Abstract
HEAT repeats correspond to tandemly arranged curlicue-like structures that appear to serve as flexible scaffolding on which other components can assemble. Using sensitive sequence analysis techniques we detected HEAT repeats in various chromosome-associated proteins, including four families of proteins associated with condensins and cohesins, which are nuclear complexes that contain structural maintenance of chromosome (SMC) proteins. Among the proteins detected were the XCAP-D2 and XCAP-G subunits of the Xenopus laevis 13S condensin complex, the Aspergillus BimD and Sordaria macrospora Spo76p proteins, the budding yeast Scc2p protein, and the related Drosophila transcriptional activator Nipped-B. Clathrin adaptor and COP-I coatomer subunits, which function in vesicle coat assembly and were previously noted to share weak sequence similarity to condensin subunits, also contain HEAT repeats. HEAT repeats were also found in the TBP-associated TIP120 protein, a global enhancer of transcription, and in the budding yeast Mot1p protein, which is a member of the SWI2/SNF2 family. SWI2/SNF2 proteins, some of which are helicases, perform diverse roles in transcription control, DNA repair, and chromosome segregation and form chromatin-remodeling complexes. HEAT repeats also were found in dis1-TOG family and cofactor D family microtubule-associated proteins, which, owing to their roles in microtubule dynamics, perform functions related to mitotic progression and chromosome segregation. Hence, our analysis predicts structural features of these proteins and suggests that HEAT repeats may play important roles in chromosome dynamics.
Structural maintenance of chromosome (SMC) proteins appear to play a dynamic role in packaging and shaping chromosomes, which is important for compacting DNA into the tight confines of the nucleus (Hirano 1999). SMC proteins are ATPases evolutionarily related to ABC transporters (Saitoh et al. 1994), but are unique inasmuch as their Walker A and B ATP-binding sites are separated by two sizable regions predicted to form α-helical coiled coils between which is inserted a putative flexible hinge. An antiparallel coiled-coil interaction between two SMC subunits has been proposed to produce a functional dimer with ATP-binding sites at each end (Melby et al. 1998). There are four classes of SMC proteins (SMC1–SMC4) that form two types of heterodimers: SMC1/SMC3 and SMC2/SMC4. SMC2/SMC4 dimers are involved in chromosome condensation and dosage compensation, whereas SMC1/SMC3 dimers are involved in sister chromatid cohesion and DNA recombination and repair. In eukaryotes, SMC dimers interact with non-SMC subunits to form condensin and cohesin complexes. In Xenopus laevis, for example, the SMC heterodimer XCAP-E/XCAP-C interacts with the non-SMC subunits XCAP-G, XCAP-D2, and XCAP-H to form a condensin complex that converts interphase chromatin into mitoticlike chromosomes (Hirano et al. 1997). SMC complexes may act as DNA crosslinkers with the flexible hinge facilitating an ATP-modulated scissorslike motion linked to changes in chromosome structure (Hirano 1999). Other biochemical and genetic interactions between SMC proteins and non-SMC proteins have also been reported (Hirano 1999).
SMC proteins also function as chromosome structural components—the chicken SMC protein ScII, for example, being the second most abundant component (after topoisomerase II) of the mitotic nuclear scaffold or matrix (Saitoh et al. 1994). Indeed, SMC proteins superficially resemble intermediate filaments inasmuch as they contain an internal α-helical heptad repeat region for coiled-coil formation flanked by globular domains. Although the biological significance of the nuclear scaffold has been controversial, current evidence suggests that the nucleus is indeed highly ordered with a high degree of structural flexibility (Lamond and Earnshaw 1998). It has been proposed, for example, that DNA replication occurs at a fixed number of “replication factories” attached to a nucleoskeleton and to which chromosomes move to initiate their replication (Jackson and Cook 1995; Cook 1999). Thus, given their dual roles as both restructuring enzymes and chromosome structural components, SMC proteins appear to play a central role in the dynamic organization of the nucleus (Saitoh et al. 1995).
