Abstract
Trax/Translin heteromers, also known as C3PO, have been proposed to activate RNA-induced silencing complex (RISC) by facilitating endonucleolytic cleavage of the siRNA passenger strand. We report on the crystal structure of hexameric Drosophila C3PO formed by truncated Translin and Trax, along with electron microscopic and mass spectrometric studies on octameric C3PO formed by full-length Translin and Trax. Our studies establish that Trax adopts the Translin fold, possesses catalytic centers essential for C3PO’s endoribonuclease activity and interacts extensively with Translin to form an octameric assembly. The catalytic pockets of Trax subunits are located within the interior chamber of the octameric scaffold. Truncated C3PO, like full-length, exhibits endoribonuclease activity leaving 3′ hydroxyl-cleaved ends. We have measured the catalytic activity of C3PO and shown it to cleave near stoichiometric amounts of substrate per second.
In the RNA interference (RNAi) pathway, small interfering RNAs (siRNAs) direct Argonaute 2 (Ago2) to cleave mRNA targets, resulting in post-transcriptional gene silencing1,2. In Drosophila, Dicer-2 initiates RNAi by processing long double-stranded RNA (dsRNA) into ~21-nucleotide (nt) long siRNA duplexes. Dicer-2 and R2D2 form a heteromeric complex, which facilitates transfer of siRNA onto Ago2, the catalytic component of the RNA-induced silencing complex (RISC)1–4. Following loading of siRNA duplexes into RISC, the passenger strand of duplex is cleaved by Ago2 and discarded, leaving the guide strand bound in the nucleic acid-binding channel5. Finally, the guide strand-bound Ago2 functions as the effector by recognizing and degrading the target mRNA1,2,6,7. Recently, a protein complex called C3PO (Component 3 Promoter of RISC) was shown to be a Mg2+-dependentendoribonuclease, which facilitates RISC activation by siRNA unwinding, as well as through removal of cleaved passenger strand8.
C3PO turns out to be the well-studied multimeric complex of Translin and Trax, where Trax acts as the catalytic subunit8. Previous studies on the Translin-Trax complex have implicated its role in diverse biological processes such as cell growth regulation, mRNA processing, spermatogenesis, neuronal development/function, regulation of genome stability and carcinogenesis, although their precise roles in some of these processes require further investigations9,10. Previous studies have shown that Translin and Trax behave distinctly, such that only Translin binds tightly to single-stranded DNA or RNA10–12, whereas only Trax possesses endoribonuclease activity8. Crystal structures of Translin have been solved from mouse (PDB code: 1KEY), human (PDB code: 1J1J) and fruit fly (PDB code: 2QRX), establishing that Translin adopts a seven α-helical bundle fold13–15, which aligns further into an octameric scaffold16. By contrast, the instability of Trax in isolation17, has hindered attempts at its structure determination. Based on modeling studies, Trax has been predicted to fold like Translin8. The lack of structural information on either Trax or C3PO has raised many unanswered questions: does Trax indeed fold like Translin? Why do they behave so differently given proposed similar folds? How does Trax interact with Translin? What is the ratio of Translin to Trax in multimeric C3PO and how does C3PO carry out its endoribonuclease activity? To begin to address these questions, we initiated a program of X-ray crystallographic, electron microscopic, and mass spectrometric studies, supplemented by cleavage assays, on reconstituted D. melanogaster C3PO.
RESULTS
Crystal structure of C3PO composed of truncated Translin and Trax
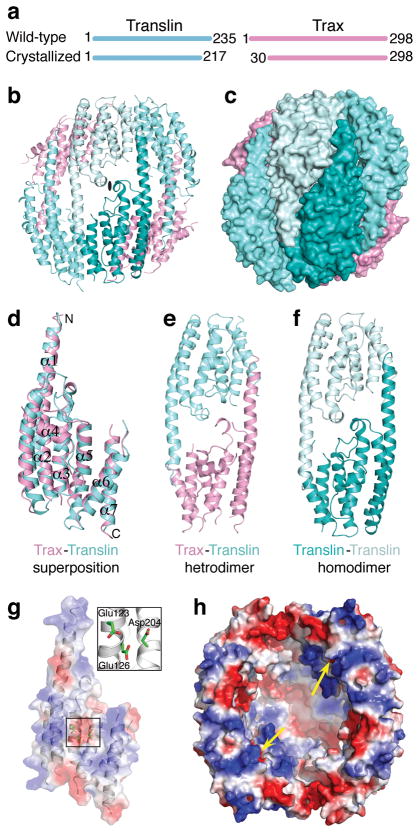
We were able to obtain crystals for reconstituted C3PO by co-expressing recombinant full-length Drosophila Translin and Trax in E. coli, but our attempts to obtain useful diffraction from these crystals was unsuccessful, even after extensive trials. To improve the diffraction, we attempted to obtain reconstituted C3PO following serial truncations of unstructured regions (based on secondary structure prediction) present at N- or C-termini in both Translin and Trax. One such C3PO, that is formed by Translin with 18-amino acids (aa) truncated at the C-terminus (Fig. 1a) and Trax with 29-aa truncated at the N-terminus (Fig. 1a), gave large diffracting crystals suitable for structural studies. Anisotropic diffraction data extending to 3.4–3.9 Å in different directions was collected on a seleno-methionine-labelled C3PO crystal and was used to solve the structure (Table 1 and Supplementary Figs. 1a). The model building and sequence assignment was greatly helped by location of Se atoms in both Translin (2 SeMet sites) and Trax (5 SeMet sites) as shown in stereo in Supplementary Fig. 1b and the known structure of Translin13. In the crystal, truncated C3PO is composed of six subunits, containing two copies of Trax and four copies of Translin (Figs. 1b and 1c). The six subunits are arranged as trimer of dimers, aligned side by side, thereby forming a concave bowl-like structure. The dimer in the middle of the hexameric assembly is formed by two copies of Translin (Translin-Translin homodimer), which in turn is bracketed on both sides by Translin-Trax heterodimers. The molecular weight (MW) of truncated hexameric C3PO present in the crystal is consistent with the estimated MW of ~150 kDa by gel filtration and multi-angle light scattering (MALS) measurements (Supplementary Fig. 2a).
Figure 1.
