Abstract
Fanconi anemia (FA) is an inherited chromosomal instability disorder characterized by childhood aplastic anemia, developmental abnormalities and cancer predisposition. One of the hallmark phenotypes of FA is cellular hypersensitivity to agents that induce DNA interstrand crosslinks (ICLs), such as mitomycin C (MMC). FA is caused by mutation in at least 14 genes which function in the resolution of ICLs during replication. The FA proteins act within the context of a protein network in coordination with multiple repair factors that function in distinct pathways. SNM1B/Apollo is a member of metallo-β-lactamase/βCASP family of nucleases and has been demonstrated to function in ICL repair. However, the relationship between SNM1B and the FA protein network is not known. In the current study, we establish that SNM1B functions epistatically to the central FA factor, FANCD2, in cellular survival after ICL damage and homology-directed repair of DNA double-strand breaks. We also demonstrate that MMC-induced chromosomal anomalies are increased in SNM1B-depleted cells, and this phenotype is not further exacerbated upon depletion of either FANCD2 or another key FA protein, FANCI. Furthermore, we find that SNM1B is required for proper localization of critical repair factors, including FANCD2, BRCA1 and RAD51, to MMC-induced subnuclear foci. Our findings demonstrate that SNM1B functions within the FA pathway during the repair of ICL damage.
INTRODUCTION
Fanconi anemia (FA) is a rare human genetic disease that is highly heterogeneous. There are currently 14 identified FA complementation groups (A, B, C, D1, D2, E, F, G, I, J, L, M, N and P) (1–4). Thirteen of the FA genes are inherited in an autosomal recessive manner; however, one gene, FANCB, is located on the X chromosome and thus exhibits an X-linked inheritance pattern. FA is a chromosomal instability disorder characterized by bone marrow failure, congenital abnormalities and increased incidence of malignancies (5). One hallmark phenotype observed in FA cells is cellular hypersensitivity to interstrand crosslink (ICL)-inducing agents, including mitomycin C (MMC) and cisplatin, and this feature of FA is frequently used as a clinical diagnostic test.
DNA ICLs represent one of the most formidable lesions, as covalent linkage of the strands of the double helix inhibits essential processes such as transcription and replication. The sensing and repair of ICLs is a complex process which involves the activities of multiple DNA repair proteins including the FA factors as well as proteins that function in nucleotide excision repair, translesion bypass synthesis and homologous recombination (5). Central to the FA protein network is the core complex composed of eight subunits (FANCA, B, C, E, F, G, L and M) that coordinately function to monoubiquitinate FANCD2 and FANCI, a critical event in FA protein-mediated ICL repair (6). The monoubiquitination signal targets the FANCD2–FANCI complex to subnuclear repair foci where it colocalizes with proteins required for homologous recombination, including BRCA1, BRCA2/FANCD1 and RAD51 (7–9). Monoubiquitinated FANCD2 (FANCD2-Ub) also interacts with a DNA nuclease, FAN1, which is recruited to sites of ICL damage to facilitate repair (10–13).
In yeast, the Snm1/Pso2 gene plays critical roles in ICL repair (14). There are three SNM1 orthologs in mammalian cells, DCLRE1A (SNM1A), DCLRE1B (SNM1B/APOLLO) and DCLRE1C (ARTEMIS), which share a highly conserved metallo-β-lactamase/βCASP domain (15,16). SNM1A functions in ICL repair in a pathway distinct from the FA pathway (17,18), and DCLRE1C/ARTEMIS acts within the non-homologous end-joining pathway of DNA double-strand break (DSB) repair (19–21). SNM1B/APOLLO, a 60 kDa protein, possesses intrinsic 5′ to 3′ exonuclease activity and has important functions in telomere maintenance and the repair of ICL and ionizing radiation (IR)-induced DNA damage (22–29).
Previous studies have demonstrated that siRNA knockdown of SNM1B in human cell lines results in hypersensitivity to ICL-inducing agents, low-to-moderate sensitivity to IR, impaired ICL-induced S phase arrest, increased chromosomal anomalies and elevated levels of ICL-induced quadriradial formation (24,26). These phenotypes are characteristic of cells derived from FA patients (1,5), thereby raising the possibility that SNM1B may function within the FA pathway. Consistent with this notion, it has been reported that SNM1B and FANCD2 can be co-immunoprecipitated from human cells (24). However, whether SNM1B functions within the FA pathway of ICL repair remains an open question. Recently, a mutant allele of SNM1B was identified in a patient with Hoyeraal–Hreidarsson syndrome, a severe form of dyskeratosis congenita, characterized by aplastic anemia, microcephaly, bone marrow failure and immunodeficiency (30). These phenotypes closely parallel those observed in FA; however, cells derived from this patient did not exhibit hypersensitivity to MMC and were primarily defective for telomere maintenance. Nonetheless, this study demonstrates that SNM1B mutations are associated with human genetic disease and emphasizes the importance of elucidating the cellular functions of SNM1B.
In the current study, we sought to test the hypothesis that SNM1B functions within the FA pathway during ICL repair processes. Using an siRNA approach in human cell lines, we demonstrate that SNM1B functions epistatically to FANCD2 in cellular ICL repair and homology-directed repair of DNA DSBs. In addition, we find that combined depletion of SNM1B and FANCD2 or FANCI does not increase levels of either spontaneous or ICL-induced chromosomal anomalies, compared with individual siRNA knockdown of each gene. We also demonstrate that SNM1B is required for efficient localization of key repair proteins, including FANCD2, BRCA1 and RAD51, to ICL-induced foci. Together, our findings provide evidence that SNM1B functions in the context of the FA network during ICL repair.
RESULTS
Depletion of SNM1B in human cells
To elucidate the roles of SNM1B in cellular responses to ICL damage, we used an siRNA approach to knockdown expression in human cell lines. We designed two specific siRNAs, siSNM1B-1, which targets mRNA sequences in a unique region of the conserved metallo-β-lactamase/βCASP domain (nt 339–359), and siSNM1B-2, which targets a sequence within the non-conserved C-terminus (nt 1248–1266) (Supplementary Material, Fig. S1A). To validate the siRNA approach, siSNM1B-1 and -2 were transfected into HeLa cells, and SNM1B transcript levels were assessed by semi-quantitative reverse-transcription polymerase chain reaction (RT–PCR). We observed that both siRNAs markedly reduced SNM1B mRNA levels at 48 h post-transfection compared with a non-silencing (NS) siRNA control (Supplementary Material, Fig. S1B). Due to sequence conservation between members of the βCASP superfamily, we also performed RT–PCR to examine transcript levels of the related SNM1A and ARTEMIS genes and confirmed that knockdown is specific to SNM1B (Supplementary Material, Fig. S1B). We also verified that SNM1B protein levels were markedly decreased upon siRNA transfection in HCT116 cells expressing a V5-epitope-tagged SNM1B protein (Supplementary Material, Fig. S1C). Thus, transient transfection of the siRNAs efficiently depleted SNM1B mRNA and significantly reduced protein levels.