Here we report the occurrence of HEAT repeats in condensin- and cohesin-associated proteins. HEAT repeats (Andrade and Bork 1995) correspond to tandemly arranged bihelical structures (Chook and Blobel 1999; Cingolani et al. 1999; Groves et al. 1999; Kobe 1999; Vetter et al. 1999) (Fig. 1a) that appear to function as flexible joints that can wrap around target substrates and as scaffolding on which other molecular components may assemble. We also report the occurrence of HEAT repeats in other proteins associated with chromosome dynamics and function, including transcription factors and microtubule-associated proteins (MAPs).
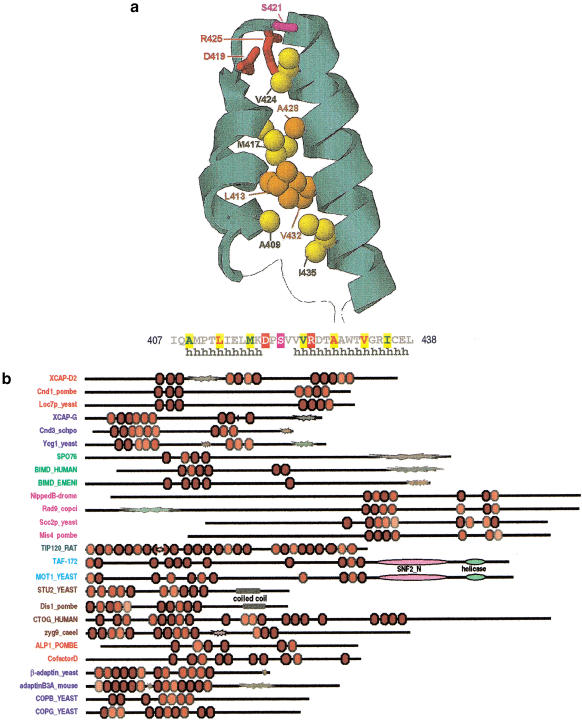
Figure 1.
Features of HEAT-repeat proteins. (a) Typical structure of a repeat. Sidechain atoms for more highly conserved positions are shown. The structure and sequence corresponds to repeat-10 of importin-β (1QGKA). (b) Schematic representation of domain architectures. HEAT repeats are colored red in proportion to their profile scores, using lighter shades for less conserved repeats. Protein names are colored by families. “Explosions” indicate compositionally biased regions and are shaded red for acidic, blue for basic, and green for uncharged polar biases. Other domains are as indicated.
RESULTS
Starting from an alignment of known HEAT repeats and using iterative search and alignment procedures (see Methods), we found previously unreported HEAT repeats in various chromosome-related proteins as shown in Figure 2. Note, however, that a statistical analysis based on Gibbs sampling (Neuwald et al. 1995) indicates that additional (weakly conserved) repeats are structurally present in many of these proteins. Similarly, a modified PSI-BLAST search procedure based on hidden Markov models, which was developed subsequent to this analysis (Neuwald and Poleksic 2000), detects and aligns regions containing additional repeats in these sequences. Since Figures 1 and 2 do not show alignments for these weakly conserved repeats owing to their statistical uncertainty, our analysis suggests a lower bound on the number of HEAT repeats present in these proteins.
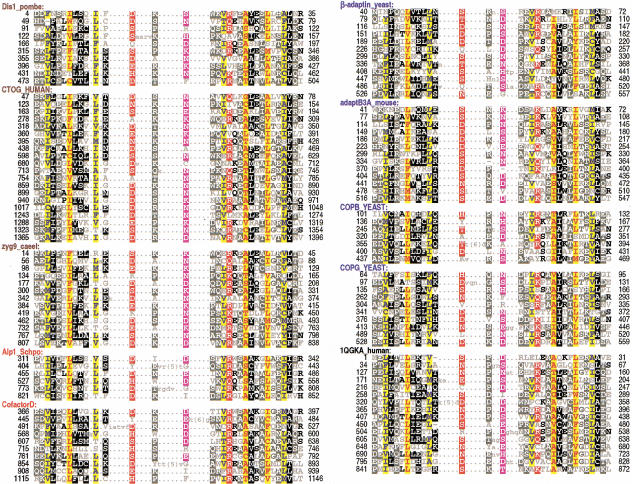
Figure 2.