Overall structure of truncated hexameric C3PO, Trax-Translin interactions and an active center in Trax for endoribonuclease activity. (a) Lengths of wild-type Translin and Trax and truncated constructs that yielded crystals of C3PO. (b) Ribbon representation of truncated C3PO with two molecules of Trax colored in pink and four molecules of Translin colored in different shades of cyan. (c) Surface representation of truncated C3PO in the same view and color-coded as in (b). (d) Structural superposition of Trax (pink) and Translin (cyan) shown in ribbon representation. (e) Heterodimer of Trax and Translin as seen in the truncated C3PO structure (f) Homodimer of Translin positioned between the two heterodimers of Trax and Translin in the truncated C3PO structure. (g) Electrostatic surface and ribbon representation of Trax showing presence of three acidic residues (shown in box) along one face in the center of the Trax molecule. An enlarged view of this region is shown in a box. (h) Electrostatic surface representation of truncated C3PO showing location of active sites within the inner concave face (indicated by yellow arrows) for the endoribonuclease activity.
Table 1.
Data collection and refinement statistics.
| Truncated C3PO (SeMet) | ||
|---|---|---|
| Data collection | ||
| Space group | P6122 | |
| Cell dimensions | ||
| a, b, c (Å) | 196.03, 196.03, 155.19 | |
| α, β, γ(°) | 90, 90, 120 | |
| Inflection | Peak | |
| Wavelength (Å) | 0.9794 | 0.9792 |
| Resolution (Å) | 30.0–3.4 (3.52–3.40) | 50.0–3.8 (3.94–3.80) |
| Rsym or Rmerge | 21.4 (86.5) | 24.0 (85.3) |
| I/σI | 11.4 (2.7) | 10.8 (2.4) |
| Completeness (%) | 99.8 (99.8) | 99.9 (99.9) |
| Redundancy | 14.4 (11.0) | 21.4 (15.8) |
| Refinement | ||
| Resolution (Å) | ||
| low resolution | 20 | |
| high Resolution (a*, b*, c*)# | 3.9, 3.9, 3.4 | |
| No. of Reflections (total/test) | 19572/999 | |
| Rwork/Rfree | 27.0/33.8 | |
| No. atoms | ||
| Protein | 4718 | |
| Ligand/ion | - | |
| Water | - | |
| B-factors | ||
| Protein | 89.4 | |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.016 | |
| Bond angles (°) | 0.864 | |
One crystal was used for both data sets.
Values for highest resolution shell are shown in parenthesis.
After anisotropic correction, high resolution data beyond these limits were excluded during refinement.
One Translin-Trax heterodimer and one Translin molecule from the middle Translin-Translin homodimer are present in the asymmetric unit and are related to rest of the complex via 2-fold crystallographic symmetry (Fig. 1b and 1c). Our structure of truncated hexameric C3PO provides the much anticipated structural information on Trax. Like Translin, the overall fold of Trax also involves a seven α-helical bundle, with structural superposition between Translin and Trax exhibits a rmsd of ~1.8 Å (Fig. 1d). For Trax, no electron density was found for 26-aa at the C-terminus and 29-aa present between α4 and α5. Electron densities for loops present between helical regions in Trax were not clear, so they were not included in the model. The overall structure of Translin in truncated hexameric C3PO is similar to that previously reported for Translin13–15. In Translin, no electron density was found for 7-aa at the N-terminus, 2-aa at the C-terminus and residues present in the loop between α6 and α7. Further, no major conformational change was observed in the Translin structure upon binding to Trax.
Views of the Translin-Trax heterodimer and the Translin-Translin homodimer in the structure of truncated hexameric C3PO are shown in Figs. 1e and 1f, respectively. Due to the similar fold of Translin and Trax, extensive complementarity exists in the surface features at the Trax-Translin interfaces, as observed between the Translin-Translin interfaces. All these interfaces are formed by a large number of non-polar and polar amino acids involved in hydrophobic and polar interactions, including salt-bridges and hydrogen bonds (Supplementary Figs. 3a, b, c, d).
Interestingly, three invariant acidic residues, Glu123 and Glu126 on α3 and Asp204 on α5, are present within 4 Å of each other, and form an acidic patch in the center of the Trax molecule (Fig. 1g). Previous studies showed that mutation of any one of these three acidic residues to Ala abolished C3PO RNA endonuclease activity and its RISC enhancer activity8. Mutations of other conserved acidic residues on Trax (E197A and E49A) or Translin (E208A and D202A) showed no such effects. In Trax, the acidic amino acid triad is expected to be the Mg2+ coordinating site that is required for RNA phosphodiester bond hydrolysis18. Trax is the first example of an RNase adopting such a fold. In our crystallographic structure of truncated hexameric C3PO, active sites for endoribonuclease activity are located on the inner concave face and the two sites are ~50 Å apart from each other (see yellow arrows, Fig. 1h).
Mass spectra of full-length C3PO
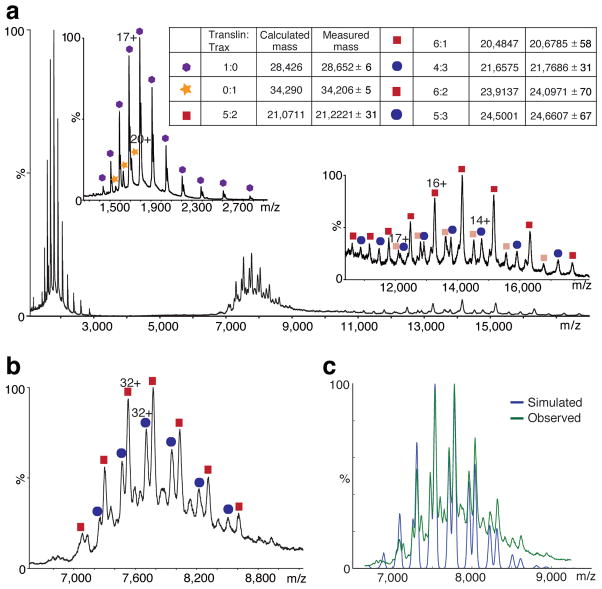
If we assume that the ratio of 4 copies of Translin to 2 copies of Trax observed in crystallized truncated C3PO still holds for full-length C3PO, we would expect a calculated MW of ~182 kDa for a supposed full-length hexameric C3PO. However, both gel filtration and MALS for full-length C3PO show a MW of ~240 kDa, which is ~55–60 kDa higher than the expected value (Supplementary Fig. 2b). In order to fully elucidate the oligomeric status of C3PO, we carried out mass-spectrometric measurements on full-length C3PO. Non-denaturing MS analysis for full-length C3PO (Fig. 2a) showed the presence of Translin and Trax in the ratio of 6:2 and 5:3, with 6:2 as the major component based on simulation analysis (Figs. 2b,c). In either case, it indicates that the full-length C3PO is an octamer (Fig. 2). Furthermore, MS analysis for truncated hexameric C3PO formed by the crystallization construct showed species with Translin:Trax ratios of 2:2, 4:2 and 6:2, with species 4:2 and 2:2 as the major components (Supplementary Fig. 4). It is conceivable that only the complex from the 4:2 species packs in an orderly manner to form crystals as observed in our crystallographic structure of truncated hexameric C3PO.