SNM1B and FANCD2 function epistatically in response to MMC-induced ICL damage
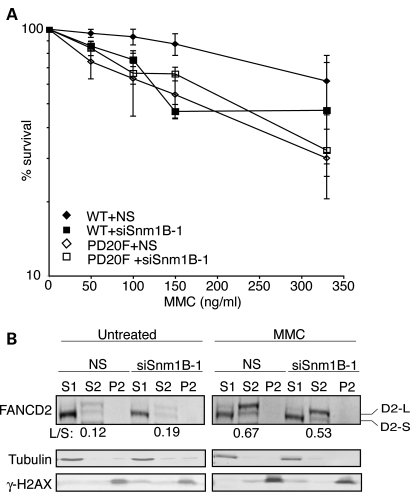
To assess the genetic interactions between SNM1B and the FA pathway during the repair of induced ICL damage, we examined the survival of cells deficient for SNM1B and FANCD2, either alone or in combination, upon exposure to the ICL-inducing agent, MMC. We depleted SNM1B in FANCD2-deficient human fibroblasts (PD20F) and PD20F complemented with WT FANCD2 cDNA (WT) (Supplementary Material, Fig. S2A; 31). We confirmed that the PD20F cells did not express detectable levels of FANCD2 protein and exhibited the hallmark-elevated levels of ICL-induced radial formation compared with WT controls (Supplementary Material, Fig. S2B). The siSNM1B- and NS-transfected PD20F and WT cells were treated with increasing amounts of MMC for 1 h, and then cellular survival was assessed relative to untreated controls. Consistent with previous studies, depletion of SNM1B or deficiency for FANCD2 led to increased sensitivity to MMC compared with the NS siRNA-transfected control cell line (Fig. 1A) (6,24,26,32–34). We found that SNM1B depletion in the FANCD2-deficient cell line did not further increase cellular sensitivity to MMC in comparison to that observed in SNM1B-depleted and FANCD2-deficient cells (Fig. 1A, Supplementary Material, Fig. S2C). These results indicate that SNM1B and FANCD2 function epistatically in cellular responses to MMC-induced DNA damage to ensure survival.
Figure 1.
SNM1B functions epistatically to FANCD2 in cellular survival to ICL damage. (A) FANCD2-deficient PD20F fibroblasts and PD20F cells complemented with FANCD2 cDNA (WT) were transfected with NS and siSNM1B-1. At 48 h post-transfection, cells were plated and treated with indicated MMC concentrations for 1 h. Percent survival compared with untreated samples was determined. Average of at least three independent transfections with standard deviation is shown. (B) FANCD2 monoubiquitination in NS- and siSNM1B-1-transfected cells. HeLa cells transfected with NS or siSNM1B-1 were treated with MMC (1 µg/ml) for 8 h at 48 h post-transfection. Cells were collected and the soluble and chromatin-associated proteins were fractionated. Western blots were probed with α-FANCD2 antibodies. Tubulin and γ-H2AX were used as controls for S1 and P2 fractions, respectively. The L/S ratios of FANCD2 are shown below blot and represent the average of three independent experiments. S1, soluble; S2, chromatin bound; P2, chromatin bound (tightly).
We next examined the impact of SNM1B depletion on FANCD2 chromatin localization. In response to ICLs, FANCD2 undergoes monoubiquitination (FANCD2-Ub), which is dependent on the FA core complex (5). FANCD2-Ub is targeted to chromatin, and this localization is required for subsequent ICL repair events, including assembly of FANCD2 into subnuclear foci and recruitment of key repair proteins (5,35,36). We exposed SNM1B-depleted and control cells to MMC and examined the cellular localization of FANCD2 by fractionation of the cytosolic (S1) and chromatin-bound (S2) proteins followed by western blotting. We observed that FANCD2 is efficiently monoubiquitinated upon MMC treatment in both control and SNM1B knockdown cells, consistent with previous reports (Fig. 1B) (24,26). The ratios of FANCD2-Ub (L) to the unmodified form (S) in the cytosolic and chromatin protein fractions were not significantly different in control and SNM1B-depleted cells (Fig. 1B). These findings indicate that SNM1B is not required for stable localization of FANCD2-Ub to the chromatin fraction.
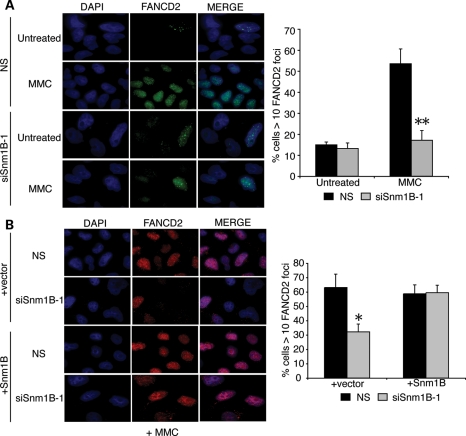
Upon MMC-induced ICL damage, FANCD2-Ub is assembled into subnuclear DNA repair foci (36) via a mechanism that can be uncoupled from the association with the chromatin fraction (37). Therefore, we examined the impact of SNM1B depletion on the efficiency of FANCD2 foci formation by immunofluorescence microscopy after treatment with MMC for 8 h. Under these experimental conditions, there is no appreciable difference in cell cycle distribution in SNM1B-depleted cells compared with controls (Supplementary Material, Fig. S3A). We observed that the percentage of MMC-treated control cells containing FANCD2 foci increased by over 3-fold compared with untreated cells, thereby indicating robust ICL-induced activation of the FA pathway (Fig. 2A). In contrast, SNM1B depletion markedly impaired induction of FANCD2 foci formation upon MMC exposure (Fig. 2A; Supplementary Material, Fig. S3B). In contrast, the percentage of MMC-treated SNM1B-depleted cells containing FANCD2 foci did not significantly increase compared with untreated SNM1B knockdown cells, and was substantially lower compared with NS siRNA-transfected cells. Taken together, these findings indicate that SNM1B is dispensable for FANCD2 monoubiquitination and chromatin localization, but it is necessary for efficient ICL-induced FANCD2 nuclear foci assembly.
Figure 2.
