Abstract
von Willebrand factor (VWF) is an essential component of hemostasis and has been implicated in thrombosis. Multimer size and the amount of circulating VWF are known to impact hemostatic function. We associated 78 VWF single nucleotide polymorphisms (SNPs) and haplotypes constructed from those SNPs with VWF antigen level in 7856 subjects of European descent. Among the nongenomic factors, age and body mass index contributed 4.8% and 1.6% of VWF variation, respectively. The SNP rs514659 (tags O blood type) contributed 15.4% of the variance. Among the VWF SNPs, we identified 18 SNPs that are associated with levels of VWF. The correlative SNPs are either intronic (89%) or silent exonic (11%). Although SNPs examined are distributed throughout the entire VWF gene without apparent cluster, all the positive SNPs are located in a 50-kb region. Exons in this region encode for VWF D2, D′, and D3 domains that are known to regulate VWF multimerization and storage. Mutations in the D3 domain are also associated with von Willebrand disease. Fifteen of these 18 correlative SNPs are in 2 distinct haplotype blocks. In summary, we identified a cluster of intronic VWF SNPs that associate with plasma levels of VWF, individually or additively, in a large cohort of healthy subjects.
Introduction
The von Willebrand factor (VWF) is an essential component of hemostasis at sites of vascular injury. This hemostatically active adhesion ligand also plays a critical role in thrombus formation at the site of a ruptured atherosclerotic plaque and in platelet aggregation induced by high fluid shear stress in areas of severe vascular stenosis. The former results in myocardial infarction when it occurs in coronary arteries, whereas the latter is a major cause of thromboembolism associated with ischemic stroke.1
VWF is the largest multimeric glycoprotein in the plasma. It is exclusively synthesized in endothelial cells and megakaryocytes initially as a monomeric glycoprotein. In the endoplasmic reticulum and Golgi apparatus of these cells, the monomers form C-terminal disulfide-linked dimers. A various number of dimers subsequently form multimers through N-terminal disulfide linkages, a process assisted by the proteolytic release of a 714-amino acid propeptide.2,3 The newly synthesized VWF multimers are either constitutively released into the circulation or stored in the Weibel-Palade bodies of endothelial cells and α-granules of megakaryocytes/platelets.4,5 On stimulation, these granules secrete the stored VWF multimers that are rich in ultra-large and hemostatically hyperactive forms.4,6 Ultra-large VWF multimers are also secreted under endothelial stress caused by inflammation but are rapidly and partially cleaved by the zinc metalloprotease ADAMTS13 (a disintegrin and metalloprotease with thrombospondin type 1 repeats) into smaller, variable-sized multimers.7–9 The hemostatic activity of VWF multimers is determined not only by multimer size,4,10 but also by the quantity of VWF antigen in the circulation. The plasma level of VWF multimers is determined by a dynamic balance between the rate of production and that of clearance. Although acquired conditions are known to affect synthesis and secretion,11 genetic factors play a major role in regulating VWF synthesis and clearance. It has been reported that genetic factors are responsible for up to 66% of variation in plasma VWF antigen level, of which 30% is related to ABO blood type.12,13
The human VWF gene is located on chromosome 12.14,15 It spans approximately 180 kb of nucleotides and contains 52 exons that encode a multidomain prepolypeptide of 2791 amino acids.14–16 The VWF gene is highly polymorphic.17 Single nucleotide polymorphisms (SNPs) of the coding and promoter sequences in the VWF gene have been extensively studied and found to influence the levels of VWF in the circulation. Furthermore, as a heavily glycosylated polypeptide,16,18 the level of circulating VWF is also affected by the structure of its carbohydrate side chains, which are associated with ABO blood group, primarily because of the presence or absence of a functional glycosyltransferase that adds either a N-acetylgalactosamine or a D-galactose to a D-galactose side chain on the H antigen.19 This structural variation in glycosylation has been reported to directly affect the rate of VWF clearance.20
Although considerable information has been learned, previous studies are limited in determining genetic impacts on VWF antigen level in the circulation as they had relatively small samples sizes and often targeted specific SNPs that are predominantly located in either exons or the promoter region. Large population-based studies are therefore needed to further investigate the relationship between VWF polymorphisms and levels of VWF antigen, especially given its risk predisposition to coronary heart disease, ischemic stroke,21–24 and von Willebrand disease (VWD).25 We have correlated VWF antigen levels in the plasma with 78 SNPs and haplotypes constructed from these SNPs in 7856 subjects of European descent in the Atherosclerosis Risk in Communities study (ARIC; www.cscc.unc.edu/aric).
Methods
Study population
ARIC is an ongoing prospective cohort study designed to assess subclinical atherosclerosis and clinical atherosclerotic events.26 Baseline samples were collected from 15 792 adults 45 to 64 years of age from 1987 to 1989 using probability sampling from Forsyth County, NC; Jackson, MS; the northwestern suburbs of Minneapolis, MN; and Washington County, MD. The cohort consists predominantly of subjects of European and African American descent, but this study was exclusively focused on subjects of European descent as information on ABO blood group was not available for African Americans when the data were analyzed. The use of human samples was approved by the National Institutes of Health Data Monitoring board and institutions.
Baseline measurements
Blood was drawn after an 8-hour or longer fasting period from an antecubital vein. VWF antigen was determined by a commercial enzyme-linked immunosorbent assay kit from American Bioproducts, and results were reported as a percentage of the Universal Coagulation Reference Plasma (Thromboscreen, Pacific Hemostasis, Curtin Matheson Scientific). The reliability coefficient (method variance plus intraindividual variance divided by total variance) obtained from repeat testing of plasma samples (from single blood draw) over several weeks was 0.68.