Figure 2.
Mass spectra and simulated spectrum of the full-length C3PO. (a) Mass spectrum of the full-length C3PO at high collision energy (80 V). The full-length C3PO undergoes gas-phase dissociation such that free monomers of translin (blue hexagons) and Trax (yellow stars) are observed at low m/z (left inset) leaving 'stripped' complexes at high m/z (right inset). Two stripped complexes for the 6:2 (translin:Trax) complex are observed because either translin (pink squares) or Trax (beige squares) can dissociate. Similarly, for the 5:3 (translin:Trax) complex, stripped complexes without translin (light blue oval) or Trax (pink squares) are observed. (b) Mass spectrum showing the m/z region of a (6,600–9,200). Translin:Trax complexes with 6:2 (red squares) and 5:3 (blue ovals) stoichiometries are identified. A similar spectrum was obtained at a collision energy of 50 V. (c) Simulated mass spectrum of the 6:2 and 5:3 translin:Trax C3PO was generated automatically using an algorithm (F. Stengel, A.J. Baldwin, M.F. Bush, H. Lioe, N. Jaya et al., unpublished data) implemented in Python, which suggests the presence of 63% and 37% occurrence of these two species, respectively. The measured spectrum obtained at collision energy 50 V is overlaid on the observed spectrum.
We performed titration of 24-mer ssRNA with full-length C3PO to monitor if it binds to one or both forms (6:2 and 5:3 Translin:Trax species) of C3PO and then analyzed the titration-products using mass-spectrometry. During titration, ssRNA concentration was varied from 0 to 20.0 μM with a fixed concentration (3.3 μM) for full-length C3PO. Peak corresponding to the 6:2 C3PO species shifted completely from free to RNA bound-form at ~1 μM concentration of ssRNA, whereas for 5:3 C3PO species peak shifted only at ~2 μM concentration of ssRNA. This suggests weaker binding of ssRNA to 5:3 C3PO compared to 6:2 C3PO species (Supplementary Fig. 5).
Negative staining electron microscopy
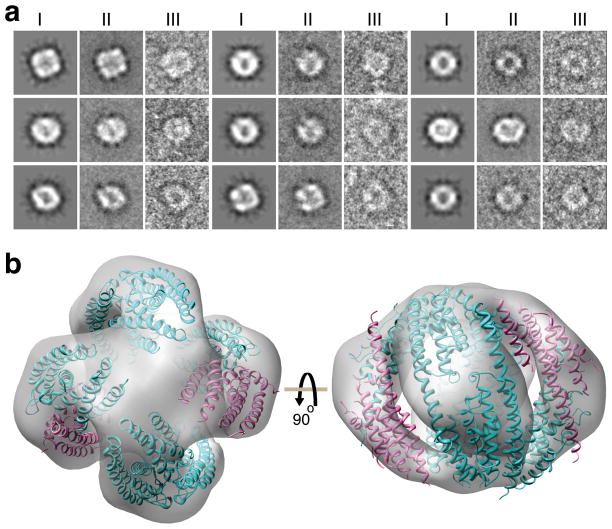
To further investigate structural details of C3PO, we used negative-staining electron microscopy and single particle analysis on the full-length C3PO and produced a three-dimensional (3D) reconstruction. Images collected from the full-length C3PO particles (Supplementary Fig. 6a) have a strong resemblance to the projections computed from the 3D reconstructions. Some of the class averages strongly indicate the presence of 4-fold symmetry in the particles (Fig. 3a). In the reconstruction, the full-length C3PO shows a “squashed nutshell” like shape (Fig. 3b) at a resolution limit of ≈ 15 Å (Supplementary Fig. 6b).
Figure 3.
Classification and averaging of negatively-stained particle images and fit of octameric C3PO into 3D EM map. (a) Montage of nine panels for comparison between reprojection of the reconstruction (image I) with the corresponding class average (image II) showing a representative particle in that class (image III). (b) Docking of the octameric full-length C3PO (Translin:Trax ratio of 6:2) model in the 3D EM map. Trax (pink) and Translin (cyan) molecules are shown in ribbon representation. Both 6:2 and 5:3 Translin:Trax models of C3PO could be fitted satisfactorily within the EM envelope. Top and side views related by a 90° rotation are shown in left and right panels, respectively.
EM reconstruction and proposed models of full-length C3PO
Visual inspection of electron microscopy density map for full-length C3PO suggested presence of four dimeric molecules of Translin-Translin and Translin/-Trax aligning side-by-side and forming a closed octameric C3PO. It thus appears likely that our crystallographic structure of truncated C3PO (Fig. 4a) corresponds to three quarters of the 3D EM map (Fig. 3b), with density corresponding to one dimer missing between the two Translin-Trax heterodimer segments. Since the C3PO particles become distorted due to the drying process associated with negatively stained samples, the EM envelope resembles a squashed-nutshell (Fig. 3b), and therefore it was necessary to decrease the curvature between Translin/Trax and Translin molecules in individual dimers, so as to accommodate them into the EM density map. In the EM envelope, we first fitted three dimers aligning them side-by-side as seen in the hexameric truncated C3PO structure. We then fitted a Translin-Translin homodimer into the remaining EM density (Fig. 3b). Given the similar fold observed for Translin and Trax, as well as the limited resolution of the electron microscope, it was not possible to differentiate between Trax and Translin in the EM map. Because of the mentioned distortions of the particles, there is an apparent gap between adjacent dimers and also around the 4-fold axis of the particles (Fig. 3b).
Figure 4.