MMC-induced FANCD2 foci formation is impaired in SNM1B-depleted cells. (A) FANCD2 foci formation in SNM1B-depleted cells. HeLa cells transfected with NS or siSNM1B-1 were treated with MMC (330 ng/ml) for 8 h at 48 h post-transfection. Cells were fixed with p-formaldehyde and stained using α-FANCD2 antibodies (green). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). The percentage of cells with greater than 10 FANCD2 foci was determined. At least 300 nuclei were scored for each sample. All images were acquired at ×100 magnification. Representative images are shown. Graph represents average percentage of foci-positive cells from three independent experiments with standard deviation. **P< 0.005, Student's two-tailed t-test. (B) Expression of siRNA-resistant SNM1B complements the ICL-induced FANCD2 foci defect in SNM1B-depleted cells. HeLa cells expressing an siSNM1B-1-resistant SNM1B cDNA were transfected with NS or siSNM1B-1. Transfected cells were treated with MMC (330 ng/ml) for 8 h. Cells were fixed with p-formaldehyde and stained using α-FANCD2 antibodies (red). Nuclei were stained with DAPI (blue). The percentage of cells with >10 FANCD2 foci was determined. At least 300 nuclei were scored for each sample. Representative images are shown. All images were acquired at ×100 magnification. Graph represents average percentage of foci-positive cells from three independent experiments with standard deviation. *P< 0.05, Student's two-tailed t-test.
We next determined whether expression of SNM1B cDNA could complement the marked defects in FANCD2 foci formation upon MMC exposure observed in SNM1B-depleted cells. Thus, we generated a lentiviral construct expressing an siRNA-resistant cDNA harboring three silent base changes within the siRNA core sequence that also contained an internal ribosomal entry site (IRES)-GFP cassette (si-1R SNM1B). Upon transduction of HeLa cells with si-1R SNM1B or a GFP control lentivirus, we sorted GFP-positive cells and performed semi-quantitative RT–PCR on total RNA isolated from cells transfected with either NS or siSNM1B-1 using two sets of primers: (i) 3′ UTR, which specifically amplifies the endogenous SNM1B transcript and (ii) exon 3/4, which amplifies both the endogenous and si-1R SNM1B transcripts. We observed substantially reduced RT–PCR products upon transfection with siSNM1B-1 in both control and si-1R SNM1B-transduced cells using the 3′ UTR primers. However, we observed substantial levels of the si-1R SNM1B transcript in knockdown cells using the exon 3/4 primers (Supplemental Material, Fig. S4A).
We also transfected si-1R SNM1B-transduced cells with siSNM1B-2, an siRNA that binds to a distinct site within the cDNA (see Supplemental Material, Fig. S1A). We found that the percentage of si-1R SNM1B-IRES-GFP expressing cells is not reduced upon transfection with si-SNM1B-1; however, si-SNM1B-2 markedly decreases the percent of GFP-positive cells compared with NS-transfected controls (Supplemental Material, Fig. S4B and C). These observations indicate that the si-1R SNM1B protein, which is translated from the same mRNA as GFP, is resistant to knockdown by siSNM1B-1.
Using the validated complementation system, we transduced cells with si-1R SNM1B or GFP control recombinant lentiviruses and sorted the GFP-positive cells. The cells were then transfected with siSNM1B-1 or the NS control, and the levels of MMC-induced FANCD2 repair foci were determined. We observed that expression of SNM1B fully complements the defect in FANCD2 foci formation in siSNM1B-1-transfected cells (Fig. 2B). These findings demonstrate that SNM1B expression is required for efficient localization of FANCD2 to sites of ICL repair.
SNM1B functions epistatically with FANCD2 and FANCI to suppress ICL-induced chromosomal anomalies
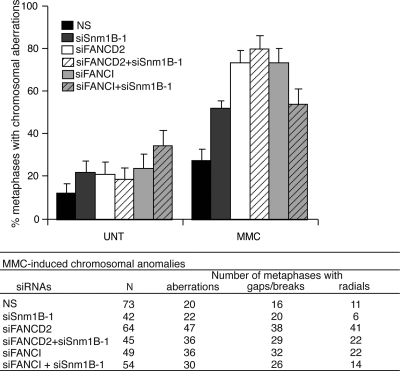
One of the hallmark phenotypes observed in FA patient cells is elevated levels of ICL-induced chromosomal anomalies, including gaps, breaks and radial structures, and quantification of chromosomal aberrations is clinically used for the diagnosis of FA (38). To further examine the genetic relationship between SNM1B and the FA pathway in ICL repair, we examined the levels of MMC-induced chromosomal aberrations in cells depleted of SNM1B or FANCD2 compared with SNM1B/FANCD2 double knockdown cells. As anticipated, we observed a significant increase in the percentage of metaphases harboring chromosomal gaps, breaks and radials in SNM1B-depleted cells, but lower levels compared with FANCD2 knockdown cells (Fig. 3; 25). Significantly, we observed that the depletion of both SNM1B and FANCD2 did not further increase the percentage of metaphases containing spontaneous or MMC-induced chromosomal aberrations (Fig. 3, Supplemental Material, Table S1).
Figure 3.
Chromosome aberrations in HeLa cells after depletion of SNM1B, FANCD2 or FANCI. (Upper panel) HeLa cells were transfected with the indicated siRNAs and treated with MMC (20 ng/ml) for 24 h. Metaphases were prepared and scored for the presence of gaps/breaks and radial chromosomes. Graph indicates the percentage of metaphases containing at least one gap/break or radial. Error bars are standard error. (Lower panel) The total number of metaphases scored for each sample, and the number of metaphases containing one or more aberrations, gaps/breaks or radials is shown. P-values for pairwise comparisons (two-tailed F-test) are shown in Supplemental Material, Table S1.
To further substantiate these findings, we determined the levels of chromosomal aberrations in cells depleted for another FA gene, FANCI, alone and in combination with SNM1B depletion (Supplemental Material, Fig. S6B). FANCI forms a complex with FANCD2 and localizes to DNA repair foci within chromatin upon ubiquitination of FANCD2–FANCI by the FA core complex (6,39,40). Consistent with previous reports (6), siRNA knockdown of FANCI resulted in markedly elevated levels of gaps, breaks and radials (Fig. 3). However, the levels of spontaneous or MMC-induced chromosomal anomalies were not further increased in cells depleted for both SNM1B and FANCI (Fig. 3, Supplemental Material, Table S1). Together, these findings indicate that SNM1B functions epistatically with FANCD2 and FANCI to suppress MMC-induced chromosomal aberrations and thus provide further supporting evidence that SNM1B functions within the FA pathway of ICL repair.