The following conditions were adjusted for as confounding factors (covariance) as they are known to affect plasma VWF antigen levels: age, body mass index (BMI), sex, hypertension, diabetes, and current smoking status.11
Genotyping and haplotype construction
SNPs generated from the Affymetrix, Version 6.0 platform in the region encompassing the VWF gene (5 928 997-6 098 180 bp on chromosome 12) were included in the data analyses. We also used the fastPHASE 1.227 program to resolve haplotypes from the unphased SNP genotype data for the downstream haplotype-based association analysis. To characterize the linkage disequilibrium (LD) pattern in the region of interest, we applied Haploview28 to determine the regions in strong LD, their cosegregation rates, and underlying haplotypes in each region.
Imputation of ABO blood group
The ABO blood groups were estimated using the SNP dosage in the ARIC genome-wide association study (GWAS) database. Four SNPs were used to serve as tag-SNPs for 4 major ABO genotypic blood groups based on LD (R2 > 0.7) in the HapMap CEU sample: rs514659 (tag O group), rs8176749 (one of the functional variants for B group), rs8176704 (tag A2 group), and rs512770 (the functional variant for O2).29,30
Data analysis
All SNPs were evaluated for Hardy-Weinberg equilibrium by χ2 statistics. Two P values were calculated for each SNP using the R function “chisq.test.” One was based on the asymptotic χ2 distribution, and the other was based on up to 20 000 permutations. Specifically, the P value of each SNP was first calculated by 2000 permutations; then, for all the SNPs with P values of < .01, 20 000 permutations were used to obtain more accurate P values.
Linear models were used to evaluate the association of actual or log VWF antigen levels with each SNP. Two sets of covariates were considered. Set A included 6 confounding factors: age, BMI, sex, hypertension, diabetes, and current smoker status. The age and BMI were continuous measurements, and the remaining 4 covariates were binary indicators. Set B included all variables in set A plus 4 surrogate genotypes for ABO blood types.
The frequencies of each haplotype across all the persons were calculated, which could take values of 0, 1, or 2. Linear models were applied with actual or log VWF antigen level as responses and haplotype frequency as a predictor while controlling for the confounding effects of covariate set A or B. They were also used to assess the significance of all haplotypes within each LD block. We used 2 common methods to map haplotypes directly with genotype data: the score tests approach,31 which is implemented in an R package R/haplo.stats, and a fully likelihood method,32 which is implemented in a stand-alone HAPSTAT program.
The statistical significance was determined as P < .0004, which was calculated based on a formula of 0.05 divided by 120 (the maximal numbers of potential tests in this study, including 78 SNPs, 34 haploptypes, and 8 LD blocks).
Results
Of the ARIC participants of European descent, 8125 were available for analysis, but 269 were excluded because of (1) failure of SNP data to meet GWAS quality control standards as previously discussed30 (n = 150), (2) missing data on VWF antigen level (n = 31), (3) missing one of the covariates used for data adjustment (n = 66), or (4) missing information on ABO blood type (n = 22). The remaining 7856 were included in the final analysis. The distribution of covariates is listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
VWF antigen level
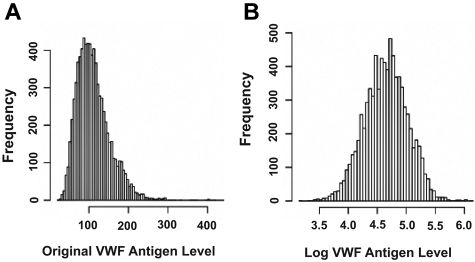
The level of plasma VWF antigen varied from 24% to 456%. The mean ± SD VWF antigen level for males (112.9% ± 42.7%) was significantly higher than that for females (111.1% ± 42.4%; Wilcoxon rank-sum test, P = .05, supplemental Table 1). Supplemental Table 2 lists the percentile of subjects in groups based on VWF antigen levels, with 45% of subjects having VWF antigen levels at 100% or less. Up to 5% of subjects have VWF levels of < 56%, but their bleeding profiles are presently unknown. As shown in Figure 1, the distribution of the VWF antigen was skewed to the right, which could potentially invalidate the normal distribution assumption of a regression model. All the data were therefore analyzed for original and log VWF antigen levels, but only data generated with log VWF antigen are presented. The VWF antigen was analyzed before and after ABO blood type adjustment. The SNP rs514659 (type O tag) has the strongest influence, contributing to 15.4% of the variance of log VWF levels, and the second strongest is rs8176704 (type A tag) at 4.1% (supplemental Table 3). Among the nongenetic covariates, age and BMI had stronger influence, accounting for approximately 4.8% and 1.6% of the variance of log VWF values. This finding is consistent with an early report from the ARIC study.11
Figure 1.
Plasma VWF levels. Levels were plotted based on actual and log values to correct a skewed distribution of actual VWF antigen, which could potentially invalidate the normal distribution assumption of a regression model.
SNP correlation with VWF antigen level
Of the 78 VWF SNPs analyzed for this study, 72 (92.3%) are intronic and 6 (7.7%) are exonic. The frequencies and locations of these SNPs are listed in supplemental Table 4. The frequency of homozygosity for rare alleles varied from 0% to 24.4% with 13 SNPs being < 1% and 4 SNPs not detected. The changes in nucleic acids and amino acids for the 6 exonic SNPs and their domain locations, constructed based on a report by Mancuso et al17 and the Sheffield VWF Database (www.VWF.group.shef.ac.uk), are listed in supplemental Table 5. Four SNPs found to be in Hardy-Weinberg disequilibrium were removed from constructing the haplotype map and haplotype specific analysis but contributed to SNP-specific P value calculations.