Multiple alignment of HEAT repeats discussed in the text. For comparison, we also include the 16 HEAT repeats that were detected (and correctly aligned) by our procedures for human importin-β, whose structure is known (Cingolani et al. 1999; pdb code 1QGKA). For each aligned column, conserved residues—elevated with binomial tail probabilities (Neuwald and Green 1994) of p ≤ .0001—and less conserved, related residues—with tail probabilities of p ≤ .0005—are indicated using an automated hierarchical scheme (Neuwald et al. 1999) as follows: ≥1.25 bits of information, red highlight; 1.25–0.75 bits of information, magenta; ≥70% conserved and hydrophobic, yellow; >66% conserved, black; 50%–66% conserved, dark gray; 33%–50% conserved, black; <33% conserved, dark gray; unconserved, light gray.
Among the sequences detected are four protein families associated with SMC ATPases (Fig. 2). Two families of SMC-associated HEAT-repeat proteins correspond to the XCAP-D2 and XCAP-G condensin subunits (Kimura et al. 1998). These subunits appear to play a regulatory role and are required for condensin's in vitro DNA reconfiguring activity (Kimura et al. 1998), but their exact function is unclear. HEAT repeats also occur in a third family of proteins that may interact with the cohesin complex, including Sordaria macrospora Spo76p, which is required for sister chromatid cohesiveness, chromosome axis morphogenesis, and chromatin condensation in meiosis (van Heemst et al. 1999). The related Aspergillus BimD protein functions in chromosome segregation and genetically interacts with the SMC protein SudA (Holt and May 1996).
A fourth family includes the budding yeast Scc2p and the fission yeast Mis4p proteins, which are also responsible for sister chromatid cohesion (Toth et al. 1999). Although Scc2p is required for association of cohesin with chromosomes, it is not a stoichiometric cohesin subunit, suggesting that it loads cohesin (Toth et al. 1999). We also detected weak similarity to HEAT repeats in a stoichiometric subunit of the cohesin complex (Toth et al. 1999), namely, yeast Scc3p (data not shown), but this similarity lacks statistical support. Other Scc2/Mis4 family members include the fungal Rad9 protein, which is required for meiotic chromosome condensation and synaptonemal complex formation (Seitz et al. 1996), and the Drosophila Nipped-B protein. Interestingly, Nipped-B participates in transcriptional activation by remote enhancers, perhaps by structurally facilitating enhancer–promoter communication (Rollins et al. 1999), which suggests that this protein family performs additional chromosome-related functions.
We also found HEAT repeats in another transcription-related protein, namely, budding yeast Mot1p—a SWI2/SNF2 family member that regulates the distribution of TATA-binding protein (TBP) between promoter and nonpromoter sites by catalyzing the ATP-dependent dissociation of TBP from DNA (Davis et al. 1992; Muldrow et al. 1999; Adamkewicz et al. 2000). Another TBP-associated HEAT repeat protein is rat TIP120, which globally enhances transcription by all three RNA polymerases (Makino et al. 1999). The TIP120 repeats appear to be contiguously distributed throughout this protein in a manner reminiscent of importin-β.
Two microtubule-associated protein (MAP) families were also found to contain HEAT repeats. MAP proteins are important for mitotic spindle assembly and chromosome movement during cell division because of their roles in microtubule dynamics. One of these, the dis1-TOG family, includes fission yeast Dis1p, human TOG, X. laevis XMAP215, budding yeast Stu2p, Caenorhabditis elegans ZYG-9, and Drosophila mini spindles (msps) (Cullen et al. 1999, and references therein). Proteins in this family are associated with the mitotic spindle and centrosomal regions in a cell-cycle-dependent manner and contain two or more ∼200-residue repetitive regions (Cullen et al. 1999). Each of these regions appear to contain five HEAT repeats, although some repeats seem to be degenerate and were not detected (Fig. 1b). HEAT repeats occur in another family of MAPs that includes mammalian tubulin-folding cofactor D and the fission yeast Alp1 proteins (Hirata et al. 1998, and references therein). These proteins function in the in vitro formation of assembly-competent α/β-tubulin heterodimers, from which microtubules are generated (for review, see Lewis et al. 1997).