Impact of SNM1B deficiency on localization of key ICL factors to subnuclear repair foci
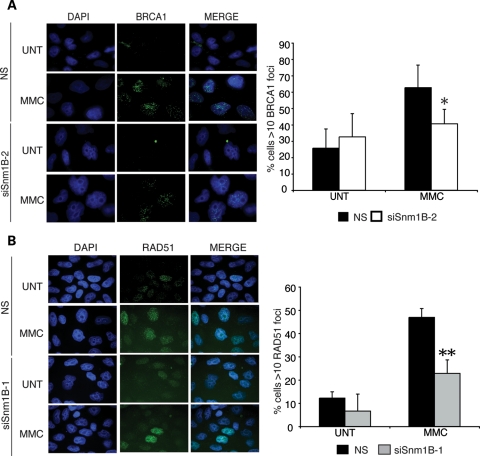
To further investigate the molecular defects in ICL repair caused by SNM1B depletion, we examined the localization of key repair factors to subnuclear foci in response to MMC treatment. During the processing of ICLs, DNA DSB intermediates are generated and repaired by the homologous recombination pathway (5). Homologous recombination facilitates replication restart by catalyzing strand invasion of the one-sided DSB between sister chromatids (41). FANCD2 co-localizes in ICL-induced foci with BRCA1, BRCA2/FANCD1 and RAD51, proteins with central roles in homologous repair of DSB intermediates (7,9,36,42–45). Previous studies have demonstrated that BRCA1 co-immunoprecipitates with the activated, monoubiquitinated form of FANCD2 and mediates FANCD2 foci formation (36). Our results demonstrating that SNM1B is required for efficient FANCD2 foci formation prompted us to examine the impact of SNM1B depletion on ICL-induced BRCA1 foci formation. We quantitated the percentage of NS siRNA-transfected control cells that contained BRCA1 foci and observed an increase of 26–63% upon MMC treatment (Fig. 4A). In comparison, SNM1B-depleted cells exhibited impaired induction and a significantly lower percentage of BRCA1 foci-positive cells in response to ICL damage compared with controls (Fig. 4A, Supplementary Material, Fig. S5A). These findings suggest that SNM1B is required for normal localization of BRCA1 to MMC-induced repair foci.
Figure 4.
MMC-induced BRCA1 and RAD51 foci formation are impaired in SNM1B-depleted cells. (A) BRCA1 foci formation in SNM1B-depleted cells. HeLa cells transfected with NS or siSNM1B-2 were treated with MMC (330 ng/ml) for 8 h at 48 h post-transfection. Cells were fixed with p-formaldehyde and stained with α-BRCA1 antibodies (green). Nuclei were stained with DAPI (blue). The percentage of cells containing >10 foci was determined. (B) RAD51 foci formation in SNM1B-depleted cells. siSNM1B-1- or NS-transfected cells were fixed with p-formaldehyde and stained with α-RAD51 antibodies (green). Nuclei were stained with DAPI (blue). The percentage of cells containing >10 foci was determined. At least 300 nuclei for each sample were scored. Representative images are shown. All images were required at ×100 magnification. Graphs represent average of three independent experiments with standard deviation. *P< 0.05; **P< 0.005.
In mammals, BRCA1 promotes efficient localization of the homologous recombination protein, RAD51, to subnuclear foci in response to ICLs (46,47). Thus, we also examined the impact of SNM1B depletion on RAD51 foci formation upon exposure to MMC. We observed that the depletion of SNM1B also impaired the assembly of ICL-induced RAD51 subnuclear foci compared with controls (Fig. 4B). We also examined RAD51 foci formation at 16 h after MMC addition, and similar to the 8 h time point, SNM1B depletion reduced the percentage of RAD51-foci-positive cells (Supplementary Material, Fig. S5B). Together, these findings suggest that SNM1B facilitates the recruitment of proteins required for homologous recombination.
SNM1B and FANCD2 function epistatically to promote homology-directed repair of DNA DSBs
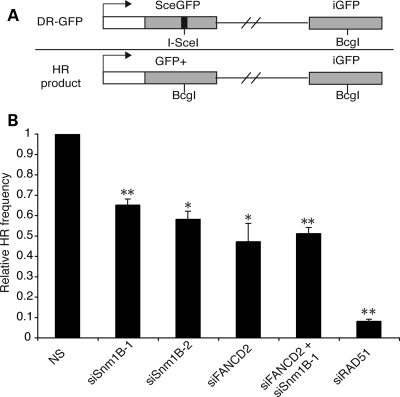
Proteins within the FA network, including FANCA, FANCG, FANCC, BRCA2/FANCD1, FANCN/PALB2, FANCJ/BACH1 and FANCD2, promote homologous recombination of chromosomal breaks (6,42,44,48,49). Our findings indicating that SNM1B functions to ensure efficient localization of critical homologous recombination proteins led us to examine the impact of SNM1B depletion on homology-directed repair. To this end, we used the characterized direct repeat green fluorescent protein (DR–GFP) reporter construct that measures levels of homologous recombination of a specific chromosomal DSB generated by the rare-cutting endonuclease, I-SceI (50) (Fig. 5A). Successful homology-directed repair of the DR–GFP substrate restores expression of a wild-type GFP cassette, and levels of homologous recombination can be assessed by flow cytometry.
Figure 5.
SNM1B promotes homology-mediated repair of DNA DSBs. (A) Schematic of DR–GFP construct containing a GFP cassette interrupted by an I-SceI recognition site. Upon I-SceI expression, a DSB is generated, and GFP expression is restored upon homologous recombinational repair using a downstream internal GFP fragment (iGFP). HeLa cells containing the chromosomally integrated DR–GFP reporter construct were transfected with the indicated siRNAs. At 24 h post-transfection, cells were infected with adenovirus expressing the I-SceI nuclease. Cells were collected after 48 h and the percentage of GFP-positive cells was determined by flow cytometry. (B) Relative HR frequencies. The relative HR frequencies of SNM1B, FANCD2, SNM1B/FANCD2 and RAD51-depleted cells were calculated based on the percentage of GFP-positive cells relative to that observed in control (NS) cells. Graph represents the average of three independent experiments. Bars are standard deviations. *P< 0.05; **P< 0.005, Student's two-tailed t-test.
We depleted either SNM1B or FANCD2 in a HeLa cell line harboring a chromosomally integrated DR–GFP construct by siRNA transfection. Knockdown of FANCD2 protein levels was verified by western blotting (Supplemental Material, Fig. S6A). At 24 h post-transfection, the cells were infected with an adenovirus expressing I-SceI to initiate homologous recombination via the generation of a specific DSB. To assess the efficiency of I-SceI transduction and site cleavage in SNM1B-depleted and control cells, we isolated genomic DNA after I-SceI transduction and PCR amplified a region flanking the I-SceI cleavage site (51). The PCR products were subsequently digested with the I-SceI enzyme, and the percentage of undigested product (i.e. I-SceI site loss) was determined (Supplemental Material, Fig. S6C–E). We observed a similar percentage of I-SceI site loss in SNM1B knockdown cells and controls (Supplemental Material, Fig. S6D and E). These findings provide evidence that comparable levels of I-SceI repairable breaks were generated in the SNM1B-depleted and control cell lines.