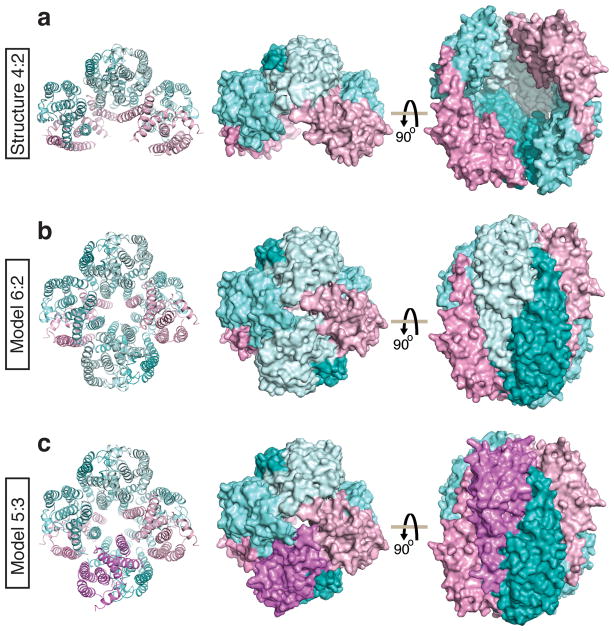
Models of full-length octameric C3PO (6:2 and 5:3 Translin:Trax) based on the crystal structure of truncated hexameric C3PO (4:2 Translin:Trax). (a) Crystal structure of truncated hexameric C3PO (4:2 Translin:Trax) with Translin in cyan and Trax in pink. (b) Model of the full-length octameric C3PO (6:2 Translin:Trax) generated by addition of a homodimer of Translin (cyan) into the crystal structure of truncated C3PO. (c) Model of the full-length octameric C3PO (5:3 Translin:Trax) generated by addition of a heterodimer of Translin (cyan) and Trax (pink) into the crystal structure of truncated C3PO.
Based on the crystallographic structure of truncated C3PO, the subunits composition from MS studies and the overall envelope density for full-length C3PO obtained from EM studies, we generated three-dimensional models for full-length octameric C3PO. Starting from the truncated hexameric C3PO crystal structure, we have modeled either a Translin-Translin homodimer resulting in a 6:2 Translin:Trax octamer (Fig. 4b), or a Translin-Trax heterodimer resulting in a 5:3 Translin:Trax octamer (Fig. 4c), into the remaining space between the two Trax-Translin heterodimers in the current crystallographic structure (Fig. 4a). For modeling, each individual set of dimers were slightly rotated in such a way that alignment of four dimers lead to a closed octameric model for full-length C3PO.
RNase activity of full-length and truncated C3PO
Recombinant full-length (fl), truncated (t), and E123A/E126A mutant (m) C3PO complexes were incubated with two 21-nt RNAs of different sequence (Fig. 5a). RNA cleavage was observed by full-length and truncated C3PO, but not mutant C3PO. At high RNA concentrations (1.5 μM), full-length C3PO complex cleaved under multiple turnover conditions at a rate of approximately 0.7 s−1, whereas truncated C3PO cleaved approximately 2 orders of magnitude slower (Supplementary Fig. 7). The cleavage reaction was also Mg2+-dependent and abrogated by addition of EDTA (data not shown).
Figure 5.
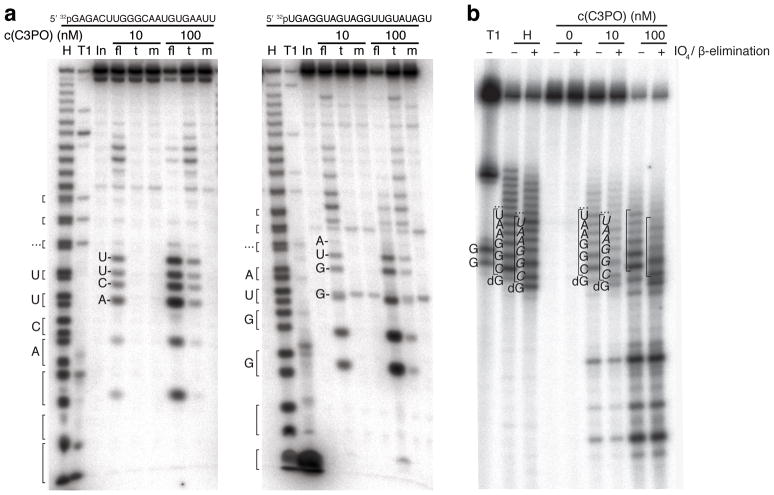
C3PO ribonuclease activity generates products with 2′ and 3′ hydroxyl termini and requires Glu123 and Glu126 residues in Trax. (a) Two 5′ 32P-labelled 21-nt oligoribonucleotides were incubated with indicated recombinant C3PO complex and protein concentration: full-length (fl), truncated (t), and the catalytic mutant C3PO (m) in which two glutamic acid residues of Trax were converted to alanine (E123A, E126A). The reactions were stopped and reaction products were separated by 18% denaturing polyacrylamide gel electrophoresis. Alkaline hydrolysis treatment of RNA substrate generates products with 2′,3′-cyclic and 2′ and 3′ monophohosphate ends, which resolve into doublets towards the bottom of the gel. Doublet bands were bracketed, and the upper band represents fragments with 2′ or 3′ phosphate and the lower band the 2′,3′-cylic phosphate product; RNase T1 generated 3′ phosphate ends only. The mobility of C3PO-digested fragments was reduced compared to their 2′ or 3′ phosphorylated derivatives. (b) A chimeric DNA/RNA oligonucleotide: 5′ 32pTATCG-AGGTGAACATCACGTACGCGGAAUACUUCGAAATGTCCGTTCGGT, containing 12 internal RNA residues (underlined), was incubated with indicated concentrations of C3PO. Half of the reaction solution was subjected to periodate oxidation and β-elimination reaction. For comparison, partial alkaline-hydrolysed RNA was first 3′-dephosphorylated using T4 polynucleotide kinase and then subjected to oxidation andβ-elimination. The identities of RNA and DNA residues are shown; italicised residues indicate that the 3′-end contains a phosphate due to β-elimination. Abbreviations: T1, partial RNase T1 digest; H, partial alkaline hydrolysis; In, input RNA, “…”, bands representing subsequent RNA bases towards the 3′ end of the oligonucleotide sequence.