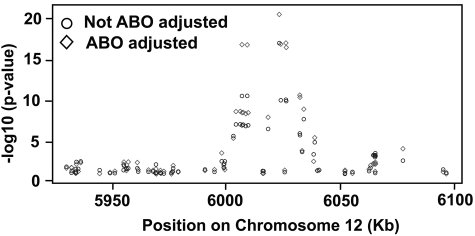
The P values for association of the 78 SNPs with VWF antigen levels are listed in supplemental Table 6. Of those, 18 (23.1%) were significantly associated with log VWF antigen levels before they were adjusted for ABO blood group (Table 1). After adjustment for ABO, the association of positive SNPs with VWF antigen levels grew stronger. One striking observation is that the 78 SNPs are distributed throughout the VWF gene without apparent hot spots, but all positive SNPs are clustered in a 50-kb region of the VWF gene (Figure 2). The positive SNPs were predominantly in introns, except 2 coding SNPs, which are located in exon 18 and exon 22 (rs1800380 and rs1063857), but neither resulted in actual changes in amino acid sequence (supplemental Table 5). The effect size of the positive SNPs was relatively small, with the strongest association (rs1063857) explaining approximately 0.9% of VWF (or log VWF) variation. However, a dosing/additive effect was detected among the positive SNPs.
Table 1.
P values of SNPs significantly associated with VWF antigen levels
| SNP | Log VWF level |
Location in the gene | Location in haplotype block | |
|---|---|---|---|---|
| Not ABO-adjusted | ABO-adjusted | |||
| rs2854871 | 6.6 × 10−6 .05 (.01) | 1.1 × 10−5 .04 (.01) | Intron 24 | Block 5 |
| rs11609815 | 9.4 × 10−8 .03 (.01) | 2.2 × 10−9 .03 (.01) | Intron 24 | Block 5 |
| rs11063995 | 8.9 × 10−8 .03 (.01) | 2.1 × 10−9 .03 (.01) | Intron 22 | Block 5 |
| rs216315 | 1.7 × 10−11 -.07 (.01) | 1.2 × 10−18 -.08 (.01) | Intron 22 | Block 5 |
| rs11612401 | 1.3 × 10−7 .03 (.01) | 3.2 × 10−9 .03 (.01) | Intron 22 | Block 5 |
| rs11610629 | 1 × 10−7 .03 (.01) | 2.8 × 10−9 .03 (.01) | Intron 22 | Block 5 |
| rs1800380 | 1.6 × 10−7 .03 (.01) | 4.4 × 10−9 .03 (.01) | Exon 22 | Block 5 |
| rs216318 | 1.7 × 10−11 -.07 (.01) | 1.3 × 10−18 -.08 (.01) | Intron 21 | Block 5 |
| rs11609728 | 1.2 × 10−7 .03 (.01) | 3.2 × 10−9 .03 (.01) | Intron 21 | Block 5 |
| rs11614912 | 4.9 × 10−7 .03 (.01) | 1.4 × 10−8 .03 (.01) | Intron 18 | Block 5 |
| rs1063857* | 3.9 × 10−19 .05 (.01) | 7.7 × 10−23 .05 (.01) | Exon 18 | Block 5 |
| rs216295 | 5.5 × 10−11 -.06 (.01) | 1 × 10−18 -.08 (.01) | Intron 17 | Block 5 |
| rs216298 | 5.7 × 10−11 -.06 (.01) | 7 × 10−19 -.08 (.01) | Intron 16 | Block 5 |
| rs216299 | 8.9 × 10−11 -.06 (.01) | 2.9 × 10−18 -.07 (.01) | Intron 16 | No |
| rs2239161 | 3.4 × 10−6 -.04 (.01) | 1.2 × 10−11 -.05 (.01) | Intron 15 | Block 6 |
| rs2239160 | 5.9 × 10−6 -.04 (.01) | 2.4 × 10−11 -.05 (.01) | Intron 15 | Block 6 |
| rs12304995 | 2.7 × 10−8 .03 (.01) | 1 × 10−9 .03 (.01) | Intron 15 | No |
| rs7139057 | 4.3 × 10−5 .03 (.01) | 9.3 × 10−6 .03 (.01) | Intron 13 | Block 7 |
| rs41388848 | .014-.04 (.02) | .00032-.05 (.01) | Intron 5 | Block 8 |
In each cell, the first row is the P value followed by the coefficient estimate (the increase of logVWF with one extra minor allele) and the corresponding SE.
Also identified in a GWAS by the CHARGE consortium.30
Figure 2.
P values of association between plasma VWF antigen and individual SNPs in the VWF gene. The data were analyzed based on log VWF antigen with or without ABO adjustment. The data show a clear cluster of positive SNPs in the 6000- to 6050-kb region, even though all SNPs analyzed in this study were distributed throughout the entire VWF gene.
Haplotype construction and correlation with VWF antigen levels
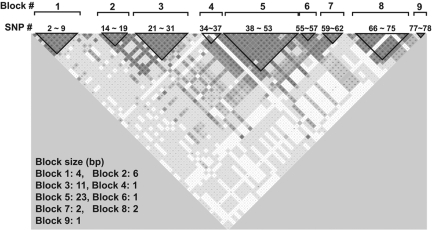
Studies in the past have identified many specific SNPs that affect plasma VWF antigen levels but had limited power to construct haplotypes because of insufficient sample sizes.33–35 Our large sample set allowed us to construct a VWF haplomap that included rare SNPs to identify specific SNPs that are transmitted together. We found 9 separate blocks that are in LD in the VWF gene (Figure 3). Figure 4 lists the common haplotypes that account for more than 98% of all haplotypes found in the cohort.