The levels of homology-directed DSB repair were determined by quantitating the percentage of GFP-positive cells at 48 h post-transduction with the I-SceI adenovirus (Fig. 5B, Supplementary Material, Fig. S6F). FANCD2 depletion reduced the frequency of homologous recombination by ∼50%; however, the defect was less severe compared with RAD51-depleted cells, as previously reported (6,13,42,49). We observed that SNM1B depletion by siSNM1B-1 and -2 also impaired homologous repair of chromosomal DSBs, and the decrease in recombination frequency was similar to that as observed in FANCD2 knockdown cells (Fig. 5B). To determine whether SNM1B and FANCD2 function epistatically in homology-directed DSB repair, we depleted both SNM1B and FANCD2 by siRNA transfection in the DR–GFP HeLa cell line. We observed that co-depletion of SNM1B and FANCD2 did not have an additive impact on the frequency of homologous recombination (Fig. 5B). These results indicate that SNM1B promotes homologous repair of chromosomal DSBs and has functions that are epistatic with the central FA protein, FANCD2.
DISCUSSION
In this study, we investigated the genetic interactions between SNM1B and the FA proteins, FANCD2 and FANCI, in the repair of highly cytotoxic DNA ICLs. The FA pathway plays integral roles in the cellular responses to and resolution of ICLs during replication. SNM1B is a DNA nuclease that is involved in the repair of ICL damage and also has important functions in cellular responses to IR-induced DSBs as well as telomere maintenance (22–28,52). Previous studies provided indirect evidence that SNM1B may act within the FA pathway; however, this question remained unanswered. In the current study, we demonstrate that SNM1B and FANCD2 function epistatically in response to MMC-induced ICL damage. We establish that SNM1B is required for efficient assembly of FANCD2 into subnuclear foci, and deficiency of both SNM1B and FANCD2 does not have an additive impact on cellular hypersensitivity to ICL damage or homologous recombinational repair of a chromosomal DSB. Furthermore, we determined that the hallmark phenotype of increased MMC-induced chromosomal anomalies observed in FANCD2 and FANCI deficiencies is not further elevated upon depletion of SNM1B. Thus, our data indicate that SNM1B has important roles during ICL repair in the context of the FA pathway.
ICLs are covalent linkages between the two strands of the DNA helix, thereby preventing strand separation. Collision of the replication machinery with an ICL leads to fork stalling and activation of the intra-S phase checkpoint (53–55). Removal of the ICL-blocking lesion requires the activities of several DNA nucleases to incise or unhook the lesion (56,57). These nucleolytic processing steps generate DSB intermediates which can be repaired via homologous recombination to restore replication (5). The MUS81-EME1 endonuclease is thought to cleave the blocked replication fork 3′ of the ICL and facilitate DSB formation, and the XPF-ERCC1 endonuclease makes a second incision to unhook the ICL lesion (24). Additional nucleases, including the SLX4 complex, MRE11-RAD50-NBS1 (MRN), SNM1A, FAN1 and FANCD2 itself have been demonstrated to function during ICL repair (10–13,55,58–63). SNM1B possesses 5′ to 3′ double-strand exonuclease activity and, thus, likely participates in nucleolytic processing of ICL repair intermediates (24,27).
In the current study, we establish that SNM1B is not required for chromatin localization of monoubiquitinated FANCD2; however, it is necessary for efficient FANCD2 foci formation in response to ICLs. Deficiency for the ubiquitin protease, USP1, leads to a similar impairment in assembly of chromatin-associated FANCD2-Ub into nuclear foci (37). It has been suggested that chromatin localization of FANCD2-Ub promotes assembly of DNA repair complexes, and USP1-mediated deubiquitination of FANCD2-Ub induces nuclear foci formation (37). The efficient localization of key repair proteins to sites of ICL damage then promotes subsequent repair events, such as homologous recombination and/or translesion synthesis (37). Based on our findings, we hypothesize that SNM1B, like USP1, has functions during an early stage of ICL repair to facilitate the stable assembly of chromatin-associated FANCD2-Ub into nuclear foci and recruitment of repair proteins.
SNM1B directly interacts with the MUS81-EME1 and MRN nucleases and is involved in replication fork collapse and formation of DSB intermediates during ICL repair (24). Interestingly, MRE11 forms a complex with FANCD2 (62), and SNM1B co-immunoprecipitates with FANCD2 via an indirect interaction (24). Thus, SNM1B likely functions in coordination with other critical DNA nucleases at an early step during ICL processing. Although the precise roles of the ICL DNA nucleases have not yet been fully elucidated, the functions of SNM1B within the FA pathway are likely independent of repair requiring SNM1A, a related member of the metallo-β-lactamase/βCASP family. In contrast to our findings, SNM1A is non-epistatic to the FA pathway. Depletion of SNM1A in FA pathway-deficient cells further increases in ICL-induced chromosomal anomalies (17), and MMC-inducible SNM1A foci formation occurs normally in FA pathway-deficient cells (18).
Our findings that FANCD2-Ub associates with chromatin but does not assemble into foci in SNM1B-depleted cells indicate that SNM1B has functions subsequent to ICL unhooking by XPF-ERCC1, which is required for FANCD2-Ub chromatin association (35), and prior to recruitment of FANCD2 and FAN1 to sites of damage. We propose that the 5′ to 3′ exonuclease activity intrinsic to SNM1B participates in generating single-strand regions at collapsed replication forks after XPF-ERCC1 unhooks the ICL lesion. A similar role for SNM1B exonucleolytic activity in 5′ to 3′ end resection during leading strand processing at telomeres has recently been proposed (22,23). Subsequent to DNA processing mediated by SNM1B, recruitment of FANCD2, which preferentially binds single-strand DNA (62), and other critical repair proteins to sites of ICL damage, such as FAN1, BRCA1 and RAD51, then participate in homologous recombinational repair of the DSB intermediates.
We conclude that SNM1B plays key roles in the context of the FA protein network of ICL repair. SNM1B has recently been identified as a human disease allele that causes Hoyeraal–Hreidarsson syndrome, thereby raising the possibility that SNM1B mutations may also be associated with phenotypes observed in FA. In further support of this notion, deletion of the SNM1B chromosomal locus has been found in nearly half of primary mediastinal B cell lymphomas analyzed in a recent study (64). Cancer predisposition, including hematological malignancies, is a hallmark phenotype associated with FA (65). The FA pathway plays critical roles in maintaining genome stability not only in response to ICL damage, but also during DNA synthesis to resolve replication stress. It has been hypothesized that the lack of proper repair of endogenous DNA damage in FA cells significantly contributes to genome instability and patient phenotypes, including bone marrow failure and cancer susceptibility (66). Therefore, it will be of interest to determine the importance of SNM1B in preventing DNA replication-associated endogenous damage, such as chromosomal gaps and breaks at common fragile sites and throughout the genome.
MATERIALS AND METHODS
Silencing RNAs
The target sequences for siRNA depletion are as follows:
siSNM1B-1: 5′-CCTCTTGCATCGTCACCTACATT-3′;
siSNM1B-2: 5′-AGTGCTGATCAATCTCAAGTT-3′ (MWG, Qiagen);
siFANCD2: 5′-CAGAGTTTGCTTCACTCTCTA-3′ (67);
siRAD51: 5′-AAGCTGAAGCGAGTTCGCCA-3′ (67);
siFANCI: 5′-CTGGCTAATCACCAAGCTTAA-3′ (6).