C3PO is an endonuclease generating 3′-OH products
Single-nucleotide resolution of the RNA cleavage products showed bands that did not co-migrate with RNA products obtained by hydrolysis (H) or RNase T1 cleavage of the starting substrate (Fig. 5a). Alkaline hydrolysis yielded 2′,3′-cyclic-phosphate and 2′ and 3′ monophosphate termini, whereas RNase T1 generated 3′ phosphate ends, suggesting that C3PO leaves 3′ hydroxyl and 5′ phosphate ends. In order to interrogate the 3′ end of the C3PO cleaved products, we performed periodate oxidation/β-elimination reactions on a chimeric substrate that contains a 12-nt RNA segment flanked by DNA residues to enhance gel resolution of cleavage products. Periodate-oxidation requires vicinal 2′,3′–hydroxyl groups. We find that C3PO-cleaved RNA fragments were substrates for oxidation/β-elimination resulting in products of 1-nt altered gel mobility (Fig. 5b), thereby demonstrating that C3PO cleavage left 2′,3′ hydroxyl termini.
Although C3PO cleavage occurred rapidly within the RNA segment of the chimeric substrate, we also observed DNA cleavage. When we compared the cleavage of 40-nt RNA and DNA single-stranded substrates, the RNA was cleaved about 76-times faster by full-length C3PO than the same sequence of DNA (data not shown), even though electrophoretic mobility shift assays showed that these substrates bound with similar affinity (Supplementary Fig. 8).
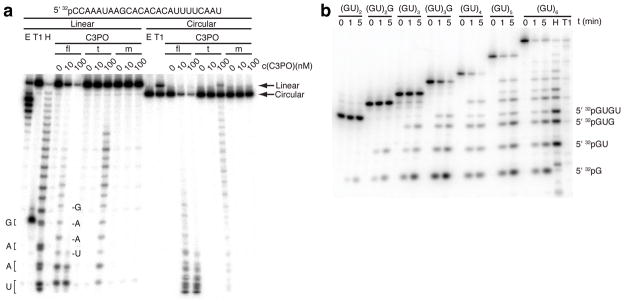
Next, we compared the cleavage activity of C3PO between linear and circularized 25-nt RNA. Linear and circular RNAs were cleaved at the same rate, except that full-length C3PO was more active than truncated C3PO (Fig. 6a). Full-length C3PO cleaved the circular RNA so rapidly that its linearized form was undetectable, also indicating that the enzyme is highly processive. Surprisingly, when we tested circular RNA as short as 13 nt, they were still cleaved as fast as linear substrates (Supplementary Fig. 9).
Figure 6.
C3PO endoribonucleolytic activity is length-dependent. (a), Recombinant C3PO complexes retain endoribonucleolytic cleavage of circularized RNA. Indicated concentrations of C3PO were incubated with a 25-nt linear or circular radiolabelled RNA containing a single guanosine residue. Circular RNA was validated by its reduced migration (arrows) upon linearising digestion by RNase T1, and its resistance to exonuclease T. (b), RNA length dependence of C3PO cleavage. Radiolabelled poly(GU) oligoribonucleotides ranging from 4 to 12 nt in length were subjected to full-length C3PO cleavage at indicated times. The identities of the shortest C3PO-digested fragments are shown. For assignment of cleavage products, the 12-nt substrate was subjected to partial alkaline hydrolysis or RNase T1 digestion, both followed by treatment with T4 PNK to remove 2′ and 3′ phosphate and 2′,3′ cyclic phosphate ends. Reactions were stopped and separated on an 18% denaturing polyacrylamide gel. Abbreviations: T1, partial RNase T1 digest; H, partial alkaline hydrolysis; E, partial Exonuclease T digest.
Finally, we determined the minimum length of RNA that could be cleaved efficiently by full-length C3PO. RNAs as short as 7 nt were readily processed by full-length C3PO, but then the rate of cleavage dropped (Fig. 6b).
DISCUSSION
C3PO’s multimeric assembly
Our study reports on the first structural characterization of the Trax protein, which adopts the seven α-helical bundle characteristic of the Translin fold8 (Fig. 1d). Recently, amino acids Glu123, Glu126 and Asp204 of Trax have been identified as key residues involved in RNase activity of C3PO based on site-directed mutagenesis experiments8. Indeed, these acidic residues are found in close proximity to each other in the crystal structure of the truncated hexameric C3PO, residing within the center of the Trax fold (Fig. 1g). An earlier amino-acid sequence analysis had predicted formation of a leucine zipper motif within the N-terminal segment (residues 73 to 108) of human Trax9,10. Based on our structure of truncated D. melanogaster C3PO, this segment encompasses the loop bridging α1-α2 helices and the α2 helix of Trax, thereby precluding leucine zipper formation for this N-terminal segment.
Truncated C3PO adopts a hexameric topology composed of four Translin and two Trax molecules in the crystal structure (Fig. 1b, c), consistent with conclusions from gel filtration and light scattering studies (Supplementary Fig. 2a). The major interactions between subunits involve formation of Translin-Translin homodimers (Fig. 1f) and our first views of Translin-Trax heterodimers (Fig. 1e), with details of the primarily hydrophobic inter-subunit interactions at four distinct interfaces shown in Supplementary Figs. 3a–d. Translin and Trax not only fold similarly (Fig. 1d), but also bind neighboring molecules using common recognition principles (Figs. 1e,f).
The extensive Translin-Trax interaction interface provides an explanation for earlier reports on the “Trax stabilizer” role played by Translin10. Several independent studies have shown that targeted deletion of Translin would lead to nearly complete loss of Trax protein in organisms like yeast, fruit flies and mice10. In all these studies, the level of Trax mRNA remained normal suggesting that loss of Trax protein occurred during the post-translational stage. Consistent with this conclusion, stand-alone Trax has been shown to be prominently unstable in vitro10, and in the absence of bound Translin, may be easily degraded in vivo. Significantly, we do not observe a Trax-Trax homodimer within the truncated hexameric C3PO topology.
Our gel filtration and light scattering (Supplementary Fig. 2), as well as mass spectroscopic (Fig. 2) and negative staining electron microscopic (Fig. 3) studies demonstrate that full-length C3PO adopts an octameric topology. Further, mass spectroscopic studies establish that the major and minor species of full-length octameric C3PO adopt Translin:Trax ratios of 6:2 and 5:3, respectively (Fig. 2). Thus, the present study for the first time suggests the presence of two different species for full-length octameric C3PO in vitro. This finding raises the following question. Does either one or both of these species exist in vivo? If both species of C3PO were to exist, do they have distinct or redundant physiological functions? Although we do not currently have answers for these questions, available MS data on ssRNA complex formation sheds some light on this issue. The 6:2 C3PO species binds to ssRNA with apparently stronger affinity than does the corresponding 5:3 C3PO species (Supplementary Fig. 5).