Figure 3.
VWF haplotypes constructed using the Haploview program. The haplotypes were constructed using the default setting that (1) ignores pairwise comparisons of markers more than 500 kb apart, (2) excludes persons with more than 50% of missing genotypes, (3) examines haplotypes found 1% or more in the population, (4) removes markers with Hardy-Weinberg disequilibrium, and (5) uses R2 to define haplotype blocks. The majority of them are found in 9 LD blocks that distributed throughout the entire VWF gene.
Figure 4.
Frequencies of VWF haplotypes constructed with the Haploview program. The interactions between haplotypes in adjacent LD blocks are indicated.
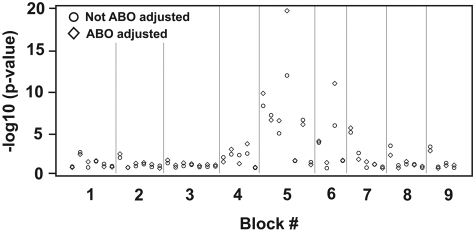
Each LD block contained a various number of haplotypes with different frequencies. Blocks 5 and 6 were significantly correlated with VWF antigen levels regardless of whether VWF antigen was expressed as the original (data not shown) or log values (Figure 5; Table 2). Block 5 had the most significant association among all the genetic variations we tested with an effect size (calculated by R2) of 1.5% for log VWF levels, much stronger than the most significant single SNP (rs10638570). The result is consistent with an additive effect among individual SNPs. Consistent with individual SNPs, the association between specific haplotype blocks and VWF antigen levels was significantly improved after the ABO adjustment (Table 2). Further analysis found that the first to fourth haplotypes in block 5 and the third haplotype in block 6 were individually associated with VWF antigen levels. These results were further verified with haplotype mapping methods that directly used the genotype data (data not shown).
Figure 5.
P values of the associations between VWF haplotypes and log VWF antigen with or without ABO adjustment, with and without ABO adjustment. There is clearly a cluster of significant associations in blocks 5 and 6.
Table 2.
P values for haplotype association with VWF levels
| Block no. | Log VWF level, % |
|
|---|---|---|
| Not ABO-adjusted | ABO-adjusted | |
| 1 | .14 | .15 |
| 2 | .54 | .16 |
| 3 | .91 | .36 |
| 4 | .0028 | .0022 |
| 5 | 2.4 × 10−24 | 1.2 × 10−32 |
| 6 | 2.6 × 10−6 | 4.6 × 10−11 |
| 7 | .0015 | .00068 |
| 8 | .038 | .15 |
| 9 | .028 | .05 |
Discussion
We have analyzed the relationship between 78 VWF SNPs and haplotypes constructed from those SNPs and plasma levels of VWF antigen in 7856 subjects of European descent in the ARIC cohort. We have found a significant association between multiple SNPs after correction for factors known to affect VWF antigen levels, including age, sex, diabetes mellitus, hypertension, BMI, tobacco use, and ABO blood group.12 Among the nongenetic confounding factors, age and BMI explains most of the VWF variation (4.8% and 1.6%, respectively), whereas the contributions from hypertension, diabetes, and smoking are very small. VWF levels are known to increase with aging, but the underlying mechanism for the BMI effect remains to be further investigated. It is unlikely that BMI affects circulating VWF antigen levels because of obesity and associated chronic systemic inflammation because other conditions known to associate with inflammation (diabetes and smoking) do not have this positive association.
Eighteen of the 78 SNPs studied (23%) were significantly associated with VWF antigen levels (Table 1). One of these positive SNPs (rs1063857) was also identified as highly correlative of VWF level in a GWAS conducted recently by the CHARGE consortium.30 (ARIC contributes to the CHARGE dataset.) After construction of a haplotype map, we identified 9 distinct haplotype blocks that are in LD (Figure 3), each containing a different number of haplotypes (Figure 4). Thirteen of the 18 SNPs (72%) with a positive association are aggregated in block 5 and 2 of 18 SNPs (11%) are in block 6. These are the only LD blocks that are significantly associated with levels of VWF antigen (Figure 5; Table 2). Together, this study yielded several interesting findings.
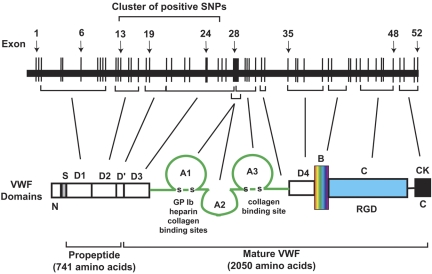
First, the positive SNPs (and positive haplotypes) are strikingly concentrated in a 50-kb region of the approximately 180-kb VWF gene on chromosome 12 (27.8% of the VWF gene; Figures 2 and 5), even though the individual SNPs included in the data analyses scattered throughout the entire VWF gene. This is a very strong signal from a localized region of the gene. The strong localized signal in this large population provides a compelling argument that this region has a strong causal relationship with plasma VWF antigen levels. Although the effect size of each positive SNP is small, it could become significant because of additive effects as demonstrated by the haplotype analysis and nongenomic factors (supplemental Table 3). The region that contains all the positive SNPs spans from exons 13 to 24 and their intron intervals (Table 1) and encodes the D2, D′, and D3 domains of VWF (Figure 6).16,18 The D2 domain and a portion of D′ constitute the major part of the VWF propeptide, which is cleaved in the Golgi to facilitate multimerization and subsequent storage in granules.36–38 Mutations in the VWF propeptide, specifically in the D2 domain, have been shown to result in decreased VWF storage, multimerization, and secretion.39 They also impact the ratio of VWF propeptide to mature VWF, which in turn affects the clearance of VWF in the circulation.40 The D3 domain is important in VWF multimerization, which could impact VWF tertiary structure and ultimately circulating levels of VWF and its clearance.39 It is also interesting to note that the rate of VWF clearance is found to be accelerated in patients with VWD Vicenza, which often have mutations, including intronic, in the D domains41–43 (we did not find positive SNPs in the exon encoding D4 and flanking intronic sequence). However, extensive biologic experiments are required to establish a mechanistic link between positive SNPs and VWF expression in general and the putative role of this 50-kb region in regulating VWF expression in particular.