The primer sequence used to generate the siSNM1 B-1-resistant SNM1B cDNA was 5′-CCTCTTACACACCGCCACCTACA-3′ (base changes within the core sequence are underlined, in bold). A NS siRNA (Qiagen All Stars negative control) was used as the NS siRNA control.
Knockdown of SNM1B expression by siRNA
The HCT116 colon cancer, HeLa cervical cancer and FANCD2 mutant (PD20F) and control (PD20F + wt FANCD2) fibroblast human cell lines were cultured as previously described (31,68). HCT116 or HeLa cells were plated at a density of 1 × 105 cells per well of 6-well dish 24 h prior to siRNA transfection. All siRNAs (25–50 nm) were transfected using Liptofectamine 2000 (Invitrogen) as per manufacturer's instructions. Co-depeletion of SNM1B and FANCD2 was performed by transfecting HeLa cells with a total of 12.5 nm siRNA followed by a transfection with a total of 25 nm siRNA 24 h after the initial transfection. PD20F and control fibroblasts were transfected with 150 nm siRNA as previously described (69). SNM1B mRNA levels were determined via RT–PCR in every experiment to verify the extent of siRNA knockdown.
Semi-quantitative RT–PCR analysis of SNM1B mRNA levels
Total RNA (1 µg) was reverse transcribed using a poly-dT(20) primer and murine leukemia virus-reverse transcriptase (Invitrogen). cDNA was amplified with SNM1B-specific primers to exons 3 and 4, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) for normalization of cDNA levels. Bands were quantitated using AlphaImager 2200 (AlphaInnotech).
Western blot analyses
Cells (1–2 × 106) were harvested and resuspended in lysis buffer (25 mm 4-(2-hydroxyethyl)-1-piperazine-ethanesulfonic acid (HEPES), pH 7.4, 10% glycerol, 200 mm KCl, 0.1% NP40, 1 mm DTT) containing phosphatase (Roche PhosSTOP) and protease inhibitors (Roche Complete Mini EDTA free). Soluble and chromatin-bound FANCD2 fractions were separated as previously described (70). Protein concentration was determined using the Bradford assay (BioRad). Lysates (50–100 µg) were analyzed by western blotting with the appropriate primary antibodies and IRdye 800 CW secondary antibodies (Li-Cor). Bands were visualized and quantitated using the Li-Cor scanner and Odyssey 2.1 software. All experiments were performed three times from at least two independent transfections.
Antibodies
α-GAPDH (clone #FL-335) and α-RAD51 (clone #S1RAD01) were from Santa Cruz. α-BRCA1 (clone #Ab-4) was from CalBiochem. α-γ-H2AX (Cat #05-636) was from Upstate. α-FANCD2 (Cat #NB100-182) was from Novus Biologicals. α-V5 was from Invitrogen. α-FANCI (Cat # ab15344) was from AbCam.
MMC survival assay
Human fibroblasts [PD20F or PD20F + FANCD2 cDNA (WT)] were transfected with siRNAs as described above. At 48 h post-transfection, cells were plated in a 12-well dish at a low density (500–2000 cells/well) and allowed to attach overnight. To ensure that an additive effect could be observed, cells were pulsed with 0, 50, 100, 150 and 330 ng/ml MMC for 1h. MMC was removed and cells were rinsed with media three to four times. Cells were allowed to grow until the untreated well-reached confluency (7–9 days). Cellular survival upon treatment with MMC was determined using a crystal violet assay, as previously described (33). Average percent survival was obtained from at least three independent experiments.
Immunofluorescence of subnuclear foci
HeLa cells (4 × 104) were seeded on cover slips in 12-well dishes 24 h prior to transfection. At 48 h post-transfection, cells were continuously exposed to MMC (330 ng/ml) for 8 h, then incubated in cold pre-extraction buffer (20 mm HEPES, 50 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 0.5% TX-100) for 5 min followed by fixation in 3.7% p-formaldehyde, 2% sucrose, 0.5% TX100 for 20 min at room temperature and washed 3× with PBS. Cells were incubated with the appropriate primary antibody for 45 min and then stained with Alexa Fluor 488 or 594 secondary antibodies (Invitrogen Molecular Probes). Cover slips were mounted on slides using ProLong Gold antifade reagent containing 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Foci were visualized using an Olympus BX61 microscope and FISHview software (Applied Spectral Imaging). All foci experiments were performed blinded. Each experiment was performed at least three independent times.
Generation of HeLa cell lines expressing siRNA-1-resistant SNM1B
siSNM1B-1-resistant (si-1R) SNM1B cDNA was generated using site-directed mutagenesis (Stratagene). Three silent-point mutations were introduced into the siRNA-1 core sequence of SNM1B cDNA. The siRNA-resistant cDNA was sequenced, and then subcloned into pLL IRES GFP lentiviral vector (UM vector core). Lentivirus expressing the si-R1 SNM1B-IRES-GFP cassette was generated by co-transfecting the recombinant lentiviral construct with packaging plasmids pLP1, pLP2 and pVSVG (ViraPower, Invitrogen) into 293T cells using calcium phosphate. The virus containing media was collected after 48 h and filtered using Millex-HV PVDF filters (Millipore). HeLa cells (2.0 × 105) were incubated with 1 ml of virus containing media containing 4 µg/ml polybrene, 1 ml Dulbecco's modified eagle medium and 10% fetal bovine serum for 24 h. Cells were harvested 24 h later, and expression of the SNM1B-IRES-GFP expression cassette was determined by flow cytometry to determine the percentage of GFP-positive cells.
GFP-positive cells were sorted (UM flow cytometry core), cultured and used for complementation experiments. HeLa cells infected with the pLL empty vector control were used for all experiments. For complementation experiments, HeLa cells expressing pLL IRES GFP or si-1R SNM1B-IRES-GFP were transfected with the siRNAs and FANCD2 foci were quantitated as described. Images were acquired at ×100 magnification at the same fluorescence intensity, and the study was performed blinded. Data represent three independent experiments with standard deviation; P-values; Student's t-test.
Chromosomal aberrations
HeLa cells (1.0 × 105) were plated in a 6-well dish, and 24 h later, the cells were transfected with 12.5 nm total siRNA using Liptofectamine 2000 then incubated overnight. The media were changed and the cells were transfected with 25 nm total siRNA and incubated overnight. Forty-eight hours after the second transfection, cells were harvested for RNA isolation or treated with MMC (20 ng/ml) for 24 h. Cells were rinsed and then incubated with colcemid (0.1 µg/ml for 3 h), and subsequently harvested and fixed. Metaphases were stained with DAPI, and then chromosomal anomalies were scored in a blinded manner. The indicated number of metaphases was scored for the presence of radials and/or gaps and breaks from at least two independent experiments. Error bars represent standard error. Statistical significance was determined using a two-tailed F-test.