All octameric C3PO particles exhibited uniformity under the electron microscope, suggesting that both 6:2 and 5:3 species could assemble in a similar octameric fashion. We therefore built three-dimensional models of full-length octameric C3PO starting with three dimers from the truncated hexameric C3PO crystallographic structure and then docked either a Translin-Translin homodimer or a Translin-Trax heterodimer to generate 6:2 (Fig. 4b) and 5:3 (Fig. 4c) Translin:Trax species of C3PO. The octameric arrangement of Translin and Trax in the model of full-length C3PO resembles the octameric quaternary fold observed in crystal structures of Translin solved previously from human15 and mouse14, in which either 2 or 3 specific Translin subunits have been replaced by Trax subunits.
Our EM-based models of the octameric fold of full-length C3PO containing 6:2 (Fig. 4b) or 5:3 (Fig. 4c) ratios of Translin:Trax require further testing, especially since we are unable to identify the positions and orientations of Translin and Trax within the EM density of full-length C3PO. It is conceivable that we could have missed details such as unanticipated subunit-subunit interactions, as well as aspects of symmetry/asymmetry characteristic of the system, that could impact on the quaternary arrangements of subunits in full-length C3PO. Therefore, we emphasize that the models have been proposed from the perspective of stimulating further investigation.
C3PO’s nucleolytic activity
C3PO exhibits rapid sequence-independent endoribonucleolytic activity generating 3′-hydroxyl and likely 5′-phosphate terminal ends. Its preferred substrate is RNA of 7-nt or longer, including short circular RNAs (Figs. 5a, 5b and Supplementary Fig. 7). The catalytic activity of C3PO resides solely with the Trax subunits as mutations of key amino acid acidic residues eliminates substrate cleavage.
We also noted that C3PO cleaved DNA, albeit at a reduced rate, which is generally consistent with a hydrolytic cleavage mechanism depending on hydroxide ions or water as nucleophiles (Fig. 5b). Gel shift analysis demonstrated at the same time that DNA and RNA oligonucleotides were bound with similar affinity to C3PO, indicating that the cleavage rate differences are likely due to placement of DNA versus RNA residues at the active site (Supplementary Fig. 8). Cleavage activity of C3PO is also length-dependent as RNA substrates less than 7 nt were cleaved slower (Fig. 6b). Together, these results suggest that nucleotides surrounding the cleavage site contribute to substrate binding and possibly also catalytic cleavage. Binding of nucleic acid substrates to C3PO is presumably mediated through Translin, and it is known that Trax can directly affect the affinity of Translin for binding DNA versus RNA19–28. Additionally, Translin has been reported to localize in the nucleus under specific cellular conditions21. Thus it is plausible that the multiple roles ascribed to C3PO also involve its newly defined nucleolytic activity.
The catalytic pockets associated with individual Trax subunits are located within the hollow interior chamber of the full-length octameric C3PO barrel-like scaffolds modeled in Figs. 4b and 4c. There appears to be no obvious gap or channel large enough to allow entry of ssRNA into the central chamber. Meanwhile, our MS data showed that full-length C3PO particle maintains the octameric arrangement upon binding to ssRNA (Supplementary Fig. 5). In the absence of a structure for the C3PO-RNA complex, one can only speculate that the octameric scaffold might undergo a conformational change, switching from a “closed state” to an “open state”, thereby providing RNA access to the catalytic pockets on Trax. As an alternative, the octameric scaffold might transiently open by partial dissociation of subunits prior to reclosure around the encapsulated RNA.
It was surprising to see short circular RNAs (13–25 nt) processed as efficiently as their linear counterparts. Given that the active site residues are positioned within the core of C3PO, these circular substrates must be able to feed through the opening pore (conformational switch model), because certainly the 13-nt circle was too small to assemble a C3PO complex around its strand (Fig. 6a and Supplementary Fig. 9). Solving a crystal structure with a nucleic acid substrate positioned inside C3PO should help clarify these remaining mechanistic questions.
ONLINE METHODS
Cloning, co-expression and purification
Full-length as well as truncated Translin (1-217) and Trax (30-298) were cloned at two different multiple cloning sites in sumo-pRSFDuet-1 vector (modified pRSFDuet-1 vector with N-terminal 6xHis and yeast sumo as fusion tag). The fusion tag (6XHis-SUMO tag) was attached only to Translin. All constructs were confirmed by DNA sequencing. Proteins were co-expressed in BL21 (DE3) E. coli cells in LB medium. The proteins were purified from the soluble fraction using Ni-chelating affinity and size-exclusion column chromatography (see Supplementary Methods for more details).
Crystallization and data collection
Crystals of C3PO obtained from SeMet-labeled truncated Translin and Trax were grown using the hanging-drop vapor-diffusion method by mixing the protein (40 mg ml−1 in 20 mM Tris-HCl pH 8.0, 50 mM KCl) with an equal volume of reservoir solution containing 0.35 M–0.7 M (NH4)2HPO4, 0.1 M Na-citrate pH 5.4–5.8 at 20 °C. For data collection, crystals were flash frozen (100 K) in the mother liquor supplemented with 30% (v/v) glycerol as cryo-protectant. The diffraction pattern obtained from the C3PO crystal showed strong diffraction anisotropy. Diffraction data sets were collected using SeMet-labeled C3PO crystal at Se peak and inflection wavelengths on X29 beamline at National Synchrotron Light Source, Brookhaven National Laboratory, with the collected data sets integrated and scaled using the HKL2000 suite30.
Structure determination and refinement
The structure of C3PO was determined using the anomalous diffraction data collected at Se peak and inflection wavelengths. A total of nine Se sites were located and refined for phasing using SHARP3131 (see Supplementary Methods for more details). The initial experimental map showed clear density for the helical regions of the complex (Supplementary Fig. 1c). The issue of strong anisotropic diffraction was addressed by anisotropic scaling and ellipsoidal truncation using the NIH Diffraction Anisotropy Server32 (www.doe-mbi.ucla.edu/~sawaya/anisoscale/anisotropy). Briefly, data residing outside an ellipse centered at the reciprocal lattice origin and having vertices at 1/3.9, 1/3.9 and 1/3.4 Å−1 along a*, b* and c*, respectively, were removed (Supplementary Fig. 1a). After this ellipsoidal truncation, anisotropic scale factors were applied and in the end, a negative isotropic B factor was used to restore the magnitude of the high-resolution reflections diminished by anisotropic scaling. During the later stage of model building and refinement, anisotropically scaled data was used, which showed improved electron density map for the model.
