Titball R. W. (2002). Tularemia. Clin. Microbiol. Rev. 15, 631–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman K. A., Enscore R. E., Lathrop S. L., Matyas B. T., McGuill M., Schriefer M. E., Stiles-Enos D., Dennis D. T., Petersen L. R., Hayes E. B. (2001). An outbreak of primary pneumonic tularemia on Martha's Vineyard. N. Engl. J. Med. 345, 1601–1606 [DOI] [PubMed] [Google Scholar]
- Filloux A., Hachani A., Bleves S. (2008). The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154, 1570–1583 10.1099/mic.0.2008/016840-0 [DOI] [PubMed] [Google Scholar]
- Forestal C. A., Malik M., Catlett S. V., Savitt A. G., Benach J. L., Sellati T. J., Furie M. B. (2007). Francisella tularensis has a significant extracellular phase in infected mice. J. Infect. Dis. 196, 134–137 [DOI] [PubMed] [Google Scholar]
- Forsman M., Sandstrom G., Sjostedt A. (1994). Analysis of 16S ribosomal DNA sequences of Francisella strains and utilization for determination of the phylogeny of the genus and for identification of strains by PCR. Int. J. Syst. Bacteriol. 44, 38–46 10.1099/00207713-44-1-38 [DOI] [PubMed] [Google Scholar]
- Francis E. (1927). Microscopic changes of tularemia in the tick Dermacentor andersoni and the bedbug Cimex lectularius. Public Health Rep. 42, 2763–277219315112 [Google Scholar]
- Golovliov I., Baranov V., Krocova Z., Kovarova H., Sjostedt A. (2003). An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 71, 5940–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S. M., Banerjee N. (1999). Mechanisms of arthropod transmission of plant and animal viruses. Microbiol. Mol. Biol. Rev. 63, 128–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann J. A., Kafatos F. C., Janeway C. A., Ezekowitz R. A. (1999). Phylogenetic perspectives in innate immunity. Science 284, 1313–1318 10.1126/science.284.5418.1313 [DOI] [PubMed] [Google Scholar]
- Hopla C. E. (1974). The ecology of tularemia. Adv. Vet. Sci. Comp. Med. 18, 25–53 [PubMed] [Google Scholar]
- Horzempa J., Carlson P. E., Jr., O'Dee D. M., Shanks R. M., Nau G. J. (2008). Global transcriptional response to mammalian temperature provides new insight into Francisella tularensis pathogenesis. BMC Microbiol. 8, 172. 10.1186/1471-2180-8-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrstka R., Stulik J., Vojtesek B. (2005). The role of MAPK signal pathways during Francisella tularensis LVS infection-induced apoptosis in murine macrophages. Microbes Infect. 7, 619–625 [DOI] [PubMed] [Google Scholar]
- Hubalek Z., Treml F., Halouzka J., Juricova Z., Hunady M., Janik V. (1996). Frequent isolation of Francisella tularensis from Dermacentor reticulatus ticks in an enzootic focus of tularaemia. Med. Vet. Entomol. 10, 241–246 [DOI] [PubMed] [Google Scholar]
- Jones J. W., Broz P., Monach D. S. (2011). Innate immune recognition of Francisella tularensis: activation of type-I interferons and the inflammasome. Front. Microbio. 2:16. 10.3389/fmicb.2011.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay B. H., Kemp D. H. (1994). Vaccines against arthropods. Am. J. Trop. Med. Hyg. 50, 87–96 [DOI] [PubMed] [Google Scholar]
- Keim P., Johansson A., Wagner D. M. (2007). Molecular epidemiology, evolution, and ecology of Francisella. Ann. N. Y. Acad. Sci. 1105, 30–66 [DOI] [PubMed] [Google Scholar]
- Kilmury S. N., Twine S. M. (2011). The Francisella tularensis proteome and its recognition by antibodies. Front. Microbio. 1:143. 10.3389/fmicb.2010.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X. H., Golovliov I., Sjostedt A. (2001). Francisella tularensis induces cytopathogenicity and apoptosis in murine macrophages via a mechanism that requires intracellular bacterial multiplication. Infect. Immun. 69, 4691–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P., Elfsmark D., Svensson K., Wikstrom P., Forsman M., Brettin T., Keim P., Johansson A. (2009). Molecular evolutionary consequences of niche restriction in Francisella tularensis, a facultative intracellular pathogen. PLoS Pathog. 5, e1000472. 10.1371/journal.ppat.1000472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey R. L., Allen L. A. (2006). Francisella tularensis LVS evades killing by human neutrophils via inhibition of the respiratory burst and phagosome escape. J. Leukoc. Biol. 80, 1224–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrumb F. R. (1961). Aerosol infection of man with Pasteurella tularensis. Bacteriol. Rev. 25, 262–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeniman C. J., Lane R. V., Cass B. N., Fong A. W., Sidhu M., Wang Y. F., O'Neill S. L. (2009). Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science 323, 141–144 10.1126/science.1165326 [DOI] [PubMed] [Google Scholar]