Figure 6.
A schematic illustration of the VWF gene and its exon correspondence to specific domains of a monomeric VWF polypeptide.
Second, the positive SNPs are predominantly found in introns. Intronic variants are increasingly recognized as a critical disease-causing genetic constituent. Several large GWAS have associated intronic SNPs with hearing impairment,44 bipolar disorder,45 asthma,46 and rheumatoid arthritis.47 A recent report also associates an intronic SNP with serum level of transferrin.48 Furthermore, chromosome 9 region p21 (9p21), which is a large stretch of noncoding sequence away from any known genes, has been shown to associate with coronary heart disease.49 Indeed, by surveying 21 429 disease-causing SNPs from 2113 publications, Chen et al50 concluded that synonymous and nonsynonymous SNPs shared similar likelihood and effect size for disease association. The exact mechanism(s) for these intronic SNPs to affect VWF antigen levels remains to be further investigated, but they could impact the form and efficiency of gene splicing and mRNA stability.51 Alternatively, one could speculate that the positively correlated introns encode for intronic miRNA52 that could potentially regulate VWF transcription. The speculation is supported by recent reports that miRNA regulates fibrinogen production53 and this regulation could determine a person's sensitivity to chronic thromboembolic pulmonary hypertension.54 Finally, these positive intronic SNPs, which are almost entirely in LD with each other, could be linked to an as yet unknown gene(s) or region(s) that impacts the synthesis, stability, and/or clearance of VWF. Consistent with this notion, several SNPs outside of the VWF gene have been found to associate with VWF antigen in a recent GWAS by the CHARGE consortium.30
Third, we found 2 haplotype blocks (blocks 5 and 6) that are significantly associated with VWF level and contain 15 (83.3%) of the 18 positive SNPs (Tables 1 and 3). The association between haplotype blocks and VWF antigen levels was stronger compared with individual SNPs, suggesting an additive effect among SNPs. This could mean that certain haplotype blocks more accurately localize the mutation, which has a causal link to VWF level or capture favorable combinations of mutations that determine VWF level when expressed together.
Table 3.
P values of association of individual haplotypes in block 5 and 6 with VWF levels
| Haplotype | Frequency | Log VWF value |
|
|---|---|---|---|
| Not ABO-adjusted | ABO-adjusted | ||
| Block 5-1 | .23 | 1.7 × 10−8 .04 (.01) | 7.8 × 10−10 .04 (.01) |
| Block 5-2 | .43 | 2.6 × 10−7 .03 (.01) | 1.4 × 10−6 -.02 (.01) |
| Block 5-3 | .07 | 4.5 × 10−5 .04 (.01) | 1.6 × 10−6 .05 (.01) |
| Block 5-4 | .08 | 5.9 × 10−12 -.07 (.01) | 7 × 10−20 -.08 (.01) |
| Block 5-5 | .06 | 0.13 .02 (.01) | 0.11 .02 (.01) |
| Block 5-6 | .04 | 1.7 × 10−6 .07 (.01) | 4.7 × 10−6 .06 (.01) |
| Block 5* | 2.4 × 10−24 | 1.2 × 10−32 | |
| Block 6-1 | .28 | .00076 .02 (.01) | .00051 .02 (.01) |
| Block 6-2 | .61 | .99 < .01 (.01) | 0.21 .01 (.01) |
| Block 6-3 | .11 | 9 × 10−6 -.04 (.01) | 4.9 × 10−11 -.05 (.01) |
| Block 6* | 2.6 × 10−6 | 4.6 × 10−11 | |
In each cell of the 2 columns at right, the first row is the P value followed by the coefficient estimate (the increase of logVWF with 1 extra haplotype) and the corresponding SE.
The P value for the entire block instead of individual haplotypes in the block.
Notably, exonic and intronic splicing mutation SNPs that have been previously identified in patients with VWD55–58 are not detected in the ARIC cohort, largely because ARIC recruited healthy subjects, who did not have bleeding manifestation or family history of bleeding based on an extensive questionnaire administered at the time of blood draws. Despite studying a healthy cohort, up to 5% of subjects have VWF antigen levels of 56% or less, (supplemental Table 2), which is a level close to that found in patients with type 1 VWD. As an epidemiologic study to investigate the etiology and natural history of atherosclerosis and clinical atherosclerotic disease, ARIC collected limited information on bleeding phenotypes. It therefore remains unclear as to whether these subjects have a bleeding tendency
Finally, it has been well established that ABO blood group has a significant impact on circulating VWF level. Subjects with O blood group have consistently lower levels of VWF, showing an average 25% reduction compared with non-O blood groups,59 potentially caused by a greater rate of clearance.12,13,20 Among the 4 SNPs that we used to type ABO blood groups, rs514659, which tags O blood type, has the strongest correlation, contributing to 15.4% of log VWF variation. The finding is consistent with previous reports. The second strongest association is found with rs8176704 (type A tag), which contributes to 4.1% of log VWF variation. The association of VWF SNPs and haplotypes with VWF levels grew stronger after ABO adjustment. The data suggest that either there is a dosing effect of multiple SNPs (as suggested by comparing the effect size between individual SNPs and haplotypes) or we have further localized the SNPs that have a causal link to VWF level.