DISCUSSION
For most of the proteins detected in our analysis the HEAT repeats occur in clusters (Fig. 1b)—an arrangement that is similar to the HEAT repeats in Huntingtin, a protein associated with a progressive neurodegenerative disorder called Huntington's disease (Huntington's Disease Collaborative Research Group 1993). By contrast, the HEAT repeats in other proteins, such as importin-β and elongation factor 3 (EF-3), form single, contiguous stretches. Interestingly, EF-3 contains two ABC-like domains in addition to the HEAT repeats, and, similarly, the EF-3-related HEAT-repeat protein GCN1 interacts with a protein containing two ABC-like domains (Marton et al. 1997), which suggests a possible mechanistic similarity to SMC complexes because they also contain both HEAT-repeat and ABC-related proteins.
Importin-β is a well characterized HEAT-repeat protein that, in conjunction with importin-α, is involved in nuclear import (for review, see Gorlich and Kutay 1999). In importin-β several HEAT repeats curl around and bind to a short, positively charged domain of importin-α (Cingolani et al. 1999). Likewise, the HEAT-repeat-containing A subunit of PP2A phosphatase appears to function as a molecular scaffold linking the catalytic subunit to various regulatory subunits, thereby facilitating dephosphorylation of diverse substrates (for review, see Millward et al. 1999). Thus, HEAT repeats in the chromosome-associated proteins described here may similarly function as flexible scaffolding that recognizes and binds target substrates, a role that is consistent with the nuclear scaffold functions performed by SMC complexes.
What potential substrates might these chromosome-associated proteins recognize? HEAT repeats may mediate binding between subunits within SMC complexes. For example, repeats in XCAP-D2 may bind to XCAP-G (or vice versa) or to the SMC proteins. The possibility of interaction between two HEAT repeat proteins is suggested by the fact that importin-α, which binds to importin-β, contains ARM repeats (Andrade and Bork 1995; Hatzfeld 1999), which are structurally similar to (Cingolani et al. 1999; Kobe et al. 1999) and appear to be evolutionarily related (Malik et al. 1997) to HEAT repeats. Another possibility is that some of the chromosome-associated HEAT repeats recognize nuclear matrix or chromatin components. For example, SMC-associated HEAT-repeat proteins may bind to histone tail domains, as was suggested to us by a recent review (Strahl and Allis 2000). This review also noted several similarities between chromatin and microtubules: Both are polymers composed of highly conserved subunits that assemble into repeating units (histones and tubulins, respectively), and both have unstructured tail domains decorated by a diverse array of posttranslational modifications. It is interesting, in this context, that we also detect HEAT repeats in two MAP families with likely roles in regulating microtubule dynamics. Thus, perhaps some of these MAP-associated HEAT repeats recognize tubulin tails. Alternatively, these HEAT repeats may recognize substrates other than tubulins. For example, the human TOG protein and its Xenopus homolog XMAP215 recently were found to associate with p34cdc2 kinase and direct it to the microtubule cytoskeleton (Charrasse et al. 2000), which suggests a targeting role similar to that of importin-β.
The HEAT repeats in Mot1p and its homologs (such as human TAF-172) are intriguing inasmuch as SWI2/SNF2 proteins share some functional similarities to SMC complexes. SWI2/SNF2 proteins perform diverse roles in transcription control, DNA repair, and chromosome segregation (Travers 1999). They often form ATP-dependent chromatin-remodeling complexes (Muchardt and Yaniv 1999), some of which are associated with the nuclear matrix (Reyes et al. 1997). For example, the SWI2/SNF2 protein BRG1 and its associated factors serve as a model system for induction of genomewide chromatin remodeling because this complex associates with chromatin upon lymphocyte antigenic activation and may be required for subsequent chromatin decondensation (Zhao et al. 1998). HEAT repeats appear to be absent, however, from most SWI2/SNF2 family members and, therefore, appear to perform a specific role in Mot1p and its close homologs.
