Abstract
UEV proteins are enzymatically inactive variants of the E2 ubiquitin-conjugating enzymes that regulate noncanonical elongation of ubiquitin chains. In Saccharomyces cerevisiae, UEV is part of the RAD6-mediated error-free DNA repair pathway. In mammalian cells, UEV proteins can modulate c-FOS transcription and the G2-M transition of the cell cycle. Here we show that the UEV genes from phylogenetically distant organisms present a remarkable conservation in their exon–intron structure. We also show that the human UEV1 gene is fused with the previously unknown gene Kua. In Caenorhabditis elegans and Drosophila melanogaster, Kua and UEV are in separated loci, and are expressed as independent transcripts and proteins. In humans, Kua and UEV1 are adjacent genes, expressed either as separate transcripts encoding independent Kua and UEV1 proteins, or as a hybrid Kua–UEV transcript, encoding a two-domain protein. Kua proteins represent a novel class of conserved proteins with juxtamembrane histidine-rich motifs. Experiments with epitope-tagged proteins show that UEV1A is a nuclear protein, whereas both Kua and Kua–UEV localize to cytoplasmic structures, indicating that the Kua domain determines the cytoplasmic localization of Kua–UEV. Therefore, the addition of a Kua domain to UEV in the fused Kua–UEV protein confers new biological properties to this regulator of variant polyubiquitination.
[Kua cDNAs isolated by RT-PCR and described in this paper have been deposited in the GenBank data library under accession nos. AF1155120 (H. sapiens) and AF152361 (D. melanogaster). Genomic clones containing UEV genes: S. cerevisiae, YGL087c (accession no. Z72609); S. pombe, c338 (accession no. AL023781); P. falciparum, MAL3P2 (accession no. AL034558); A. thaliana, F26F24 (accession no. AC005292); C. elegans, F39B2 (accession no. Z92834); D. melanogaster, AC014908; and H. sapiens, 1185N5 (accession no. AL034423). Accession numbers for Kua cDNAs in GenBank dbEST: M. musculus, AA7853; T. cruzi, AI612534. Other Kua-containing sequences: A. thaliana genomic clones F10M23 (accession no. AL035440), F19K23 (accession no. AC000375), and T20K9 (accession no. AC004786).]
Described recently as a class of proteins structurally related to the ubiquitin-conjugating enzymes (E2), a distinctive feature of UEV proteins is that they are inactive variants of E2 enzymes, lacking a recognizable catalytic center (Koonin and Abagyan 1997; Ponting et al. 1997; Sancho et al. 1998). These proteins are well conserved in sequence and structure in all eukaryotic organisms, and this results in the sharing of specific functions, such as protection of cells from DNA damaging agents (Broomfield et al. 1998; Thomson et al. 1998) and enhancement of transcription from the c-FOS promoter (Xiao et al. 1998), by UEV proteins from distant organisms. The biochemical mode of action of UEV proteins in the yeast Saccharomyces cerevisiae has been established by Hoffman and Pickart (1999). The S. cerevisiae UEV protein, also known as Mms2, interacts with the E2 enzyme Ubc13p, and the resulting heterodimer is competent for the elongation of polyubiquitin chains. A novel feature of the polyubiquitin chains thus formed is that Lys 63, instead of the canonical Lys 48, is used for the Gly–Lys isopeptide bonds between ubiquitin moieties (Hoffman and Pickart 1999). Modification of proteins by this variant polyubiquitin chain may be reversible, and could modulate the function of target proteins, without directing them for degradation (Spence et al. 2000).
In S. cerevisiae, UEV genes are part of the error-free DNA repair pathway regulated by RAD6 (Broomfield et al. 1998; Hoffman and Pickart 1999). In human cells, UEV1 (Rothofsky and Lin 1997) and UEV2/Mms2 (Xiao et al. 1998) promote the transcriptional activity of c-FOS, possibly through interactions with as yet unidentified DNA-binding transcriptional regulators (Xiao et al. 1998). Overexpression of UEV1 in human colon cancer cells induces the accumulation of cells in G2-M and poliploidy, apoptosis, and inhibition of cell differentiation (Sancho et al. 1998). How the participation of UEV proteins in all these processes relate to the activity of UEV proteins as coeffectors in the polyubiquitination of target proteins has yet to be determined. In humans, there are two different UEV proteins encoded by separate genes, UEV1 or CROC1 (Rothofsky and Lin 1997; Sancho et al. 1998), and UEV2 or MMS2 (Sancho et al. 1998; Xiao et al. 1998). The UEV1 gene codes for two isoforms generated by alternative splicing, which share a common phylogenetically conserved UEV domain (Sancho et al. 1998). The isoform UEV1B contains a unique 82-residue amino-terminal extension, the B domain (Sancho et al. 1998).
The ubiquitin-conjugating enzymes are a large group of proteins, of which many variants exist in all eukaryotic organisms (for review, see Hershko and Ciechanover 1998). Although a common protein sequence and structural theme is shared between all E2 enzymes, attempts to assign primordial ancestors as the origin of one or more branches have not been met with success, mainly due to the great interspecies variability of functionally equivalent proteins. Being a new family of proteins with strong structural and functional links to the long-known ubiquitin-conjugating enzymes, UEV proteins and their genes could be useful to study the origins of the E2 proteins and their genes. Here we have analyzed the structure of the UEV genes in a number of organisms, and found that it is very conserved between phylogenetically distant organisms. As a relevant consequence of this analysis, we have found that the human UEV1 gene is part of a hybrid gene that results from the fusion of UEV1 with a second, previously unknown gene, which we have named Kua. We also show that, in humans, Kua and UEV1 can be expressed either as independent transcriptional units and proteins, or as a hybrid Kua–UEV transcript and protein. In contrast, in flies and worms the gene for Kua is unlinked to the gene for the corresponding UEV protein, and Kua and UEV are always expressed as separate proteins.
RESULTS
Precise Conservation of the Positions of Introns in UEV Genes from Distant Organisms
We have analyzed the architecture of UEV genes in those organisms for which the complete sequences of the loci are available. Exons were predicted by applying gene prediction algorithms (Guigó et al. 1992; Solovyev et al. 1994; Burge and Karlin 1997) to DNA sequences from genomic cosmid clones, and validated by stringent alignments with expressed sequence tags. This analysis yielded the exon arrangements schematically depicted in Figure 1A, confirming the structure predicted for the human UEV1 gene (Sancho et al. 1998), with three exons coding for the common domain of the protein (C domain), one for the domain specific of isoform UEV1A (A domain), and two for the domain specific of isoform UEV1B (B domain).
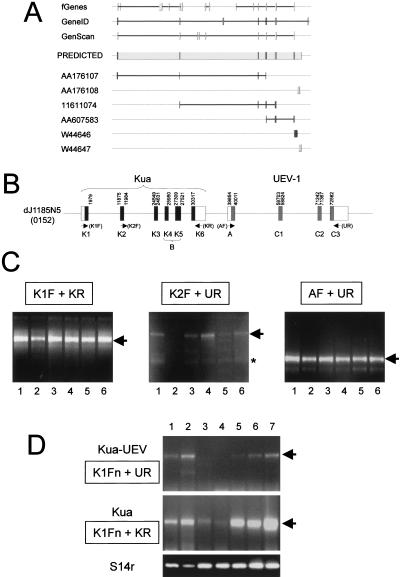
Figure 1.