In conclusion, we have analyzed the subjects of European descent within the ARIC cohort. We found a significant number of SNPs and derivative haplotypes that have a significant association with levels of circulating VWF antigen. These SNPs are almost exclusively localized to a 50-kb region of the VWF gene that codes for domains involved in encoding propeptide and regulating VWF multimerization. Our data suggest that this region plays an important role in regulating VWF antigen in the circulation by either controlling the rate of gene splicing, miRNA regulation, or serving as markers for other regulatory elements in and beyond the VWF gene.
Supplementary Material
Acknowledgments
The authors thank the staff and participants of the ARIC study for their important contributions.
The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute (contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, and N01-HC-55022). This work was supported by the ARIC contract and the National Heart, Lung, and Blood Institute (grant HL71895).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.C. and A.R.F. designed the study, analyzed the data, and wrote the manuscript; W.S. and C.B. analyzed the data and wrote the manuscript; F.Y. constructed and analyzed the haplotype and wrote the manuscript; M.B. performed VWF SNP screen and identification; W.T. performed imputation of ABO information; L.E.C. developed the hypothesis and analyzed the data; K.K.W. wrote the manuscript; E.B. developed the hypothesis, designed the study, and wrote the manuscript; and J.D. developed the hypothesis, designed the study, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jing-fei Dong, Section of Cardiovascular Sciences, Department of Medicine, BCM286, N1319, One Baylor Plaza, Baylor College of Medicine, Houston, TX 77030; e-mail: jfdong@bcm.tmc.edu.
References
- 1.Lloyd-Jones D, Adams R, Carnethon M, et al. Heart disease and stroke statistics—2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119(3):480–486. doi: 10.1161/CIRCULATIONAHA.108.191259. [DOI] [PubMed] [Google Scholar]
- 2.Isaacs NW. Cystine knots. Curr Opin Struct Biol. 1995;5(3):391–395. doi: 10.1016/0959-440x(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 3.Wagner DD, Lawrence SO, Ohlsson-Wilhelm BM, Fay PJ, Marder VJ. Topology and order of formation of interchain disulfide bonds in von Willebrand factor. Blood. 1987;69(1):27–32. [PubMed] [Google Scholar]
- 4.Sporn LA, Marder VJ, Wagner DD. Inducible secretion of large, biologically potent von Willebrand factor multimers. Cell. 1986;46(2):185–190. doi: 10.1016/0092-8674(86)90735-x. [DOI] [PubMed] [Google Scholar]
- 5.Brouland JP, Egan T, Roussi J, et al. In vivo regulation of von Willebrand factor synthesis: von Willebrand factor production in endothelial cells after lung transplantation between normal pigs and von Willebrand factor-deficient pigs. Arterioscler Thromb Vasc Biol. 1999;19(12):3055–3062. doi: 10.1161/01.atv.19.12.3055. [DOI] [PubMed] [Google Scholar]
- 6.Moake JL, Chow TW. Increased von Willebrand factor (vWf) binding to platelets associated with impaired vWf breakdown in thrombotic thrombocytopenic purpura. J Clin Apher. 1998;13(3):126–132. doi: 10.1002/(sici)1098-1101(1998)13:3<126::aid-jca6>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 7.Furlan M, Robles R, Lammle B. Partial purification and characterization of a protease from human plasma cleaving von Willebrand factor to fragments produced by in vivo proteolysis. Blood. 1996;87(10):4223–4234. [PubMed] [Google Scholar]
- 8.Tsai HM. Physiologic cleavage of von Willebrand factor by a plasma protease is dependent on its conformation and requires calcium ion. Blood. 1996;87(10):4235–4244. [PubMed] [Google Scholar]
- 9.Levy GG, Nichols WC, Lian EC, et al. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature. 2001;413(6855):488–494. doi: 10.1038/35097008. [DOI] [PubMed] [Google Scholar]
- 10.Sporn LA, Marder VJ, Wagner DD. von Willebrand factor released from Weibel-Palade bodies binds more avidly to extracellular matrix than that secreted constitutively. Blood. 1987;69(5):1531–1534. [PubMed] [Google Scholar]
- 11.Conlan MG, Folsom AR, Finch A, et al. Associations of factor VIII and von Willebrand factor with age, race, sex, and risk factors for atherosclerosis: the Atherosclerosis Risk in Communities (ARIC) Study. Thromb Haemost. 1993;70(3):380–385. [PubMed] [Google Scholar]
- 12.Orstavik KH, Magnus P, Reisner H, et al. Factor VIII and factor IX in a twin population: evidence for a major effect of ABO locus on factor VIII level. Am J Hum Genet. 1985;37(1):89–101. [PMC free article] [PubMed] [Google Scholar]
- 13.Shima M, Fujimura Y, Nishiyama T, et al. ABO blood group genotype and plasma von Willebrand factor in normal individuals. Vox Sang. 1995;68(4):236–240. doi: 10.1111/j.1423-0410.1995.tb02579.x. [DOI] [PubMed] [Google Scholar]
- 14.Ginsburg D, Handin RI, Bonthron DT, et al. Human von Willebrand factor (vWF): isolation of complementary DNA (cDNA) clones and chromosomal localization. Science. 1985;228(4706):1401–1406. doi: 10.1126/science.3874428. [DOI] [PubMed] [Google Scholar]