Since SMC proteins are major components of the nuclear scaffold, the associated HEAT-repeat proteins may mediate nuclear scaffold assembly. In fact, several other HEAT-repeat proteins perform scaffold assembly functions. For example, HEAT repeats occur in the CpcE and CpcF subunits of cyanobacterium phycocyanin lyase (A.F. Neuwald, unpubl.), which are involved in the assembly of phycobilisomes, a scaffold of membrane-associated photosynthetic antenna proteins (MacColl 1998). CpcEF attaches bilin chromophores to bilin-binding subunits—a function that appears to be important for phycobilisome assembly (for review, see Anderson and Toole 1998). Furthermore, a related (HEAT-repeat) protein, Nb1B, is involved in phycobilisome degradation (Dolganov and Grossman 1999). Still other HEAT-repeat proteins function in vesicle coat assembly. Vesicle transport occurs through membrane budding by a mechanism involving coat proteins that assemble into scaffold structures. Three coated-vesicle classes—clathrin, COP-I, and COP-II—are well characterized, and each is associated with distinct coat protein complexes (Schekman and Orci 1996). Clathrin-associated adaptor subunits were noted as possible candidates in the initial description of HEAT repeats (Andrade and Bork 1995), and our analysis confirms their presence in these subunits and in related COP-I coatomer subunits (Figs. 1b, 2). In light of this, it is also interesting that the HEAT-repeat Huntingtin protein influences endocytic and secretory vesicle transport through associations with clathrin-coated vesicles (Velier et al. 1998). It was recently suggested (Sutani et al. 1999), based on weak sequence similarity, that proteins related to XCAP-G and XCAP-D2 mediate SMC subunit assembly in a manner resembling clathrin assembly by the adaptor AP3-β. The apparent scaffold role of HEAT repeats and their presence in these proteins now strengthens this hypothesis.
In summary, our findings suggest important roles for HEAT repeats in chromosome dynamics and provide information regarding the structures of these chromosome-related proteins. Our findings also suggest that these proteins may perform substrate targeting and scaffold assembly roles. Further studies are needed to explore these possibilities.
METHODS
HEAT repeats were detected using iterative search and Gibbs sampling alignment procedures (Neuwald et al. 1995, 1997, 1999). These procedures were initialized using an alignment of known HEAT repeats. Detection of new HEAT repeats is based on the following criteria. First, profiles constructed from known HEAT repeats (Neuwald et al. 1997) find these proteins (p < .01). Conversely, profiles of repeats constructed from each of these families find known HEAT repeats (p < .01). Repeat profiles were constructed by first using an ungapped Gibbs sampling procedure (Neuwald et al. 1995) to detect and align repeats. Then these alignments were refined using gapped Gibbs sampling procedure (Neuwald et al. 1999; Neuwald and Poleksic 2000). Note that these procedures are designed to only align those sequence regions showing some statistical evidence for conserved patterns (Neuwald et al. 1997). Third, PSI-BLAST searches (Altschul et al. 1997) using the repeat regions from these proteins as queries detect known HEAT repeats among the significant hits (p ≤ .01) or among the highest scoring nonsignificant hits. Finally, the significant patterns (Neuwald and Green 1994) and predicted structural features (King et al. 1997) for repeat regions within each family were examined for consistency with known HEAT repeats.
Acknowledgments
This work was supported by grants from the NIH (LM06747 to A.F.N. and GM53926 to T.H.) and by grants from the Pew Scholars Program in the Biomedical Sciences and the Human Frontier Science Program.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL neuwald@cshl.org; FAX (516) 367-8461.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.147400.