Conservation of introns in UEV genes from distant organisms. (A) Diagrammatic representation (not to scale) of the relative exon–intron arrangement of the UEV gene in Saccharomyces cerevisiae, Schizosaccharomyces pombe, P. falciparum, Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, and Homo sapiens. Inset diagram for the two major isoforms described for human UEV1. Putative 5′ untranslated (UTR) segments are represented as open boxes. Introns are designated by numbers that are specific for each UEV gene. Vertical dotted lines within exons define segments corresponding to the exons in S. pombe UEV. (B) Alignment of UEV genes and proteins showing precise conservation of the position of the second and third introns interrupting the C domain of UEV proteins. Splice donor and acceptor sequences in intron boundaries are in lower case. The positions of introns are marked by arrowheads. Predictions of secondary structure are shown below the sequences as rods (helices) or arrows (strands).
The common domain of the UEV protein is encoded by one exon in S. cerevisiae, four in Schizosaccharomyces pombe, two in Plasmodium falciparum, three in Arabidopsis thaliana, two in Caenorhabditis elegans, three in Drosophila melanogaster and three in Homo sapiens (Fig. 1A). These exons are preceded by an initial exon (exon 1; exon A in H. sapiens), not conserved in sequence between different organisms. The exon structure for the common domain of UEV is identical in H. sapiens, A. thaliana, and D. melanogaster. S. pombe is the organism in which the UEV gene has the most exons (five). S. cerevisiae is the only organism in which the common domain of UEV is not interrupted by introns. Introns 2 and 3 of S. pombe UEV have equivalents in all other organisms, except S. cerevisiae. The positions of these introns within the corresponding UEV genes are identical, regardless of the organism (Fig. 1). These introns lie between codons, and are thus in phase zero. Also, they correspond to boundaries between structural domains within the UEV protein (Fig. 1). Therefore, they have features predicted for “early” introns (Gilbert 1987; de Souza et al. 1998). Intron 4 of S. pombe UEV is unique to this organism, and has no correspondence in any other organism. This intron is in phase 2, and it interrupts a sequence coding for an α-helical domain within the protein (Fig. 1). Therefore, this intron would be more compatible with being a “late” intron (Gilbert 1987; de Souza 1998).
In contrast to the strong sequence conservation for the rest of the gene, the first exon of the UEV genes (exon A in H. sapiens) is not conserved to a significant degree between different organisms. The size of this initial exon varies from 4 bp (1 and 1/3 codons), in C. elegans, to 88 bp (29 and 1/3 codons) in A. thaliana, or 91 bp (30 and 1/3 codons) in H. sapiens. Intron 1, separating the first exon and the exons for the common domain of UEV, is placed in different phases in different organisms: phase 0 in S. cerevisiae and P. falciparum, phase 2 in S. pombe, and phase 1 in C. elegans, A. thaliana, D. melanogaster, and H. sapiens (Fig. 1). The 3′ boundaries set by this intron impose different 5′ ends for the common domain of UEV in different organisms, which results in one to three codon differences between UEV proteins from these organisms at the amino-end of this domain (Fig. 1). Similarly, there are no sequences resembling the exons coding for the B domain of human UEV1 in the vicinity of the UEV genes of any other organism. It has been suggested that the B domain of human UEV1B could confer specific functions to this isoform (Sancho et al. 1998). We thus set our efforts to explore the evolutionary origin of the sequences coding for the B domain of UEV1B.
A New Gene in C. elegans and D. melanogaster Coding for a Protein Containing a UEV1 B Domain-Like Sequence
A search for B domain-like sequences in genomic DNA databases yielded two small fragments of significant similarity within C. elegans clone Y53C10 and D. melanogaster clone DS00863 (Fig. 2A). In the C. elegans genome, the segment included in Y53C10 is in chromosome 1, ∼2.5 Mb away from the location of the UEV gene in clone F39B2, also in chromosome 1 (http://www.sanger.ac.uk). We have found no evidence for UEV-like sequences within C. elegans Y53C10 (86 Kb) or D. melanogaster DS00863 (78 Kb) genomic clones. Therefore, in contrast to the B domain sequences in H. sapiens, the B domain-like sequences in C. elegans and D. melanogaster do not appear to be part of the corresponding UEV genes.
Figure 2.
A new gene in Caenorhabditus elegans and Drosophila melanogaster coding for B domain-like sequences. (A) Alignment of the human UEV1 B domain with TBLASTN-identified segments from C. elegans Y53C10 and D. melanogaster DS00863 genomic clones. The aligned segments are discontinuous in Y53C10, as indicated by the positions of nucleotides from the database entry. (B) Dot-plot analysis of reciprocal TBLASTX comparisons of genomic clones Y53C10-B (C. elegans) and DS00863-B (D. melanogaster). (Bottom) Scores of sequence identities for each diagonal of conserved segments, shown as horizontal bars. (C) Predicted structure of the new gene in C. elegans (middle) supported by gene prediction algorithms (top) and alignment with ESTs (bottom). (D) Predicted C. elegans protein containing a B domain-like segment (shaded). (E) Predicted structure of the new gene in D. melanogaster (middle), supported by gene prediction algorithms (top) and alignment with ESTs (bottom). (F) Predicted D. melanogaster protein containing a B domain-like segment (shaded). (G) Expression analysis by RT–PCR, with forward primers corresponding to exons 1, 2, 3, and 4 of the predicted gene, and a reverse primer corresponding to exon 5, using as templates embryo mRNAs. (H) Cytogenetic assignment of D. melanogaster UEV gene to chromosome 3 segment 64D. Digoxigenin-labeled cDNA probes were used for hybridization on wild-type Drosophila polytene chromosomes.
We hypothesized that, in C. elegans and D. melanogaster, these sequences belonged to a second gene, unrelated to UEV. Mapping of known cDNAs and ESTs onto the genomic sequence indicated the presence of known genes upstream from position 17,000 and downstream from position 29,000 in Y53C10, and upstream from position 54,500 and downstream from position 62,400 in DS00863 (data not shown). Reciprocal TBLASTX searches (Altschul et al. 1990) and dot-plot analyses between the segments in Y53C10 and DS00863 that lie within these positions (denoted here Y53C10-B and DS00863-B) indicated that they shared other conserved segments in the vicinity of the B domain-like sequence, and delineated a tentative exonic structure for a gene in Y53C10-B and DS00863-B (Fig. 2B). The exonic structure of the putative gene in Y53C10-B was further refined by means of stringent alignments with ESTs from a nonredundant database, and computational gene identification programs (Fig. 2C). A recent addition to GenBank of a C. elegans EST (accession no. AV182903) provided strong support for this analysis, and a confirmation of the existence of a gene in this region. Thus, we predict in C. elegans Y53C10-B a new gene consisting of seven exons, with the potential to encode a 319-amino acid protein (Fig. 2D). This prediction is compatible with that made available at EMBL for AL033536, a genomic contig that includes Y53C10.
Similar procedures were used to refine the exonic structure of the putative new gene containing a B domain-like sequence gene in DS00863-B. Analysis of DS00863-B with gene identification algorithms produced a consistent gene structure, compatible with the structure delinated by the regions conserved with C. elegans Y53C10-B (Fig. 2E). This tentative structure was supported by a number of ESTs with distant, but significative, similarity to sequence segments in DS00863-B (Fig. 2E). The exonic structure thus predicted was highly compatible with that delineated by the conserved sequence segments shared with Y53C10-B for the 3′ end of the putative gene. The final analysis resulted in a five-exon gene, with the potential to encode a 326-amino acid protein (Fig. 2F).
The C. elegans EST confirmed that the gene predicted is indeed expressed. However, evidence of this kind was lacking for the D. melanogaster gene. Therefore, we designed primers specific for the exons predicted for the D. melanogaster gene, for use in RT–PCR reactions. All sets of primers yielded specific amplification products from adult and larval RNA (Fig. 2G). Sequencing of the products showed that they correspond to processed RNA, formed by joining of the predicted exons, of which exons 4 and 5 are the B domain-like segments (Fig 2F). These experiments confirmed the exon structure for the new the gene, as predicted by computational methods. We have given the name Kua to this new gene (see Acknowledgments).