- 15.Collins CJ, Underdahl JP, Levene RB, et al. Molecular cloning of the human gene for von Willebrand factor and identification of the transcription initiation site. Proc Natl Acad Sci U S A. 1987;84(13):4393–4397. doi: 10.1073/pnas.84.13.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Titani K, Kumar S, Takio K, et al. Amino acid sequence of human von Willebrand factor. Biochemistry. 1986;25(11):3171–3184. doi: 10.1021/bi00359a015. [DOI] [PubMed] [Google Scholar]
- 17.Mancuso DJ, Tuley EA, Westfield LA, et al. Structure of the gene for human von Willebrand factor. J Biol Chem. 1989;264(33):19514–19527. [PubMed] [Google Scholar]
- 18.Sadler JE. Biochemistry and genetics of von Willebrand factor. Annu Rev Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins PV, O'Donnell JS. ABO blood group determines plasma von Willebrand factor levels: a biologic function after all? Transfusion. 2006;46(10):1836–1844. doi: 10.1111/j.1537-2995.2006.00975.x. [DOI] [PubMed] [Google Scholar]
- 20.Gallinaro L, Cattini MG, Sztukowska M, et al. A shorter von Willebrand factor survival in O blood group subjects explains how ABO determinants influence plasma von Willebrand factor. Blood. 2008;111(7):3540–3545. doi: 10.1182/blood-2007-11-122945. [DOI] [PubMed] [Google Scholar]
- 21.Gottesman RF, Cummiskey C, Chambless L, et al. Hemostatic factors and subclinical brain infarction in a community-based sample: the ARIC study. Cerebrovasc Dis. 2009;28(6):589–594. doi: 10.1159/000247603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danesh J, Wheeler JG, Hirschfield GM, et al. C-reactive protein and other circulating markers of inflammation in the prediction of coronary heart disease. N Engl J Med. 2004;350(14):1387–1397. doi: 10.1056/NEJMoa032804. [DOI] [PubMed] [Google Scholar]
- 23.Bongers TN, de Maat MP, van Goor ML, et al. High von Willebrand factor levels increase the risk of first ischemic stroke: influence of ADAMTS13, inflammation, and genetic variability. Stroke. 2006;37(11):2672–2677. doi: 10.1161/01.STR.0000244767.39962.f7. [DOI] [PubMed] [Google Scholar]
- 24.Frankel DS, Meigs JB, Massaro JM, et al. Von Willebrand factor, type 2 diabetes mellitus, and risk of cardiovascular disease: the Framingham Offspring Study. Circulation. 2008;118(24):2533–2539. doi: 10.1161/CIRCULATIONAHA.108.792986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadler JE, Ginsburg D. A database of polymorphisms in the von Willebrand factor gene and pseudogene: for the Consortium on von Willebrand Factor Mutations and Polymorphisms and the Subcommittee on von Willebrand Factor of the Scientific and Standardization Committee of the International Society on Thrombosis and Haemostasis. Thromb Haemost. 1993;69(2):185–191. [PubMed] [Google Scholar]
- 26.ARIC investigators. The Atherosclerosis Risk in Communities (ARIC) study: design and objectives. Am J Epidemiol. 1989;129(4):687–702. [PubMed] [Google Scholar]
- 27.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78(4):629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.Olsson ML, Chester MA. Polymorphism and recombination events at the ABO locus: a major challenge for genomic ABO blood grouping strategies. Transfus Med. 2001;11(4):295–313. doi: 10.1046/j.1365-3148.2001.00320.x. [DOI] [PubMed] [Google Scholar]
- 30.Smith NL, Chen MH, Dehghan A, et al. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: the CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation. 2010;121(12):1382–1392. doi: 10.1161/CIRCULATIONAHA.109.869156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin D, Zheng D. Likelihood-based inference on haplotype effects in genetic association studies. J Am Statist Assoc. 2006;101(473):89–104. [Google Scholar]
- 33.Davies JA, Collins PW, Hathaway LS, Bowen DJ. Effect of von Willebrand factor Y/C1584 on in vivo protein level and function and interaction with ABO blood group. Blood. 2007;109(7):2840–2846. doi: 10.1182/blood-2006-07-035105. [DOI] [PubMed] [Google Scholar]
- 34.Keightley AM, Lam YM, Brady JN, Cameron CL, Lillicrap D. Variation at the von Willebrand factor (vWF) gene locus is associated with plasma vWF:Ag levels: identification of three novel single nucleotide polymorphisms in the vWF gene promoter. Blood. 1999;93(12):4277–4283. [PubMed] [Google Scholar]
- 35.Harvey PJ, Keightley AM, Lam YM, Cameron C, Lillicrap D. A single nucleotide polymorphism at nucleotide −1793 in the von Willebrand factor (VWF) regulatory region is associated with plasma VWF:Ag levels. Br J Haematol. 2000;109(2):349–353. doi: 10.1046/j.1365-2141.2000.02000.x. [DOI] [PubMed] [Google Scholar]
- 36.Mayadas TN, Wagner DD. Vicinal cysteines in the prosequence play a role in von Willebrand factor multimer assembly. Proc Natl Acad Sci U S A. 1992;89(8):3531–3535. doi: 10.1073/pnas.89.8.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haberichter SL, Jacobi P, Montgomery RR. Critical independent regions in the VWF propeptide and mature VWF that enable normal VWF storage. Blood. 2003;101(4):1384–1391. doi: 10.1182/blood-2002-07-2281. [DOI] [PubMed] [Google Scholar]