REFERENCES
- Adamkewicz JI, Mueller CG, Hansen KE, Prud'homme WA, Thorner J. Purification and enzymic properties of mot1 ATPase, a regulator of basal transcription in the yeast Saccharomyces cerevisiae. J Biol Chem. 2000;275:21158–21168. doi: 10.1074/jbc.M002639200. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LK, Toole CM. A model for early events in the assembly pathway of cyanobacterial phycobilisomes. Mol Microbiol. 1998;30:467–474. doi: 10.1046/j.1365-2958.1998.01081.x. [DOI] [PubMed] [Google Scholar]
- Andrade MA, Bork P. HEAT repeats in the Huntington's disease protein. Nat Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- Charrasse S, Lorca T, Doree M, Larroque C. The Xenopus XMAP215 and its human homologue TOG proteins interact with cyclin B1 to target p34cdc2 to microtubules during mitosis. Exp Cell Res. 2000;254:249–256. doi: 10.1006/excr.1999.4740. [DOI] [PubMed] [Google Scholar]
- Chook YM, Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature. 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Petosa C, Weis K, Muller CW. Structure of importin-β bound to the IBB domain of importin-α. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- Cullen CF, Deak P, Glover DM, Ohkura H. mini spindles. A gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J Cell Biol. 1999;146:1005–1018. doi: 10.1083/jcb.146.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JL, Kunisawa R, Thorner J. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolganov N, Grossman AR. A polypeptide with similarity to phycocyanin α-subunit phycocyanobilin lyase involved in degradation of phycobilisomes. J Bacteriol. 1999;181:610–617. doi: 10.1128/jb.181.2.610-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Groves MR, Hanlon N, Turowski P, Hemmings BA, Barford D. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell. 1999;96:99–110. doi: 10.1016/s0092-8674(00)80963-0. [DOI] [PubMed] [Google Scholar]
- Hatzfeld M. The armadillo family of structural proteins. Int Rev Cytol. 1999;186:179–224. doi: 10.1016/s0074-7696(08)61054-2. [DOI] [PubMed] [Google Scholar]
- Hirano T. SMC-mediated chromosome mechanics: A conserved scheme from bacteria to vertebrates? Genes & Dev. 1999;13:11–19. doi: 10.1101/gad.13.1.11. [DOI] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Hirata D, Masuda H, Eddison M, Toda T. Essential role of tubulin-folding cofactor D in microtubule assembly and its association with microtubules in fission yeast. EMBO J. 1998;17:658–666. doi: 10.1093/emboj/17.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CL, May GS. An extragenic suppressor of the mitosis-defective bimD6 mutation of Aspergillus nidulans codes for a chromosome scaffold protein. Genetics. 1996;142:777–787. doi: 10.1093/genetics/142.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Cook PR. The structural basis of nuclear function. Int Rev Cytol. 1995;162A:125–149. doi: 10.1016/s0074-7696(08)61230-9. [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano M, Kobayashi R, Hirano T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- King RD, Saqi M, Sayle R, Sternberg MJ. DSC: Public domain protein secondary structure predication. Comput Appl Biosci. 1997;13:473–474. doi: 10.1093/bioinformatics/13.4.473. [DOI] [PubMed] [Google Scholar]
- Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nat Struct Biol. 1999;6:388–397. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- Kobe B, Gleichmann T, Horne J, Jennings IG, Scotney PD, the T. Turn up the HEAT. Structure Fold Des. 1999;7:R91–R97. doi: 10.1016/s0969-2126(99)80060-4. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lewis SA, Tian G, Cowan NJ. The α- and β-tubulin folding pathways. Trends Cell Biol. 1997;7:479–484. doi: 10.1016/S0962-8924(97)01168-9. [DOI] [PubMed] [Google Scholar]
- MacColl R. Cyanobacterial phycobilisomes. J Struct Biol. 1998;124:311–334. doi: 10.1006/jsbi.1998.4062. [DOI] [PubMed] [Google Scholar]