Clone DS00863, harboring the Drosophila Kua gene, is localized to segment 38B2–38C1, on chromosome 2 (Hartl et al. 1994; http://flybase.bio.indiana.edu/). There is no experimental data for the cytogenetic localization of the Drosophila UEV gene. Therefore, we performed in situ hybridization on polytene chromosomes, using a probe specific for Drosophila UEV. This permitted the cytogenetic assignment for the D. melanogaster UEV gene to 64D, on chromosome 3 (Fig. 2H). With posteriority to this analysis, the sequence of the D. melanogaster genome was released, confirming our assignment of D. melanogaster UEV to 64D (Adams et al. 2000).
In conclusion, in both C. elegans and in D. melanogaster, the “B domain”-like sequences contained in clones Y53C10 and DS00863, respectively, correspond to exons located within a new gene, Kua, expressed in worms and in flies. In both organisms, the genes Kua and UEV are in widely separated loci.
H. sapiens: Fusion of UEV1 with Kua
The entire human UEV1 gene, including exons coding for the B domain, is contained within the genomic PAC clone dJ1185N5 from chromosome 20. Conserved segments spanning a 32-Kb region within this clone were found to correspond to most of D. melanogaster and C. elegans Kua. Identical or strongly related mouse and human EST matches fully covered this region. This, together with the application of gene prediction programs allowed us to predict a six-exon human Kua gene (Fig. 3A) with the potential to code for a 270-amino acid protein. Exon 1 is predicted to contain a 5′ untranslated region (UTR) of unknown size, and exon 6 is predicted to have a 3′ UTR extending 1268 bp from the stop codon up to the first potential polyadenylation signal (Fig. 3A,B). The exons predicted for the Kua gene lie 5′ to the exons for the common- and A-domains of UEV1 (Sancho et al. 1998; Fig. 3B).
Figure 3.
A hybrid Kua–UEV gene in Homo sapiens genomic PAC clone dJ1185N5. (A) Prediction of the exon structure of human Kua (middle), supported by gene prediction algorithms (top), and alignments with expressed sequences (bottom). (B) Schematic representation of the relative arrangement of exons in the human Kua–UEV locus, based on predictions for clone dJ1185N5. (C) RT–PCR analysis showing expression of human Kua (K1F + KR), a hybrid Kua–UEV transcript (K2F + UR), and UEV1A (A + UR) in six cell lines. RT–PCR was performed with primers for the putative 5′ UTR (K1F), the 3′ UTR (KR), or the 5′ UTR of Kua (KR), and the end of exon C3 of UEV1 (UR), using as templates RNAs from the human cell lines HT-29 (lane 1), SW480 (lane 2), HeLa (lane 3), SK—PC-3 (lane 4), T24 (lane 5), and HEL (lane 6). (Arrows) Specific amplification products; (asterisk) nonspecific amplification products. The faster-migrating band in the right panel corresponds to an alternative form of UEV1A (Sancho et al. 1998). (D) Expression analysis by RT–PCR of Kua–UEV hybrid transcripts and Kua-only transcripts on RNAs from cell lines Jurkat (lane 1), SK–NS-H (lane 2), N2A (lane 3), PZ–HPV7, (lane 4), CA–HPV10 (lane 5), PC-3 (lane 6), and SK–PC-1 (lane 7). Amplification with primers for transcripts corresponding to the housekeeping S14r ribosomal protein was used to normalize for input RNA.
Next we designed primers corresponding to the predicted exons of human Kua (Fig. 3B) for use in RT–PCR reactions (Fig. 3C). An amplification product was present in all of the cell lines analyzed, of the size expected (964 bp) for the joining of exons 1–6 of Kua (Fig. 3C). Sequencing of the amplification products confirmed that they contain all of the predicted exons, including those encoding the B domain of UEV1B (Fig. 4A,B). In the original reports (Rothofsky and Lin 1997; Sancho et al. 1998), the 5′ end of the transcript for UEV1B was described as a 5′ UTR. In our analysis, it is part of exon 4 of Kua, the B domain being encoded by exons 4 and 5. RT–PCR with primers specific for UEV1A yielded two products in all samples (Fig. 3C, right), corresponding to alternatively spliced isoforms described previously (Sancho et al. 1998).
Figure 4.
Sequences of human Kua and Kua–UEV cDNAs and predicted proteins. (A) Sequence shared by Kua and Kua–UEV transcripts. (B) Sequence specific for Kua-only transcripts. This sequence is continuous with A in Kua-only transcripts, and corresponds to exon K6 (Fig. 3). (C) UEV C domain sequence from Kua–UEV transcripts. This sequence is continuous with A in Kua–UEV transcripts, and results from joining exons C1, C2, and C3 (Fig. 3B). Exon boundaries, as predicted in Fig. 3, are marked by vertical bars. The sequences of primers used for RT–PCR in Fig. 3 are boxed. The B domain is shaded.
The UEV1B isoform of UEV1 was isolated independently by two groups using either RACE or RT–PCR (Rothofsky and Lin 1997; Sancho et al. 1998). Our analysis shows that what was then described as a domain specific for the UEV1B isoform of UEV1 is encoded by two exons that are also part of a second gene, located upstream from UEV1, which we now call Kua. Thus, we hypothesized the existence of continuous transcriptional units joining the exons predicted for Kua and UEV1. Shown in Figure 3C are RT–PCR experiments performed with primers for the predicted exon 2 of Kua (forward primer) and exon C3 of UEV (reverse primer) yielding a specific product in the majority of the cell lines analyzed. Sequencing confirmed that these products correspond to the joining of exons 2, 3, 4, and 5 of Kua, and exons C1, C2, and C3 of UEV1 (Fig. 4). RT–PCR reactions with primers corresponding to regions in Kua that were not supported by our gene prediction analyses to be part of exons did not yield any specific amplification products (data not shown). Therefore, this analysis demonstrates the existence of a hybrid Kua–UEV trancript containing all exons from Kua and UEV1, except exon 6 of Kua and exon A of UEV1. This transcript has the potential to encode a protein with two distinct domains, Kua at its amino half, and UEV at its carboxyl half.
To determine the relative levels of Kua and Kua–UEV transcripts, RT–PCR was performed under nonsaturating conditions on RNAs from seven different cell lines. For normalization, amplification was performed with primers for transcripts for the ribosomal protein S14r. In these analyses, the ratio of Kua–UEV to Kua amplification products ranged from 0.1 (samples 1 and 2) to 0.02 (samples 5 and 7), in those samples with visible Kua–UEV amplification products (Fig. 3D). Two samples did not yield measurable levels of Kua–UEV hybrid amplification products under these conditions (30 cycles), although products were observed with 35 amplification cycles or more (data not shown).
Kua: A New Family of Conserved Proteins
Several organisms express Kua transcripts, including H. sapiens, Mus musculus, D. melanogaster, C. elegans, Trypanosoma cruzi, each of which was predicted to express one Kua protein, and the plant A. thaliana, in which we predict three different Kua proteins corresponding to distinct genes (Fig. 5A). The three animal Kua proteins show identities of 94%, 67%, and 61% with respect to the human protein (comparison with the sequence between positions 83 and 238 of the human protein), whereas the three plant Kua proteins, strongly related to each other, show 29%–34% identities with respect to the human protein. Although yeasts have UEV proteins (Broomfield et al. 1998; Sancho et al. 1998), they do not have potential Kua proteins.