- 38.Wagner DD, Saffaripour S, Bonfanti R, et al. Induction of specific storage organelles by von Willebrand factor propolypeptide. Cell. 1991;64(2):403–413. doi: 10.1016/0092-8674(91)90648-i. [DOI] [PubMed] [Google Scholar]
- 39.Haberichter SL, Allmann AM, Jozwiak MA, Montgomery RR, Gill JC. Genetic alteration of the D2 domain abolishes von Willebrand factor multimerization and trafficking into storage. J Thromb Haemost. 2009;7(4):641–650. doi: 10.1111/j.1538-7836.2009.03290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gadisseur A, Berneman Z, Schroyens W, Michiels JJ. Laboratory diagnosis of von Willebrand disease type 1/2E (2A subtype IIE), type 1 Vicenza and mild type 1 caused by mutations in the D3, D4, B1-B3 and C1-C2 domains of the von Willebrand factor gene: role of von Willebrand factor multimers and the von Willebrand factor propeptide/antigen ratio. Acta Haematol. 2009;121(2):128–138. doi: 10.1159/000214853. [DOI] [PubMed] [Google Scholar]
- 41.Castaman G, Tosetto A, Rodeghiero F. Reduced von Willebrand factor survival in von Willebrand disease: pathophysiologic and clinical relevance. J Thromb Haemost. 2009;7(suppl 1):71–74. doi: 10.1111/j.1538-7836.2009.03381.x. [DOI] [PubMed] [Google Scholar]
- 42.Michiels JJ, Berneman Z, Gadisseur A, et al. Laboratory diagnosis and molecular basis of mild von Willebrand disease type 1. Acta Haematol. 2009;121(2):85–97. doi: 10.1159/000214847. [DOI] [PubMed] [Google Scholar]
- 43.Sztukowska M, Gallinaro L, Cattini MG, et al. Von Willebrand factor propeptide makes it easy to identify the shorter von Willebrand factor survival in patients with type 1 and type Vicenza von Willebrand disease. Br J Haematol. 2008;143(1):107–114. doi: 10.1111/j.1365-2141.2008.07311.x. [DOI] [PubMed] [Google Scholar]
- 44.van Laer L, Huyghe JR, Hannula S, et al. A genome-wide association study for age-related hearing impairment in the Saami. Eur J Hum Genet. 2010;18(6):685–693. doi: 10.1038/ejhg.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi KH, Higgs BW, Wendland JR, et al. Gene expression and genetic variation data implicate PCLO in bipolar disorder. Biol Psychiatry. 2010;69(4):353–359. doi: 10.1016/j.biopsych.2010.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Howard TD, Zheng SL, et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol. 2010;125(2):328–335. doi: 10.1016/j.jaci.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orozco G, Hinks A, Eyre S, et al. Combined effects of three independent SNPs greatly increase the risk estimate for RA at 6q23. Hum Mol Genet. 2009;18(14):2693–2699. doi: 10.1093/hmg/ddp193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benyamin B, McRae AF, Zhu G, et al. Variants in TF and HFE explain approximately 40% of genetic variation in serum-transferrin levels. Am J Hum Genet. 2009;84(1):60–65. doi: 10.1016/j.ajhg.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen R, Davydov EV, Sirota M, Butte AJ. Non-synonymous and synonymous coding SNPs show similar likelihood and effect size of human disease association. PLoS ONE. 2010;5(10):e13574. doi: 10.1371/journal.pone.0013574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harker LA, Ross R, Slichter SJ, Scott CR. Homocystine-induced arteriosclerosis: the role of endothelial cell injury and platelet response in its genesis. J Clin Invest. 1976;58(3):731–741. doi: 10.1172/JCI108520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ying SY, Lin SL. Intron-mediated RNA interference and microRNA biogenesis. Methods Mol Biol. 2009;487:387–413. doi: 10.1007/978-1-60327-547-7_19. [DOI] [PubMed] [Google Scholar]
- 53.Chen Z, Nakajima T, Tanabe N, et al. Susceptibility to chronic thromboembolic pulmonary hypertension may be conferred by miR-759 via its targeted interaction with polymorphic fibrinogen alpha gene. Hum Genet. 2010;128(4):443–452. doi: 10.1007/s00439-010-0866-8. [DOI] [PubMed] [Google Scholar]
- 54.Fort A, Borel C, Migliavacca E, et al. Regulation of fibrinogen production by microRNAs. Blood. 2010;116(14):2608–2615. doi: 10.1182/blood-2010-02-268011. [DOI] [PubMed] [Google Scholar]
- 55.Goodeve A. Genetics of type 1 von Willebrand disease. Curr Opin Hematol. 2007;14(5):444–449. doi: 10.1097/MOH.0b013e32826f4b41. [DOI] [PubMed] [Google Scholar]
- 56.Lillicrap D. Genotype/phenotype association in von Willebrand disease: is the glass half full or empty? J Thromb Haemost. 2009;7(suppl 1):65–70. doi: 10.1111/j.1538-7836.2009.03367.x. [DOI] [PubMed] [Google Scholar]
- 57.Castaman G, Plate M, Giacomelli SH, Rodeghiero F, Duga S. Alterations of mRNA processing and stability as a pathogenic mechanism in von Willebrand factor quantitative deficiencies. J Thromb Haemost. 2010;8(12):2736–2742. doi: 10.1111/j.1538-7836.2010.04060.x. [DOI] [PubMed] [Google Scholar]
- 58.Gallinaro L, Sartorello F, Pontara E, et al. Combined partial exon skipping and cryptic splice site activation as a new molecular mechanism for recessive type 1 von Willebrand disease. Thromb Haemost. 2006;96(6):711–716. [PubMed] [Google Scholar]
- 59.Gill JC, Endres-Brooks J, Bauer PJ, Marks WJ, Jr, Montgomery RR. The effect of ABO blood group on the diagnosis of von Willebrand disease. Blood. 1987;69(6):1691–1695. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.