Analysis of homologous recombination frequencies
HeLa cells were transfected with the DR–GFP Puro homologous recombination substrate plasmid (generous gift from Maria Jasin, Sloan-Kettering Institute) (35), and single colonies were expanded after puromycin selection. HeLa cells harboring the stably integrated DR–GFP plasmid were transfected with siSNM1B-1 or -2, NS, siFANCD2 and siRAD51 siRNAs, as above. At 24 h post-transfection, cells were infected with either an adenovirus expressing the I-SceI endonuclease or a negative control adenovirus at 100–200 PFU/cell. Cells were trypsinized at 48 h post-infection, and GFP-positive cells were determined by flow cytometry using a C6 cytometer (Accuri) and CFlow software. Averages were obtained from at least three independent experiments.
I-SceI site-loss assay
Genomic DNA was isolated from siRNA-transfected and control cells that have been transduced with the I-SceI adenovirus to induce repair events within the integrated DR–GFP substrate. The region flanking the I-SceI site was PCR amplified from 2 µg of genomic DNA from each sample (51). The PCR products were digested with purified I-SceI enzyme (Roche) overnight at 37°C. Products were run on a 1.2% agarose gel, and the fluorescence intensity of the bands was quantitated using Alphaimager 2200 (AlphaInnotech). The percentage of I-SceI site loss was determined as previously described (51). Products amplified from HeLa cells infected with an empty adenovirus were used as positive controls for complete I-SceI digestion. Experiments were performed on DNA isolated from two independent experiments.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by Public Health Service grant AI063058 from the National Institutes of Health (NIAID) and Pew Scholar's Award (Pew Foundation) to J.M.S. and T32-GM-07544 (NIGMS) to J.M.M.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs David Ferguson, Christine Canman and John Moran for helpful discussions and Stephanie Kraftson, Laurel Chadde, Neil Patel and Anandi Karumbati for technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Moldovan G.L., D'Andrea A.D. How the fanconi anemia pathway guards the genome. Annu. Rev. Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim Y., Lach F.P., Desetty R., Hanenberg H., Auerbach A.D., Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat. Genet. 2011;43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stoepker C., Hain K., Schuster B., Hilhorst-Hofstee Y., Rooimans M.A., Steltenpool J., Oostra A.B., Eirich K., Korthof E.T., Nieuwint A.W., et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat. Genet. 2011;43:138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 4.Crossan G.P., van der Weyden L., Rosado I.V., Langevin F., Gaillard P.H., McIntyre R.E., Gallagher F., Kettunen M.I., Lewis D.Y., Brindle K., et al. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat. Genet. 2011;43:147–152. doi: 10.1038/ng.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson L.H., Hinz J.M. Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: mechanistic insights. Mutat. Res. 2009;668:54–72. doi: 10.1016/j.mrfmmm.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E.R., 3rd, Hurov K.E., Luo J., Ballif B.A., Gygi S.P., Hofmann K., D'Andrea A.D., et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussain S., Wilson J.B., Medhurst A.L., Hejna J., Witt E., Ananth S., Davies A., Masson J.Y., Moses R., West S.C., et al. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum. Mol. Genet. 2004;13:1241–1248. doi: 10.1093/hmg/ddh135. [DOI] [PubMed] [Google Scholar]
- 8.Taniguchi T., D'Andrea A.D. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107:4223–4233. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- 9.Wang X., Andreassen P.R., D'Andrea A.D. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell Biol. 2004;24:5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu T., Ghosal G., Yuan J., Chen J., Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 11.Smogorzewska A., Desetty R., Saito T.T., Schlabach M., Lach F.P., Sowa M.E., Clark A.B., Kunkel T.A., Harper J.W., Colaiacovo M.P., et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol. Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kratz K., Schopf B., Kaden S., Sendoel A., Eberhard R., Lademann C., Cannavo E., Sartori A.A., Hengartner M.O., Jiricny J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 13.MacKay C., Declais A.C., Lundin C., Agostinho A., Deans A.J., MacArtney T.J., Hofmann K., Gartner A., West S.C., Helleday T., et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X., Moses R.E. The beta-lactamase motif in Snm1 is required for repair of DNA double-strand breaks caused by interstrand crosslinks in S. cerevisiae. DNA Repair (Amst.) 2003;2:121–129. doi: 10.1016/s1568-7864(02)00192-1. [DOI] [PubMed] [Google Scholar]
- 15.Callebaut I., Moshous D., Mornon J.-P., de Villartay J.-P. Metallo-β-lactamase fold within nucleic acids processing enzymes: the β-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dronkert M.L., de Wit J., Boeve M., Vasconcelos M.L., van Steeg H., Tan T.L., Hoeijmakers J.H., Kanaar R. Disruption of mouse SNM1 causes increased sensitivity to the DNA interstrand cross-linking agent mitomycin C. Mol. Cell Biol. 2000;20:4553–4561. doi: 10.1128/mcb.20.13.4553-4561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemphill A.W., Bruun D., Thrun L., Akkari Y., Torimaru Y., Hejna K., Jakobs P.M., Hejna J., Jones S., Olson S.B., et al. Mammalian SNM1 is required for genome stability. Mol. Genet. Metab. 2008;94:38–45. doi: 10.1016/j.ymgme.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang K., Moldovan G.L., D'Andrea A.D. RAD18-dependent recruitment of SNM1A to DNA repair complexes by a ubiquitin-binding zinc finger. J. Biol. Chem. 2010;285:19085–19091. doi: 10.1074/jbc.M109.100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y., Pannicke U., Schwarz K., Lieber M.R. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 20.Moshous D., Callebaut I., de Chasseval R., Corneo B., Cavazzana-Calvo M., Le Deist F., Tezcan I., Sanal O., Bertrand Y., Philippe N., et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 21.Rooney S., Alt F.W., Lombard D., Whitlow S., Eckersdorff M., Fleming J., Fugmann S., Ferguson D.O., Schatz D.G., Sekiguchi J. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J. Exp. Med. 2003;197:553–565. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam Y.C., Akhter S., Gu P., Ye J., Poulet A., Giraud-Panis M.J., Bailey S.M., Gilson E., Legerski R.J., Chang S. SNM1B/Apollo protects leading-strand telomeres against NHEJ-mediated repair. EMBO J. 2010;29:2230–2241. doi: 10.1038/emboj.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu P., van Overbeek M., Rooney S., de Lange T. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol. Cell. 2010;39:606–617. doi: 10.1016/j.molcel.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae J.B., Mukhopadhyay S.S., Liu L., Zhang N., Tan J., Akhter S., Liu X., Shen X., Li L., Legerski R.J. Snm1B/Apollo mediates replication fork collapse and S Phase checkpoint activation in response to DNA interstrand cross-links. Oncogene. 2008;27:5045–5056. doi: 10.1038/onc.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demuth I., Bradshaw P.S., Lindner A., Anders M., Heinrich S., Kallenbach J., Schmelz K., Digweed M., Meyn M.S., Concannon P. Endogenous hSNM1B/Apollo interacts with TRF2 and stimulates ATM in response to ionizing radiation. DNA Repair (Amst.) 2008;7:1192–1201. doi: 10.1016/j.dnarep.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demuth I., Digweed M., Concannon P. Human SNM1B is required for normal cellular response to both DNA interstrand crosslink-inducing agents and ionizing radiation. Oncogene. 2004;23:8611–8618. doi: 10.1038/sj.onc.1207895. [DOI] [PubMed] [Google Scholar]