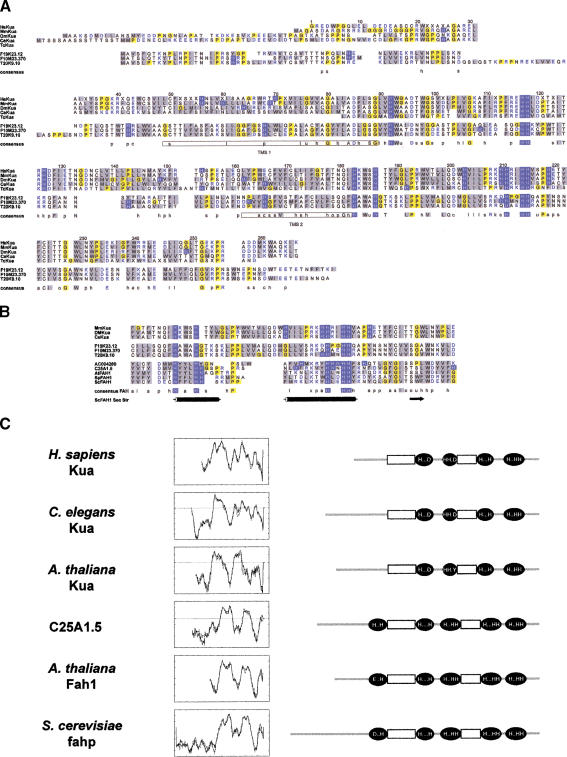
Figure 5.
Kua, a new family of conserved proteins. (A) Alignment of Kua proteins predicted from PCR-generated cDNAs (Homo sapiens and Drosophila melanogaster), ESTs (Mus musculus and T. cruzi), or genomic sequences (Caenorhabditis elegans and Arabidopsis thaliana, F19K23.12, F10M23.370, and T20K9.10). The prediction for the A. thaliana F10M23.370 genomic sequence is supported by EST AA712589. Color codes: blue background, His; yellow background, Gli and Pro; grey background, hydrophobic residues; blue, charged residues. Key for consensus sequences: a, aromatic (F, H, W, Y); c, charged (D, E, H, K, R); h, hydrophobic (F, L, M, V, W, Y, I); l, aliphatic (I, L, V); o, alcohol (S, T); p, polar (C, D, E, H, K, N, Q, R, S, T); s, small (A, C, D, G, N, P, S, T); u, tiny (A, G, S); x, helix breaking (G, P);−, negative (D, E); +, positive (H, K, R). Potential transmembrane domains are boxed in the consensus sequence. (B) Alignment of the segments of proteins containing the two histidine-rich motifs detected by PHI–BLAST search of GenBank nr database with the profile H-x-(YWF)-x-H-x(8,25)-(RK)-x(2)-H-x(2)-H-H, generated after the consensus sequence for Kua proteins in this region. Shown are fatty acid hydroxylases from D. melanogaster (AC004280) C. elegans (C25A1.5), A. thaliana (AtFAH1), Schizosaccharomyces pombe (SpFAH1) and Saccharomyces cerevisiae (ScFAH1). (Bottom) Secondary structure predictions for yeast FAH (cylinders, alpha helix; arrow, beta sheet). (C) Transmembrane domain predictions and compared topological models for Kua proteins and fatty acid hydroxylases. Left panel, predictions of transmembrane domains for human (AF155120), worm (Y53C10A.5), and plant (F19K23.12), Kua and worm (C25A1.5), and plant (At Fah1) fatty acid hydroxylases. Right panel, diagrams representing the positions of the histidine-rich motifs in the same proteins, relative to the predicted transmembrane domains. Diagrams are not drawn to scale.
A pattern of histidine residues conserved in all Kua proteins is reminiscent of certain histidine-rich motifs in which specific residues are coordinated by transition metals (Shanklin et al. 1994). Membrane-bound fatty acid desaturases and hydroxylases contain several histidine-rich motifs, with the general pattern H-x(2,3)-(x-H)-H (Fox et al. 1993; Mitchell and Martin 1997). Kua proteins contain two of these motifs (Fig. 5A). Although all Kua proteins contain fatty acid hydroxylase-like His-rich motifs, only plant Kua proteins show a marginal degree of homology (E values in the order of 10−2) to fatty acid hydroxylases within the segment of the protein containing such motifs. Nevertheless, a PHI–BLAST search of a nonredundant database with a sequence profile generated based on the consensus sequence for Kua proteins yielded bonafide fatty acid hydroxylases (Fig. 5B).
In fatty acid hydroxylases and desaturases, the protein segments containing these motifs are located on the cytoplasmic face of the ER membrane, such that the relevant histidine residues become closely apposed for the coordination of a di-iron cluster (Mitchell and Martin 1997). A prediction of trasmembrane domains for the Kua proteins indicates an overall topology similar to that of fatty acid hydroxylases (Mitchell and Martin 1997) (Fig. 5C, left panel), especially when comparing the transmembrane prediction for the plant Kua proteins (e.g., F19K23.12) with that for A. thaliana Fah1 (Fig. 5C, left panel). The positions of the histidine-rich motifs relative to the predicted transmembrane domains of Kua proteins and fatty acid hydroxylases are diagrammatically represented in Figure 5C (right panel).
Subcellular Localization of UEV1A, Kua, and Kua–UEV
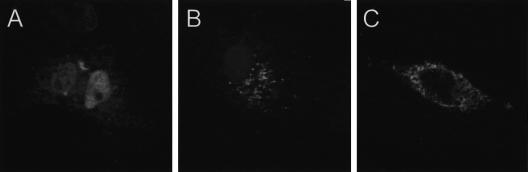
A partial UEV1 protein has been reported to localize to the nucleus of mammalian cells (Rothofsky and Lin 1997). The above analysis of the polypeptide sequence of Kua predicted its localization to endomembranes. To experimentally determine the subcellular localization of Kua and Kua–UEV, we generated constructs for the expression of Kua, Kua–UEV, and UEV1A in COS-7 cells, bearing in-frame a hemagluttinin tag at their carboxyl termini. The epitope-tagged full-lentgh UEV1A isoform of human UEV1 showed a nuclear localization, with a uniform pattern and nucleolar exclusion (Fig. 6A). The full-length Kua protein localized mainly to cytoplasmic structures, with a pattern compatible with its association with the endoplasmic reticulum (Fig. 6B), thus providing experimental evidence supporting the predicted localization of Kua to endomembranes. Finally, the hybrid Kua–UEV protein was also associated with cytoplasmic structures, with a clear nuclear exclusion, in a pattern very similar to that displayed by Kua (Fig. 6C). In conclusion, two alternative forms of UEV1 are targeted to distinct subcellular localizations, nucleus, or cytoplasm. This differential targeting is determined by the sequence present at the amino terminus of UEV1, with the Kua domain in Kua–UEV directing its localization to cytoplasmic structures.
Figure 6.
Subcellular localization of UEV1A (A), Kua (B), and Kua–UEV (C) proteins tagged with a hemagluttinin epitope. Liposome-mediate transient transfection was performed on COS-7 cells with plasmids engineered for the expression of the corresponding proteins bearing a hemagluttinin epitope at their carboxyl termini, and processed for indirect immunofluorescent confocal microscopy (× 400).
DISCUSSION
The analysis performed in this study allows us to postulate an evolutionary history of the genes for the polyubiquitination coeffector UEV, and how they have become fused, in humans, to a second gene, which codes for a new class of proteins.
Evolution of UEV Gene Introns
The strict conservation in the positions of introns interrupting the coding sequences for the common domain of UEV in organisms as phylogenetically distant as yeasts and humans is a remarkable fact. The localization of introns in positions that are phylogenetically invariant has been observed in genes such as globin or alcohol dehydrogenase, with the development of additional insertions occurring as primordial genes have diverged (Naito et al. 1991; Sherman et al. 1992). For a given conserved protein, a certain degree of variation is frequently present in the location of introns relative to protein sequence, which is taken as supporting the “introns-late” theory (Cavalier-Smith 1991), as well as the occurrence of sliding and mutations in equivalent exon–intron junctions between genes from different organisms (Long et al. 1995; Gilbert et al. 1997). The position of introns 2 and 3 of S. pombe UEV is strictly invariant in distant organisms, P. falciparum (intron 3), A. thaliana (introns 2 and 3), C. elegans (intron 2), D. melanogaster (introns 2 and 3), and H. sapiens (introns 2 and 3). The maintenance of the positions of these introns through ∼5 × 108 years of evolution suggests an early insertion of these introns, an argument further supported by the observations that these introns are in phase zero, and placed between structural domains (Gilbert 1987; de Souza et al. 1998).