The number and location of the selenium positions played a key role in identifying Translin (2 Se atoms) and Trax (5 Se atoms) molecules in the asymmetric unit (Supplementary Fig. 1b). The final model has been refined to an Rwork of 27.0% and an Rfree of 33.8%. The final data collection and refinement statistics are given in Table 1. Also see in Supplementary methods.
Multi-angle light scattering (MALS)
Molar mass values of purified full-length C3PO and truncated C3PO were determined by gel filtration and MALS as described in Supplementary methods.
Mass spectrometry (MS)
For mass spectrometric analysis, full-length and truncated C3PO complexes were buffer exchanged into 1 M ammonium acetate at pH 8.0, using Amicon Ultra-0.5, Ultracel-10 Membrane (Millipore). For the ssRNA titration experiments, concentration of ssRNA was varied from 0–2.0 μM at full-length C3PO concentration of 3.3 μM. The 24-mer ssRNA for MS analysis was purchased from Integrated DNA technologies. Nanoflow electrospray ionization (nESI-MS) was conducted on a high mass Q-ToF2 mass spectrometer (Waters)33 (see Supplementary Methods for more details).
Negative stain electron microscopy
The purified full-length C3PO (protein concentration: 10 μg/ml) was applied to a glow discharged, carbon coated grids. The grids were washed with three drops of water, followed by three drops of uranyl acetate, and dried. Images were collected under low dose conditions on a JEOL2100F electron microscope operating at an accelerating voltage of 200 kV (Supplementary Fig. 6a), using under focus values between 1 and 2 μm and a pixel size of 3.06 Å. For random conical tilt reconstruction, image pairs were collected with a first image at a tilt of 55°, and a second at 0°.
The Fourier shell correlation curve shows good correlation of two independently reconstructed models, up to spatial frequencies that correspond to spacings of ~15 Å, according to the 0.5 correlation criterion (Supplementary Fig. 6b). A plot of the angular coverage of the data shows that all the views needed for a reconstruction were represented by the data (Supplementary Fig. 6c). EM image processing and labeling/tagging experiments were carried out as described in Supplementary Methods. Electron microscopy maps were displayed with the UCSF CHIMERA software package34, which was also employed to manually fit the Translin-Translin homodimer and Translin-Trax heterodimer in the EM map.
5′ end radiolabelling of oligonucleotides
Oligonucleotides were 5′ end radiolabelled using T4 polynucleotide kinase (PNK) (New England Biolabs) and γ-32P ATP (see Supplementary Methods for more details). Circular RNA was radiolabelled similarly, as linear RNA, prior to circularization using T4 Rnl1 (Tuschl lab). Radiolabeled RNAs were denatured with 1 volume of stop buffer (8 M urea, 10 mM EDTA, and bromophenol blue), incubated for 30 sec at 95°C and immediately chilled on ice, then separated by denaturing polyacrylamide gel. Radiolabelled bands were excised, gel extracted overnight in 0.4 M NaCl, ethanol precipitated and resuspended in H2O.
Partial alkaline hydrolysis, RNase T1, and exonuclease T treatment of oligonucleotides
RNA partial alkaline hydrolysis was performed by incubating 100 nM oligonucleotide with 1.2 μg/μl yeast tRNA and 0.1 M Na2CO3, pH 9.2 for 30 sec at 95°C then chilling on ice. Partial RNase T1 or exonuclease T digestion of oligonucleotides were similarly treated: 100 nM oligonucleotide is incubated in a 5 μl reaction containing yeast tRNA and 0.2 unit/μl RNase T1 or 5 units/μl exonuclease T. RNase T1 and exonuclease T digestions were incubated at 24°C for 1 and 10 min, respectively.
C3PO nuclease reactions
Unless otherwise indicated, reactions were performed in 10 μl with 100 nM C3PO and 100 nM oligonucleotide in reaction buffer (20 mM Tris-HCl (pH 8.0), 50 mM KCl, 5 mM MgCl2 and 10% glycerol) for 5 min at 24°C. Reactions were stopped with 1 volume of stop buffer. 2–4 μl of the reaction was separated on a denaturing sequencing-type polyacrylamide gel in 0.5 × TBE buffer at 60 W for 2 hrs. The gels were dried on Whatman paper, visualized using a phosphorimager screen (Fujifilm FLA-5000) and analyzed using ImageGauge v4.1 software (Fujifilm).
Oxidation and β-elimination reactions
Periodate oxidation and β-elimination was performed as described35,36. C3PO-digested oligonucleotides were recovered by phenol-chloroform and ethanol precipitation, then dried and resuspended in borax buffer, pH 8.6. NaIO4 (28.6 mM final) was added and the reactants were incubated for 10 min in the dark at 24°C, followed by addition of glycerol (5% final) for 10 more min. The samples were concentrated in a SpeedVac (Eppendorf) to 5 μl. Borax buffer, pH 9.5 was added and the samples were incubated for 90 min at 45°C (See Supplementary methods for more details). Samples were ethanol precipitated twice and resuspended in H2O and an equal volume of stop buffer. 4 μl of each sample were separated by denaturing polyacrylamide gel electrophoresis.
Kinetic determination of the catalytic activities of C3PO was carried out as outlined in Supplementary Methods. Procedure for Removal of 3′ and 2′,3′-cyclic phosphates from partially alkaline-hydrolysed oligonucleotides is described in Supplementary Methods.
Supplementary Material
Acknowledgments
We thank Dr. Haitao Li from Sloan-Kettering Institute for assistance with synchrotron data collection, Dr. Hao Wu from Weill Cornell Medical College for access to and assistance with MALS experiments, and Dr. Paula Upla from New York Structural Biology Center for help with the electron microscopy of labeled proteins. We are grateful to the staff of X-29 beamline at the National Synchrotron Light Source, Brookhaven National Laboratory and ID-24-E beamline at the Advanced Photon Source, Argonne National Laboratory for their help with data collection. D.J.P. is supported by NIH grant AI068776. T.T. is supported by funds from HHMI and NIH. A.Y.P and C.V.R acknowledge funding from the BBSRC/the Royal Society and Walter’s Kundert Trust, respectively.