- Makino Y, Yogosawa S, Kayukawa K, Coin F, Egly JM, Wang Z, Roeder RG, Yamamoto K, Muramatsu M, Tamura T. TATA-Binding protein-interacting protein 120, TIP120, stimulates three classes of eukaryotic transcription via a unique mechanism. Mol Cell Biol. 1999;19:7951–7960. doi: 10.1128/mcb.19.12.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Eickbush TH, Goldfarb DS. Evolutionary specialization of the nuclear targeting apparatus. Proc Natl Acad Sci. 1997;94:13738–13742. doi: 10.1073/pnas.94.25.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton MJ, Vazquez de Aldana CR, Qiu H, Chakraburtty K, Hinnebusch AG. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2alpha kinase GCN2. Mol Cell Biol. 1997;17:4474–4489. doi: 10.1128/mcb.17.8.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: Long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millward TA, Zolnierowicz S, Hemmings BA. Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem Sci. 1999;24:186–191. doi: 10.1016/s0968-0004(99)01375-4. [DOI] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M. ATP-dependent chromatin remodelling: SWI/SNF and co. are on the job. J Mol Biol. 1999;293:185–197. doi: 10.1006/jmbi.1999.2999. [DOI] [PubMed] [Google Scholar]
- Muldrow TA, Campbell AM, Weil PA, Auble DT. MOT1 can activate basal transcription in vitro by regulating the distribution of TATA binding protein between promoter and nonpromoter sites. Mol Cell Biol. 1999;19:2835–2845. doi: 10.1128/mcb.19.4.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Neuwald AF, Green P. Detecting patterns in protein sequences. J Mol Biol. 1994;23:698–712. doi: 10.1006/jmbi.1994.1407. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Poleksic A. PSI-BLAST searches using hidden Markov models of structural repeats: prediction of an unusual sliding DNA clamp and of β-propellers in UV-damaged DNA binding protein. Nucleic Acids Res. 2000;28:3570–3580. doi: 10.1093/nar/28.18.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Liu JS, Lawrence CE. Gibbs motif sampling: Detection of bacterial outer membrane protein repeats. Protein Sci. 1995;4:1618–1632. doi: 10.1002/pro.5560040820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Liu JS, Lipman DJ, Lawrence CE. Extracting protein alignment models from the sequence database. Nucleic Acids Res. 1997;25:1665–1677. doi: 10.1093/nar/25.9.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JC, Muchardt C, Yaniv M. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J Cell Biol. 1997;137:263–274. doi: 10.1083/jcb.137.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N, Goldberg IG, Wood ER, Earnshaw WC. ScII: An abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J Cell Biol. 1994;127:303–318. doi: 10.1083/jcb.127.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N, Goldberg I, Earnshaw WC. The SMC proteins and the coming of age of the chromosome scaffold hypothesis. Bioessays. 1995;17:759–766. doi: 10.1002/bies.950170905. [DOI] [PubMed] [Google Scholar]
- Schekman R, Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- Seitz LC, Tang T, Cummings WJ, Zolan ME. The rad9 gene of Coprinus cinereus encodes a proline-rich protein required for meiotic chromosome condensation and synapsis. Genetics. 1996;142:1105–1117. doi: 10.1093/genetics/142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M. Fission yeast condensin complex: Essential roles of non-SMC subunits for condensation and cdc2 phosphorylation of Cut3/SMC4. Genes & Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes & Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. An engine for nucleosome remodeling. Cell. 1999;96:311–314. doi: 10.1016/s0092-8674(00)80543-7. [DOI] [PubMed] [Google Scholar]
- van Heemst D, James F, Poggeler S, Berteaux-Lecellier V, Zickler D. Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell. 1999;98:261–271. doi: 10.1016/s0092-8674(00)81020-x. [DOI] [PubMed] [Google Scholar]
- Velier J, Kim M, Schwarz C, Kim TW, Sapp E, Chase K, Aronin N, DiFiglia M. Wild-type and mutant huntingtins function in vesicle trafficking in the secretory and endocytic pathways. Exp Neurol. 1998;152:34–40. doi: 10.1006/exnr.1998.6832. [DOI] [PubMed] [Google Scholar]
- Vetter IR, Arndt A, Kutay U, Gorlich D, Wittinghofer A. Structural view of the Ran–Importin β interaction at 2.3 Å resolution. Cell. 1999;97:635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]