Conversely, intron 4 of S. pombe UEV interrupts an α-helical domain in the S. pombe UEV protein, which would be more consistent with a late insertion of this intron (de Souza et al. 1998). The absence of the equivalent of this intron in all other organisms suggests either the removal through splicing and reinsertion of processed and reverse-transcribed RNA (Fink 1987), or an insertional event specific to S. pombe, not transmitted evolutionarily. Absence of other S. pombe UEV introns in other organisms (all introns in S. cerevisiae, intron 2 in P. falciparum, intron 3 in C. elegans) could also be due to retrotransposition, or the result of independent origins and evolution of each intron. The conservation in S. cerevisiae of the 5′-most intron, in C. elegans of the two 5′-most introns, or the loss of the 3′-most intron in A. thaliana, D. melanogaster, and H. sapiens, would also be consistent with a scenario of retrotranscription followed by homologous recombination as a mechanism for the loss of introns in the evolution of the UEV genes (Fink 1987). In any case, it appears that both early and late scenarios (Trotman 1998) could apply to different UEV gene introns.
The analysis of intron 1 of the UEV genes suggests that it originated as a result of a process distinct from the ones discussed above for the introns that interrupt the common domain. Intron 1 is inserted in different phases in different organisms, and it generates different carboxyl- and amino-ends in the flanking exons. Also, exon 1 (exon A in humans), placed 5′ to this intron, is different for all organisms. This suggests that exon 1 evolved separately from the rest of the UEV gene.
Fusion of the Kua and UEV Genes
We also show that, in humans, one of the two UEV genes in this organism, UEV1, is adjacent to an unrelated gene, which we have named Kua, and can be expressed as a hybrid Kua–UEV transcript and protein. In contrast, in D. melanogaster and C. elegans, Kua and UEV are independent genes, coding for separate transcripts and proteins. Therefore, the combination of computational and experimental approaches used here show how two genes, which are unrelated and located in separate loci in worms and insects, converge into a hybrid gene and protein in humans. The identification of Kua in C. elegans and D. melanogaster was possible without any prior knowledge of expressed sequences, applying ab initio biocomputational methods on genomic sequences. Of these methods, the use of dot-plot analyses of reciprocal TBLASTX alignments was sufficient to infer the structure of Kua in both organisms, later confirmed with alignments with ESTs and experimental data. Therefore, this could be a very useful tool for the identification of genes when expression data are not available.
New genes are thought to originate through events such as gene duplications (Ohno 1970; Ohta 1989), exon shuffling (Gilbert 1978), or the generation of processed genes (McCarrey and Thomas 1987). In metazoans, fusion of genes generally involves a number of intermediate processes, such as duplication and shuffling of exons (Bazan et al. 1989; Simmer et al. 1990; Chen et al. 1997; Coppock et al. 1998). Secondary events, such as retrotransposition with exon capture (Long and Langley 1993) or genetic hitchhiking associated with selective sweeps can generate chimeric genes (Nurminsky et al. 1998; Long et al. 1999). These events often involve extensive refashioning of coding and noncoding regions (Nurminsky et al. 1998). The Kua–UEV fusion does not appear to involve such promiscuous changes, and it rather suggests the occurrence of a direct fusion of loci. The likely scenario in the Kua–UEV fusion would be a two-step process, duplication of the UEV gene, followed by fusion of the duplicated gene to Kua. This model is supported by the fact that, in humans, there are two UEV genes, of which the UEV1 gene is fused to Kua in chromosome 20, whereas the UEV2 gene is on chromosome 8, without any evidence for this type of fusion (B. Cormand and T.M. Thomson, unpubl.). Therefore, UEV2 would correspond to the gene in the original locus, and UEV1 to the duplicated gene, which would undergo subsequent rearrangement with a head-to-tail fusion to Kua.
The generation of three different classes of transcripts from the Kua–UEV locus represents a unique strategy aimed at the modular expression of two genes, coding either for two separate polypeptides, or as a combination of both to yield a single two-domain polypeptide. The generation of a Kua–UEV hybrid transcript could be the result either of cis-splicing directed by canonical splice sites, or trans-splicing. Trans-splicing has been shown to occur in mammalian cells, either in artificial (Bruzik and Maniatis 1992) or natural (Caudevilla et al. 1998; Kingzette et al. 1998; Akopian et al. 1999; Li et al. 1999; Zaphiropoulos 1999) settings. Although trans-splicing in mammalian cells usually occurs between transcripts from genes in separate loci or chromosomes, it has been reported to occur also between transcripts from clustered genes (Zaphiropoulos 1999). It has also been shown that both cis- and trans-splicing can be concomitant mechanisms for the generation of hybrid transcripts from the same genes in mammalian cells (Eul et al. 1995). Our observations do not provide sufficient information to infer the splicing mechanism prevalent in the generation of hybrid Kua–UEV transcripts. However, there are indirect arguments against trans-splicing as the major mechanism for the generation of these transcripts. First, trans-splicing in mammalian transcripts appears to be regulated by sequences at the acceptor exon, with the consensus GAAGAAG(G/C) (Caudevilla et al. 1998). Sequences fully compatible with this consensus are present in exon C1 of UEV1, and also at equivalent positions in exon C1 of UEV2, but only UEV1, and not UEV2, is involved in hybrid Kua–UEV transcripts (T.M. Thomson, unpubl.). A more speculative argument would be based on the teleological nature of the fusion of Kua and UEV from the standpoint of the evolution of these genes. The driving force for the evolutionary rearrangement and fusion of Kua and UEV into a single locus would be stronger for cis-splicing being a major mechanism for the generation of hybrid transcripts than it would be for a trans-splicing mechanism.
The close proximity of Kua to UEV1 could raise the question whether the mere juxtaposition of two genes with the same transcriptional direction is sufficient for the generation of detectable run-off transcription from the upstream gene. To test whether this is a common situation, we have performed a survey of all genes on human chromosome 22 with a distance between genes of ≤25 Kb. Of 546 genes annotated on this chromosome, 221 correspond to pairs that are within a distance of ≤25 Kb and with the same transcriptional direction. Five of these pairs correspond to overlapping genes. Of the remaining 216 gene pairs, BLASTN searches of EST databases have identified two with transcripts matching both genes in the pair, that could correspond to transcripts bridging the two genes. For one of these two gene pairs, PNUTL1 and GP1BB, experimental evidence for the existence of hybrid transcripts, as well as single-gene transcripts, has been reported (Zieger et al. 1997; Yagi et al. 1998). The second gene pair with a potential hybrid transcript has not been characterized, and corresponds to genes predicted for a hypothetical protein (transcript dJ1194E15.3) and an EST cluster (dJ1104E15.5). The PNUTL1–GP1BB fusion transcript is predicted to contain two open reading frames (ORFs), and appears to result from defective truncation of the upstream PNUTL1 transcript due to an imperfect polyadenylation signal sequence (Zieger et al. 1997). Therefore, the approach used in this analysis can detect potential gene-bridging hybrid transcripts in 1% of the gene pairs analyzed. This is probably an underestimate of all instances of gene fusions expressing hybrid transcripts, because not all fused gene pairs will be represented by ESTs matching both genes in expression databases. Also, some of the annotated genes could have been misrepresented as single genes, especially if they have been predicted on the basis of matching ESTs. One conclusion of this type of analysis, relevant to the present study, is that transcript fusions between two adjacent genes, although not infrequent, are observed only in a subset of closely associated gene pairs.