- Meibom K. L., Charbit A. (2010). Francisella tularensis metabolism and its relation to virulence. Front. Microbio. 1:140. 10.3389/fmicb.2010.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morner T. (1992). The ecology of tularaemia. Rev. Sci. Tech. 11, 1123–1130 [PubMed] [Google Scholar]
- Moule M. G., Monack D. M., Schneider D. S. (2010). Reciprocal analysis of Francisella novicida infections of a Drosophila melanogaster model reveal host-pathogen conflicts mediated by reactive oxygen and imd-regulated innate immune response. PLoS Pathog. 6, e1001065. 10.1371/journal.ppat.1001065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano F. E., Schmerk C. (2007). The Francisella pathogenicity island. Ann. N. Y. Acad. Sci. 1105, 122–137 [DOI] [PubMed] [Google Scholar]
- Nigrovic L. E., Wingerter S. L. (2008). Tularemia. Infect. Dis. Clin. North Am. 22, 489–504, ix. [DOI] [PubMed] [Google Scholar]
- Oyston P. C., Sjostedt A., Titball R. W. (2004). Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2, 967–978 [DOI] [PubMed] [Google Scholar]
- Parker R. R., Spencer R. R., Francis E. (1924). Tularaemia infection in ticks of species Dermacentor andersoni Stiles in the Bitterroot Valley, Montana. Public Health Rep. 1057–107319314929 [Google Scholar]
- Parola P., Raoult D. (2001). Ticks and tickborne bacterial diseases in humans: an emerging infectious threat. Clin. Infect. Dis. 32, 897–928 [DOI] [PubMed] [Google Scholar]
- Penn R. L. (2005). “Francisella tularensis (tularemia),” in Principles and Practice of Infectious Diseases, eds Mandell G. L., Bennett J. E., Dolin R. (Oxford: Churchill Livingstone; ), 2674–2685 [Google Scholar]
- Petersen J. M., Mead P. S., Schriefer M. E. (2009). Francisella tularensis: an arthropod-borne pathogen. Vet. Res. 40, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J. M., Schriefer M. E. (2005). Tularemia: emergence/re-emergence. Vet. Res. 36, 455–467 [DOI] [PubMed] [Google Scholar]
- Qin A., Scott D. W., Thompson J. A., Mann B. J. (2009). Identification of an essential Francisella tularensis subsp. tularensis virulence factor. Infect. Immun. 77, 152–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read A., Vogl S. J., Hueffer K., Gallagher L. A., Happ G. M. (2008). Francisella genes required for replication in mosquito cells. J. Med. Entomol. 45, 1108–1116 [DOI] [PubMed] [Google Scholar]
- Reese S. M., Dietrich G., Dolan M. C., Sheldon S. W., Piesman J., Petersen J. M., Eisen R. J. (2010). Transmission dynamics of Francisella tularensis subspecies and clades by nymphal Dermacentor variabilis (Acari: Ixodidae). Am. J. Trop. Med. Hyg. 83, 645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M., Lewis E. B. (2000). A brief history of Drosophila's contributions to genome research. Science 287, 2216–2218 10.1126/science.287.5461.2216 [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Yandell M. D., Wortman J. R., Gabor Miklos G. L., Nelson C. R., Hariharan I. K., Fortini M. E., Li P. W., Apweiler R., Fleischmann W., Cherry J. M., Henikoff S., Skupski M. P., Misra S., Ashburner M., Birney E., Boguski M. S., Brody T., Brokstein P., Celniker S. E., Chervitz S. A., Coates D., Cravchik A., Gabrielian A., Galle R. F., Gelbart W. M., George R. A., Goldstein L. S., Gong F., Guan P., Harris N. L., Hay B. A., Hoskins R. A., Li J., Li Z., Hynes R. O., Jones S. J., Kuehl P. M., Lemaitre B., Littleton J. T., Morrison D. K., Mungall C., O'Farrell P. H., Pickeral O. K., Shue C., Vosshall L. B., Zhang J., Zhao Q., Zheng X. H., Lewis S. (2000). Comparative genomics of the eukaryotes. Science 287, 2204–2215 10.1126/science.287.5461.2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M., Akimana C., Asare R., Kouokam J. C., Atay S., Kwaik Y. A. (2009). Intracellular fate of Francisella tularensis within arthropod-derived cells. Environ. Microbiol. 11, 1473–1481 [DOI] [PubMed] [Google Scholar]
- Santic M., Al-Khodor S., Abu Kwaik Y. (2010). Cell biology and molecular ecology of Francisella tularensis. Cell. Microbiol. 12, 129–139 [DOI] [PubMed] [Google Scholar]
- Santic M., Asare R., Skrobonja I., Jones S., Abu Kwaik Y. (2008). Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect. Immun. 76, 2671–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M., Molmeret M., Abu Kwaik Y. (2005a). Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-gamma. Cell. Microbiol. 7, 957–967 10.1111/j.1462-5822.2005.00529.x [DOI] [PubMed] [Google Scholar]
- Santic M., Molmeret M., Klose K. E., Jones S., Kwaik Y. A. (2005b). The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7, 969–979 10.1111/j.1462-5822.2005.00526.x [DOI] [PubMed] [Google Scholar]