Footnotes
PDB accession codes
The atomic coordinates and structure factors of the D. melanogaster truncated hexameric C3PO complex have been deposited in the Protein Data Bank with accession code 3RIU.
Author’s contributions
Y.T. designed and performed the experiments leading to crystallization of C3PO and undertook initial structural characterization, while D.K.S. improved the density map, built the model and finished the refinement under the supervision of D.J.P.; M.A. and S.J. performed the cleavage assays under the supervision of T.T.; R.D. and W.J.R. performed the EM studies; A.Y.P performed the MS studies under the supervision of C.V.R.; Q.Y. and Y.T. performed the MALS studies. All authors participated in writing their contributions to the paper.
References
- 1.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 3.Liu Q, et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–5. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 4.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–80. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–13. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35:368–76. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–61. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, et al. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750–3. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaendling A, McFarlane RJ. Biological roles of translin and translin-associated factor-X: RNA metabolism comes to the fore. Biochem J. 2010;429:225–34. doi: 10.1042/BJ20100273. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Wu Y, Baraban JM. The Translin/Trax RNA binding complex: clues to function in the nervous system. Biochim Biophys Acta. 2008;1779:479–85. doi: 10.1016/j.bbagrm.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eliahoo E, et al. Mapping of interaction sites of the Schizosaccharomyces pombe protein Translin with nucleic acids and proteins: a combined molecular genetics and bioinformatics study. Nucleic Acids Res. 2010;38:2975–89. doi: 10.1093/nar/gkp1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lluis M, Hoe W, Schleit J, Robertus J. Analysis of nucleic acid binding by a recombinant translin-trax complex. Biochem Biophys Res Commun. 2010;396:709–13. doi: 10.1016/j.bbrc.2010.04.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta GD, Makde RD, Rao BJ, Kumar V. Crystal structures of Drosophila mutant translin and characterization of translin variants reveal the structural plasticity of translin proteins. FEBS J. 2008;275:4235–49. doi: 10.1111/j.1742-4658.2008.06571.x. [DOI] [PubMed] [Google Scholar]
- 14.Pascal JM, Hart PJ, Hecht NB, Robertus JD. Crystal structure of TB-RBP, a novel RNA-binding and regulating protein. J Mol Biol. 2002;319:1049–57. doi: 10.1016/S0022-2836(02)00364-9. [DOI] [PubMed] [Google Scholar]
- 15.Sugiura I, et al. Structure of human translin at 2.2 A resolution. Acta Crystallogr D Biol Crystallogr. 2004;60:674–9. doi: 10.1107/S0907444904002549. [DOI] [PubMed] [Google Scholar]
- 16.VanLoock MS, Yu X, Kasai M, Egelman EH. Electron microscopic studies of the translin octameric ring. J Struct Biol. 2001;135:58–66. doi: 10.1006/jsbi.2001.4383. [DOI] [PubMed] [Google Scholar]
- 17.Claussen M, Koch R, Jin ZY, Suter B. Functional characterization of Drosophila Translin and Trax. Genetics. 2006;174:1337–47. doi: 10.1534/genetics.106.063016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang W. An equivalent metal ion in one- and two-metal-ion catalysis. Nat Struct Mol Biol. 2008;15:1228–31. doi: 10.1038/nsmb.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aharoni A, Baran N, Manor H. Characterization of a multisubunit human protein which selectively binds single stranded d(GA)n and d(GT)n sequence repeats in DNA. Nucleic Acids Res. 1993;21:5221–8. doi: 10.1093/nar/21.22.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aoki K, Suzuki K, Ishida R, Kasai M. The DNA binding activity of Translin is mediated by a basic region in the ring-shaped structure conserved in evolution. FEBS Lett. 1999;443:363–6. doi: 10.1016/s0014-5793(99)00010-1. [DOI] [PubMed] [Google Scholar]
- 21.Aoki K, et al. A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat Genet. 1995;10:167–74. doi: 10.1038/ng0695-167. [DOI] [PubMed] [Google Scholar]
- 22.Gupta GD, et al. Co-expressed recombinant human Translin-Trax complex binds DNA. FEBS Lett. 2005;579:3141–6. doi: 10.1016/j.febslet.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Kasai M, et al. The translin ring specifically recognizes DNA ends at recombination hot spots in the human genome. J Biol Chem. 1997;272:11402–7. doi: 10.1074/jbc.272.17.11402. [DOI] [PubMed] [Google Scholar]
- 24.Laufman O, Ben Yosef R, Adir N, Manor H. Cloning and characterization of the Schizosaccharomyces pombe homologs of the human protein Translin and the Translin-associated protein TRAX. Nucleic Acids Res. 2005;33:4128–39. doi: 10.1093/nar/gki727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sengupta K, Rao BJ. Translin binding to DNA: recruitment through DNA ends and consequent conformational transitions. Biochemistry. 2002;41:15315–26. doi: 10.1021/bi026378m. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Boja ES, Oubrahim H, Chock PB. Testis brain ribonucleic acid-binding protein/translin possesses both single-stranded and double-stranded ribonuclease activities. Biochemistry. 2004;43:13424–31. doi: 10.1021/bi048847l. [DOI] [PubMed] [Google Scholar]
- 27.Wu XQ, Gu W, Meng X, Hecht NB. The RNA-binding protein, TB-RBP, is the mouse homologue of translin, a recombination protein associated with chromosomal translocations. Proc Natl Acad Sci U S A. 1997;94:5640–5. doi: 10.1073/pnas.94.11.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu XQ, Xu L, Hecht NB. Dimerization of the testis brain RNA-binding protein (translin) is mediated through its C-terminus and is required for DNA- and RNA-binding. Nucleic Acids Res. 1998;26:1675–80. doi: 10.1093/nar/26.7.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stengel F, et al. Rapid client protection by small heat shock proteins leads to variable complex morphology. in preparation. [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallographica Section D-Biological Crystallography. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 32.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–5. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sobott F, Hernandez H, McCammon MG, Tito MA, Robinson CV. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal Chem. 2002;74:1402–7. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 34.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Alefelder S, Patel BK, Eckstein F. Incorporation of terminal phosphorothioates into oligonucleotides. Nucleic Acids Res. 1998;26:4983–8. doi: 10.1093/nar/26.21.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akbergenov R, et al. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 2006;34:462–71. doi: 10.1093/nar/gkj447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.