In contrast to the Kua–UEV fusion, the PNUTL1–GP1BB gene fusion does not result in a fusion of proteins (Zieger et al. 1997). A second difference is that the mature PNUTL1–BP1BB fusion includes the terminal exon from the upstream gene, with its transcriptional truncation signal. Because this signal is apparently inefficient, the PNUTL1–BP1BB hybrid transcript could be the consequence of a genuine transcriptional run-off. In contrast, the mature processed transcripts of the Kua–UEV fusion have spliced out the terminal exon of the upstream gene, Kua, as well as the first exon of the downstream gene, UEV1A, which contains a 5′ untranslated sequence. RT–PCR experiments aimed at detecting fused transcripts that contain these two exons have failed to yield any amplification products. In the Kua–UEV fusion, therefore, a continuous primary transcript between both genes is subjected to specific splicing events that allow the expression of a two-domain protein. This also implies that a truncation of transcripts at the terminal exon of Kua must proceed at a rate that is sufficiently slow to allow the subsequent splicing events for the maturation of the Kua–UEV transcript. A carefully orchestrated balance between splicing and truncation of transcripts has been shown to occur in lower organisms (Ull et al. 1993).
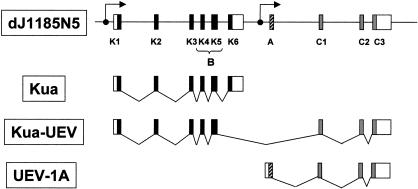
A summary of the different classes of transcripts generated at the Kua–UEV locus is depicted in Figure 7. The locus for H. sapiens UEV1, on chromosome 20q13.2, contains two contiguous genes, Kua and, immediately downstream, UEV1. We predict that each gene has its own promoter, an initial exon with a 5′ UTR, and a terminal exon with a 3′ UTR. The genes Kua and UEV1 can be expressed as separate transcriptional units from their independent promoters, yielding, respectively, the proteins Kua and UEV1A. By means of alternative splicing, the transcript originating from the Kua promoter can generate a hybrid Kua–UEV transcript, with the potential to code for a two-domain Kua–UEV protein. Productive processing of the latter transcript to include the exons coding for the common domain of UEV requires removal of exon K6 by splicing, before truncation occurs, signalled by the polyadenylation and truncation signal present in that exon. For such a transcript to produce an uninterrupted reading frame yielding a two-domain Kua–UEV protein, exon A of UEV1 must be spliced out. In this scheme, the isoform UEV1B (or CROC1B), as originally described, corresponds to a partial, rather than a full-length, transcript and the B domain is a segment of Kua.
Figure 7.
Diagrammatic representation summarizing the three major classes of transcriptional units generated at the Kua–UEV locus. Transcription from the promoter located upstream from the first exon of Kua can eventually yield either a Kua-only transcript ending in K6 (Kua), or a Kua–UEV hybrid transcript. The maturation of the longer transcript is possible only after removal of the K6 exon, which contains signals for truncation of the primary transcript. Transcription from the promoter located upstream from the A exon yields UEV1A. (Black boxes) Kua exons; (light grey boxes) UEV “common domain” exons; (shaded box) UEV “A” exon; (open boxes) 5′ and 3′ UTRs; (filled circles and arrows) putative transcription initiation sites.
It has been shown that evolutionary fusion of proteins often indicates a functional interaction between them (Enright et al. 1999; Marcotte et al. 1999). In our hands, yeast two-hybrid experiments have not provided unequivocal evidence for direct interaction between Kua and UEV proteins from any of the organisms studied (N. Loukili and T.M. Thomson, unpubl.). Nevertheless, it remains possible that these two proteins perform functions in the same biochemical pathway, such that a requirement for functional interaction would provide a driving force for their evolutionary fusion. One of the functional consequences of the fusion of Kua with UEV in humans is that the domain regulating polyubiquitination is redirected for localization to cytoplasmic structures, rather than the nucleus. As a consequence of their different subcellular localizations, nuclear (UEV1A) and cytoplasmic (Kua–UEV) forms of UEV1 could target different substrates for variant (K63) polyubiquitination. Thus, an endomembrane-associated form of UEV1 could preferentially direct the variant polyubiquitination of substrates closely associated with the cytoplasmic face of the ER, possibly, although not necessarily, in conjunction with membrane-bound ubiquitin-conjugating enzymes (Sommer and Jentsch 1993). The substrates could be either proteins specifically targeted for ubiquitination by these UEV–E2 complexes, or misfolded ER-associated proteins that are dislocated into the cytoplasm for subsequent ubiquitination (Kopito 1997).
METHODS
Biocomputational Analysis
Putative conserved coding regions between genomic sequences were identified with reciprocal TBLASTX (Altschul et al. 1990), the output processed with MSPCrunch (Sonnhammer and Durbin 1994), and conserved segments visualized with aplot (http://www1.imim.es/∼jabril/GFFTOOLS/APLOT.html). For prediction of protein coding genes, three different ab initio gene prediction programs were used, Genscan (Burge and Karlin 1997), Geneid (Guigó et al. 1992), and Fgenes (Solovyev et al. 1994). To support the resulting exonic structures, BLASTN searches were performed against the EST division of GenBank (Benson et al. 2000). A minimum subset of ESTs covering the matches was selected, all other EST matches being either identical or included therein. Each of these ESTs was aligned with the matching genomic clone sequence using the program est_genome (Mott 1997). Putative coding domains were recorded using the gff format (http://www.sanger.ac.uk/Software/formats/GFF/GFF_Spec.shtml), and plotted with gff2ps (available at http://www1.imim.es/∼jabril/GFFTOOLS/GFF2PS.html). Protein sequences were aligned with ClustalW (Thompson et al. 1994), and secondary structures predicted with the PHD package (Rost 1996). Transmembrane domains were predicted with TMpred (http://www.ch.embnet.org/software/TMPRED_form.html). Searches for distant homologies were performed with PSI–BLAST (Altschul et al. 1997).
Expression Analysis by RT–PCR
RNAs from human cell lines and mouse tissues were isolated by the acid phenol procedure (Chomczynski and Sacchi 1987). RNAs from D. melanogaster embryos were isolated by guanidium isothiocyanate extraction and CsCl gradients, and enriched for mRNA on oligo dT-cellulose columns (Sambrook et al. 1989). RNAs were resuspended in diethyl pyrocarbonate-treated H2O, and 20 μg were treated with 1 unit RNase-free RQ1 DNase I (Promega, Madison, WI), in a reaction containing 10 mM Tris-HCl at pH 8.0, 5 mM MgCl2, and 40 units RNasin (Promega). For Drosophila samples, RT–PCR was done by a single-tube procedure (Life Technologies, Barcelona), with forward primers 1 (5′-AATGACATCAACGAACGTC-3′), 2 (5′-CTTAGTTTCGACTTCTCCGCGAT-3′), 3 (5′-CCTGTG CGGCATTATAACGG-3′), or 4 (5′-TGGCATACCAATTCTCG GCTA-3′), and reverse primer 5′-CGATGATGAGCTCGAAT GTTGA-3′ (see Fig. 2G). For human samples, a two-step RT–PCR procedure was used, with reverse transcription of 1 μg RNA in a reaction containing 1× first-strand buffer, 200 μM dNTPs, 500 ng oligo-dT(12–18), and 200 units RNase H(−) RT (Life Technologies) at 42°C for 1 h. Aliquots were used as templates for hot-start PCR in 25-μl reactions containing 1× buffer, 130 μM dNTPs, 5 pmol of each primer and 0.2 units Taq polymerase (Ecogen, Barcelona). Amplification products were gel purified and aliquots used for nested or seminested PCR, and products sequenced from both strands by cycle sequencing and resolution in a ABI Prism 310 automatic sequencer (Applied Biosystems). Forward primers used for RT–PCR and nested PCR were K1F (5′-GTCATTGGGCGTGATCT-3′) or KF1n (5′-GAGCTGGACGAGGACGAG-3′) for exon 1 of human Kua, K2F (5′-CAGGCTCATCGCCCACAACC-3′) for exon 2, and K4F (5′-ATGGCCTACAAGTTCCGCACC-3′), for exon 4. Reverse primers were KR (5′-GGCAGATGGCTTCG GTTTGG-3′), for exon 6 of Kua, or UR (5′-CTAAGGG GAGAAGGCAGAGA-3′), for exon C3 of UEV1 (see Fig. 3B). Amplification products were gel purified and sequenced as above. To determine relative levels of amplification of Kua–UEV and UEV transcripts, RT–PCR products were electrophoresed in ethidium bromide-containing agarose gels, and intensities (arbitrary units) determined for specific bands, normalized relative to the intensity of RT–PCR amplification products of the same RNAs with primers for the ribosomal protein gene S14r.