- Santic M., Molmeret M., Abu Kwaik Y. (2007). Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-gamma. Cell. Microbiol. 9, 2314. [DOI] [PubMed] [Google Scholar]
- Santic M., Molmeret M., Klose K. E., Abu Kwaik Y. (2006). Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol. 14, 37–44 10.1016/j.tim.2005.11.008 [DOI] [PubMed] [Google Scholar]
- Saslaw S., Carlisle H. N. (1961). Studies with tularemia vaccines in volunteers. IV. Brucella aggiutinins in vaccinated and nonvaccinated volunteers challenged with Pasteurella tularensis. Am. J. Med. Sci. 242, 166–172 [DOI] [PubMed] [Google Scholar]
- Schmerk C. L., Duplantis B. N., Howard P. L., Nano F. E. (2009). A Francisella novicida pdpA mutant exhibits limited intracellular replication and remains associated with the lysosomal marker LAMP-1. Microbiology 155, 1498–1504 10.1099/mic.0.025445-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan T. G., Piesman J., Golde W. T., Dolan M. C., Rosa P. A. (1995). Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. U.S.A. 92, 2909–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer S. A., Harvey M. D., Goodlett D. R., Ernst R. K. (2007). Structural heterogeneity and environmentally regulated remodeling of Francisella tularensis subspecies novicida lipid A characterized by tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 18, 1080–1092 10.1016/j.jasms.2007.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnvik A., Priebe H. S., Grunow R. (2004). Tularaemia in Europe: an epidemiological overview. Scand. J. Infect. Dis. 36, 350–355 [DOI] [PubMed] [Google Scholar]
- Telford S. R., Goethert H. K. (2011). Towards an understanding of the perpetuation of the agent of tularemia. Front. Microbio. 1:150. 10.3389/fmicb.2010.00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel R., Lai X. H., Crosa L., Kozlowicz B., Heffron F. (2006). Attenuated Francisella novicida transposon mutants protect mice against wild-type challenge. Infect. Immun. 74, 5095–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K., Krum J. G., Bestor A., Jewett M. W., Grimm D., Bueschel D., Byram R., Dorward D., Vanraden M. J., Stewart P., Rosa P. (2006). Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 74, 3554–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus R. G., Bishop J. V., Mejia J. S. (2006). The immunomodulatory factors of arthropod saliva and the potential for these factors to serve as vaccine targets to prevent pathogen transmission. Parasite Immunol. 28, 131–141 [DOI] [PubMed] [Google Scholar]
- Triebenbach A. N., Vogl S. J., Lotspeich-Cole L., Sikes D. S., Happ G. M., Hueffer K. (2010). Detection of Francisella tularensis in Alaskan mosquitoes (Diptera: Culicidae) and assessment of a laboratory model for transmission. J. Med. Entomol. 47, 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonkavaara M., Telepnev M. V., Ryden P., Sjostedt A., Stoven S. (2008). Drosophila melanogaster as a model for elucidating the pathogenicity of Francisella tularensis. Cell. Microbiol. 10, 1327–1338 [DOI] [PubMed] [Google Scholar]
- Vyrostekova V. (1993). [Transstadial transmission of Francisella tularensis in the tick, Ixodes ricinus, infected during the larval stage]. Cesk. Epidemiol. Mikrobiol. Imunol. 42, 71–75 [PubMed] [Google Scholar]
- Wehrly T. D., Chong A., Virtaneva K., Sturdevant D. E., Child R., Edwards J. A., Brouwer D., Nair V., Fischer E. R., Wicke L., Curda A. J., Kupko J. J., III, Martens C., Crane D. D., Bosio C. M., Porcella S. F., Celli J. (2009). Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell. Microbiol. 11, 1128–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D. S., Brotcke A., Henry T., Margolis J. J., Chan K., Monack D. M. (2007a). In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U.S.A. 104, 6037–6042 10.1073/pnas.0609675104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D. S., Henry T., Monack D. M. (2007b). Francisella tularensis: activation of the inflammasome. Ann. N. Y. Acad. Sci. 1105, 219–237 10.1196/annals.1409.005 [DOI] [PubMed] [Google Scholar]
- Yu J. J., Raulie E. K., Murthy A. K., Guentzel M. N., Klose K. E., Arulanandam B. P. (2008). The presence of infectious extracellular Francisella tularensis subsp. novicida in murine plasma after pulmonary challenge. Eur. J. Clin. Microbiol. Infect. Dis. 27, 323–325 [DOI] [PubMed] [Google Scholar]
- Zogaj X., Klose K. E. (2011). Genetic manipulation of Francisella tularensis. Front. Microbio. 1:142. 10.3389/fmicb.2010.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]