In Situ Hybridization on Drosophila Polytene Chromosomes
Drosophila polytene chromosome spreads were obtained from third instar wild-type larvae (Canton S strain) salivary glands. cDNA for D. melanogaster Kua was generated by RT–PCR with specific primers (primers 1 and reverse; see above and Fig. 2G), in single-tube RT–PCR reactions (Life Sciences) using as a template mRNA from Drosophila embryos. cDNA for D. melanogaster UEV corresponded to the insert in IMAGE clone LD23138 (Research Genetics), cloned in pOT2 (plasmid pOT2/DmUEV). DNA probes were labeled by random-priming (Boehringer-Mannheim). Chromosomes were denatured in 70 mM NaOH for 2 min, rinsed in 2× SSC and dehydrated in graded ethanols. Hybridization was done at 58°C overnight. Biotinylated probes were detected with streptavidin-HRP and diaminobenzidine (Sigma). Chromosomes were counterstained with Giemsa (Pardue 1994).
Expression Constructs and Transient Transfection Experiments
Full-length Kua, Kua–UEV, and UEV1A were amplified by RT–PCR using as a template total RNA from the cell lines HT-29 or Jurkat, and Expand High Fidelity polymerase (Boehringer-Mannheim), and the products subcloned in pGEM-T (Promega). The resulting inserts were amplified with primers for subcloning into pGEM11Z-HA, bearing sequences coding for the hemagluttining epitope, such that this sequence was placed in-frame at the carboxyl termini of the cDNAs. The resulting HA-tagged cDNAs were subcloned into pcDNA3.1 (Invitrogen). For transient transfection, 1 μg of plasmid DNA was transfected with Lipofectamine Plus (Life Sciences) into COS-7 cells grown on glass coverslips. As a control, pcDNA3.1 vector DNA was used. Twenty-four hours after transfection, cells were washed, fixed in 4% paraformaldehyde/PBS, and permeabilized with 1% saponin/2% BSA/PBS. Cells were incubated with rat monoclonal anti-HA antibody (Boehringer-Mannheim) for 2 h, washed, and further incubated for 1 h with FITC-conjugated goat anti-rat Ig (Dako), washed, mounted in Immuno-Fluore (ICN), and observed under a Leica confocal microscope (Wetzlar, Germany). Transfection efficiencies ranged from 10%–15%.
Acknowledgments
We thank J. Rozas for critically reviewing the manuscript, and C. Harvey for helpful contributions. This work was funded by grant PB97-1170 of the Ministerio de Educación y Ciencia (to T.M.T), and supported in part by grants BIO98-0443-C02-01 (to R.G.) and PB96-1253 of the MEC (to F.S. and M.C.), and grant 18/99 from the Fundación Marqués de Valdecilla, Santander, Spain (to J.M.). R.C., M.V., V.M.D., J.A., and M.B. were supported by fellowships from the Fundación Marqués de Valdecilla, the Fundació per a la Recerca Vall d'Hebrón, the Ministerio de Educación y Ciencia, the Instituto de Salud Carlos III (99/9345), and the Ministerio de Educación y Ciencia (FP95-38817943), respectively. The designation Kua is after the Catalan word cua, meaning “queue” or “tail”.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL tthomson@hg.vhebron.es; FAX 34-93-489-4064.
Article published online before print: Genome Res., 10.1101/gr.140500.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.140500.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Akopian AN, Okuse K, Souslova V, England S, Ogata N, Wood JN. Trans-splicing of a voltage-gated sodium channel is regulated by nerve growth factor. FEBS Lett. 1999;445:177–182. doi: 10.1016/s0014-5793(99)00126-x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman D. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan JF, Fletterick RJ, Pilis SJ. Evolution of a bifunctional enzyme: 6-phosphofructo-2-kinase/fructose-2,6bisphosphatase. PNAS. 1989;86:9642–9646. doi: 10.1073/pnas.86.24.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. GenBank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broomfield S, Chow BL, Xiao W. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. PNAS. 1998;95:5678–5683. doi: 10.1073/pnas.95.10.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzik JP, Maniatis T. Spliced leader RNAs from lower eukaryotes are trans-spliced in mammalian cells. Nature. 1992;360:692–695. doi: 10.1038/360692a0. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Caudevilla C, Serra D, Miliar A, Codony C, Asins G, Bach M, Hegardt FG. Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. PNAS. 1998;95:12185–12190. doi: 10.1073/pnas.95.21.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Intron phylogeny: A new hypothesis. Trends Genet. 1991;7:145–148. [PubMed] [Google Scholar]
- Chen JJ, Janssen BJ, Williams A, Sinha N. A gene fusion at a homeobox locus: Alterations in leaf shape and implications for morphological evolution. Plant Cell. 1997;9:1289–1304. doi: 10.1105/tpc.9.8.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Coppock DL, Cina-Poppe D, Gilleran S. The quiescin Q6 gene (QSCN6) is a fusion of two ancient gene families: Thioredoxin and ERV1. Genomics. 1998;54:460–468. doi: 10.1006/geno.1998.5605. [DOI] [PubMed] [Google Scholar]
- de Souza SJ, Long M, Klein RJ, Lin S, Gilbert W. Toward a resolution of the introns early/late debate: Only phase zero introns are correlated with the structure of ancient proteins. PNAS. 1998;95:5094–5099. doi: 10.1073/pnas.95.9.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Illopoulos I, Kyrpides NC, Ousounis CA. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- Eul J, Graessmann M, Graessmann A. Experimental evidence for RNA trans-splicing in mammalian cells. EMBO J. 1995;14:3226–3235. doi: 10.1002/j.1460-2075.1995.tb07325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink GR. Pseudogenes. Yeast? Cell. 1987;49:5–6. doi: 10.1016/0092-8674(87)90746-x. [DOI] [PubMed] [Google Scholar]
- Fox BG, Shanklin J, Somerville CR, Munck E. Stearoyl-acyl carrier protein Δ9 desaturase from Ricinus communis is a diiron-oxo protein. PNAS. 1993;90:2486–2490. doi: 10.1073/pnas.90.6.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- ————— The exon theory of genes. Cold Spring Harbor Symp Quant Biol. 1987;52:901–905. doi: 10.1101/sqb.1987.052.01.098. [DOI] [PubMed] [Google Scholar]
- Gilbert W, de Souza SJ, Long M. Origin of genes. PNAS. 1997;94:7698–7703. doi: 10.1073/pnas.94.15.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigó R, Knudsen S, Drake N, Smith T. Prediction of gene structure. J Mol Biol. 1992;226:141–157. doi: 10.1016/0022-2836(92)90130-c. [DOI] [PubMed] [Google Scholar]
- Hartl DL, Nurminsky DI, Jones RW, Lozovskaya ER. Genome structure and evolution in Drosophila: Applications of the framework P1 map. PNAS. 1994;9:6824–6829. doi: 10.1073/pnas.91.15.6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hoffman RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- Kingzette M, Spieker-Polet H, Yam PC, Zhai SK, Knight KL. Trans-chromosomal recombination within the Ig heavy chain switch region in B lymphocytes. PNAS. 1998;95:11840–11845. doi: 10.1073/pnas.95.20.11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E, Abagyan RA. TSG101 may be the prototype of a class of dominant negative ubiquitin regulators. Nat Genet. 1997;16:3331–3341. doi: 10.1038/ng0897-330. [DOI] [PubMed] [Google Scholar]
- Kopito RR. ER quality control: The cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- Li BL, Li XL, Duan ZJ, Lee O, Lin S, Ma ZM, Chang CC, Yang XY, Park JP, Mohandas TK, et al. Human acyl-CoA:cholesterol acyltransferase-1 (ACAT-1) gene organization and evidence that the 4.3-kilobase ACAT-1 mRNA is produced from two different chromosomes. J Biol Chem. 1999;274:11060–11071. doi: 10.1074/jbc.274.16.11060. [DOI] [PubMed] [Google Scholar]
- Long M, Langley CH. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science. 1993;260:91–95. doi: 10.1126/science.7682012. [DOI] [PubMed] [Google Scholar]
- Long M, Rosenberg C, Gilbert W. Intron phase correlations and the evolution of the intron/exon structure of genes. PNAS. 1995;92:12495–12499. doi: 10.1073/pnas.92.26.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Wang W, Zhang J. Origin of new genes and source for N-terminal domain of the chimerical gene, jingwei, in Drosophila. Gene. 1999;238:135–141. doi: 10.1016/s0378-1119(99)00229-2. [DOI] [PubMed] [Google Scholar]
- Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- McCarrey JR, Thomas K. Human testis-specific PGK gene lacks introns and possesses characteristics of a processed gene. Nature. 1987;326:501–505. doi: 10.1038/326501a0. [DOI] [PubMed] [Google Scholar]
- Mitchell AG, Martin CE. Fah1p, a Saccharomyces cerevisiae cytochrome b5 fusion protein, and its Arabidopsis thaliana homolog that lacks the cytochrome b5 domain both function in the alpha-hydroxylation of sphingolipid-associated very long chain fatty acids. J Biol Chem. 1997;272:28281–28288. doi: 10.1074/jbc.272.45.28281. [DOI] [PubMed] [Google Scholar]
- Mott R. EST_GENOME: A program to align spliced DNA sequences to unspliced genomic DNA. Comput Appl Biosci. 1997;13:477–478. doi: 10.1093/bioinformatics/13.4.477. [DOI] [PubMed] [Google Scholar]
- Naito Y, Riggs CK, Vanderbon TL, Riggs AF. Origin of a “bridge” intron in the gene for a two-domain globin. PNAS. 1991;88:6672–6676. doi: 10.1073/pnas.88.15.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurminsky DI, Nurminskaya MV, De Aguiar D, Hartl DL. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature. 1998;396:572–575. doi: 10.1038/25126. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Berlin: Springer-Verlag; 1970. [Google Scholar]
- Ohta T. Role of gene duplication in evolution. Genome. 1989;31:304–310. doi: 10.1139/g89-048. [DOI] [PubMed] [Google Scholar]
- Rothofsky ML, Lin SL. CROC-1 encodes a protein which mediates transcriptional activation from the human FOS promoter. Gene. 1997;195:141–149. doi: 10.1016/s0378-1119(97)00097-8. [DOI] [PubMed] [Google Scholar]
- Pardue M-L. Looking at polytene chromosomes. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster. Practical uses in cell and molecular biology. San Diego, CA: Academic Press; 1994. pp. 333–351. [Google Scholar]
- Ponting CP, Cai Y-D, Bork P. The breast cancer gene product TSG101: A regulator of ubiquitination? J Mol Med. 1997;75:467–469. [PubMed] [Google Scholar]
- Rost B. PHD: Predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sancho E, Vila MR, Sánchez-Pulido L, Lozano JJ, Paciucci R, Nadal M, Fox M, Harvey C, Bercovich B, Loukili N, et al. Role of UEV1, an inactive variant of the E2 ubiquitin-conjugating enzymes, in in vitro differentiation and cell cycle behavior of HT-29-M6 cells. Mol Cell Biol. 1998;18:576–589. doi: 10.1128/mcb.18.1.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J, Whittle E, Fox BG. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry. 1994;33:12686–12694. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- Sherman DR, Kloek AP, Krishnan BR, Guinn B, Goldberg DE. Ascaris hemoglobin gene: Plant-like structure reflects the ancestral globin gene. PNAS. 1992;89:11696–11700. doi: 10.1073/pnas.89.24.11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer JP, Kelly RE, Rinker AG, Jr, Scully JL, Evans DR. Mammalian carbamyl phosphate synthetase (CPS). DNA sequence and evolution of the CPS domain of the Syrian hamster multifunctional protein CAD. J Biol Chem. 1990;265:10395–10402. [PubMed] [Google Scholar]
- Solovyev VV, Salamov AA, Lawrence CB. Predicting internal exons by oligonucleotide composition and discriminant analysis of spliceable open reading frames. Nucleic Acids Res. 1994;22:5156–5163. doi: 10.1093/nar/22.24.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T, Jentsch S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature. 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Durbin RA. A workbench for large-scale sequence homology analysis. Comput Appl Biosci. 1994;10:301–307. doi: 10.1093/bioinformatics/10.3.301. [DOI] [PubMed] [Google Scholar]
- Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson TM, Khalid H, Sancho E, Ariño J. Role of UEV1A, a homologue of the tumor suppressor protein TSG101, in protection from DNA damage. FEBS Lett. 1998;423:49–52. doi: 10.1016/s0014-5793(98)00060-x. [DOI] [PubMed] [Google Scholar]
- Trotman CNA. Introns-early: Slipping lately? Trends Genet. 1998;14:132–134. doi: 10.1016/s0168-9525(98)01400-0. [DOI] [PubMed] [Google Scholar]
- Ull E, Matthews KR, Tschudi C. Temporal order of RNA-processing reactions in trypanosomes: Rapid trans splicing precedes polyadenylation of newly synthesized tubulin transcripts. Mol Cell Biol. 1993;13:720–725. doi: 10.1128/mcb.13.1.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Lin SL, Broomfield S, Chow BL, Wei YF. The products of the yeast MMS2 and two human homologs (hMMS2 and CROC-1) define a structurally and functionally conserved Ubc-like protein family. Nucleic Acids Res. 1998;26:3908–3914. doi: 10.1093/nar/26.17.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi M, Zieger B, Roth GJ, Ware J. Structure and expression of the human septin gene HCDCREL-1. Gene. 1998;212:229–236. doi: 10.1016/s0378-1119(98)00146-2. [DOI] [PubMed] [Google Scholar]
- Zaphiropoulos PG. RNA molecules containing exons originating from different members of the cytochrome P450 2C gene subfamily (CYP2C) in human epidermis and liver. Nucleic Acids Res. 1999;27:2585–2590. doi: 10.1093/nar/27.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieger B, Hashimoto Y, Ware J. Alternative expression of platelet glycoprotein Ibβ mRNA from an adjacent 5′ gene with an imperfect polyadenylation signal sequence. J Clin Invest. 1997;99:520–525. doi: 10.1172/JCI119188. [DOI] [PMC free article] [PubMed] [Google Scholar]