- 27.Lenain C., Bauwens S., Amiard S., Brunori M., Giraud-Panis M.J., Gilson E. The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr. Biol. 2006;16:1303–1310. doi: 10.1016/j.cub.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 28.Ye J., Lenain C., Bauwens S., Rizzo A., Saint-Leger A., Poulet A., Benarroch D., Magdinier F., Morere J., Amiard S., et al. TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell. 2010;142:230–242. doi: 10.1016/j.cell.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 29.Akhter S., Lam Y.C., Chang S., Legerski R.J. The telomeric protein SNM1B/Apollo is required for normal cell proliferation and embryonic development. Aging Cell. 2010;9:1047–1056. doi: 10.1111/j.1474-9726.2010.00631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Touzot F., Callebaut I., Soulier J., Gaillard L., Azerrad C., Durandy A., Fischer A., de Villartay J.P., Revy P. Function of Apollo (SNM1B) at telomere highlighted by a splice variant identified in a patient with Hoyeraal-Hreidarsson syndrome. Proc. Natl Acad. Sci. USA. 2010;107:10097–10102. doi: 10.1073/pnas.0914918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timmers C., Taniguchi T., Hejna J., Reifsteck C., Lucas L., Bruun D., Thayer M., Cox B., Olson S., D'Andrea A.D., et al. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol. Cell. 2001;7:241–248. doi: 10.1016/s1097-2765(01)00172-1. [DOI] [PubMed] [Google Scholar]
- 32.Godthelp B.C., van Buul P.P., Jaspers N.G., Elghalbzouri-Maghrani E., van Duijn-Goedhart A., Arwert F., Joenje H., Zdzienicka M.Z. Cellular characterization of cells from the Fanconi anemia complementation group, FA-D1/BRCA2. Mutat. Res. 2006;601:191–201. doi: 10.1016/j.mrfmmm.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi T., Garcia-Higuera I., Xu B., Andreassen P.R., Gregory R.C., Kim S.T., Lane W.S., Kastan M.B., D'Andrea A.D. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- 34.Ho G.P., Margossian S., Taniguchi T., D'Andrea A.D. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol. Cell Biol. 2006;26:7005–7015. doi: 10.1128/MCB.02018-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhagwat N., Olsen A.L., Wang A.T., Hanada K., Stuckert P., Kanaar R., D'Andrea A., Niedernhofer L.J., McHugh P.J. XPF-ERCC1 participates in the Fanconi anemia pathway of cross-link repair. Mol. Cell Biol. 2009;29:6427–6437. doi: 10.1128/MCB.00086-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M.S., Timmers C., Hejna J., Grompe M., D'Andrea A.D. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 37.Kim J.M., Parmar K., Huang M., Weinstock D.M., Ruit C.A., Kutok J.L., D'Andrea A.D. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev. Cell. 2009;16:314–320. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Auerbach A.D., Wolman S.R. Susceptibility of Fanconi's anaemia fibroblasts to chromosome damage by carcinogens. Nature. 1976;261:494–496. doi: 10.1038/261494a0. [DOI] [PubMed] [Google Scholar]
- 39.Yuan F., El Hokayem J., Zhou W., Zhang Y. FANCI protein binds to DNA and interacts with FANCD2 to recognize branched structures. J. Biol. Chem. 2009;284:24443–24452. doi: 10.1074/jbc.M109.016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sims A.E., Spiteri E., Sims R.J., 3rd, Arita A.G., Lach F.P., Landers T., Wurm M., Freund M., Neveling K., Hanenberg H., et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 41.Gudmundsdottir K., Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- 42.Nakanishi K., Yang Y.G., Pierce A.J., Taniguchi T., Digweed M., D'Andrea A.D., Wang Z.Q., Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl Acad. Sci. USA. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taniguchi T., Garcia-Higuera I., Andreassen P.R., Gregory R.C., Grompe M., D'Andrea A.D. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 44.Niedzwiedz W., Mosedale G., Johnson M., Ong C.Y., Pace P., Patel K.J. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 45.Vandenberg C.J., Gergely F., Ong C.Y., Pace P., Mallery D.L., Hiom K., Patel K.J. BRCA1-independent ubiquitination of FANCD2. Mol. Cell. 2003;12:247–254. doi: 10.1016/s1097-2765(03)00281-8. [DOI] [PubMed] [Google Scholar]
- 46.Bhattacharyya A., Ear U.S., Koller B.H., Weichselbaum R.R., Bishop D.K. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 2000;275:23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 47.Huber L.J., Yang T.W., Sarkisian C.J., Master S.R., Deng C.X., Chodosh L.A. Impaired DNA damage response in cells expressing an exon 11-deleted murine Brca1 variant that localizes to nuclear foci. Mol. Cell Biol. 2001;21:4005–4015. doi: 10.1128/MCB.21.12.4005-4015.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F.J., Livingston D.M. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 49.Moldovan G.L., Madhavan M.V., Mirchandani K.D., McCaffrey R.M., Vinciguerra P., D'Andrea A.D. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol. Cell Biol. 2010;30:1088–1096. doi: 10.1128/MCB.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pierce A.J., Jasin M. Measuring recombination proficiency in mouse embryonic stem cells. Methods Mol. Biol. 2005;291:373–384. doi: 10.1385/1-59259-840-4:373. [DOI] [PubMed] [Google Scholar]
- 51.Weinstock D.M., Nakanishi K., Helgadottir H.R., Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Overbeek M., de Lange T. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr. Biol. 2006;16:1295–1302. doi: 10.1016/j.cub.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 53.Mladenov E., Tsaneva I., Anachkova B. Activation of the S phase DNA damage checkpoint by mitomycin C. J. Cell Physiol. 2007;211:468–476. doi: 10.1002/jcp.20957. [DOI] [PubMed] [Google Scholar]
- 54.Costanzo V., Shechter D., Lupardus P.J., Cimprich K.A., Gottesman M., Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell. 2003;11:203–213. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 55.Pichierri P., Rosselli F. The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. EMBO J. 2004;23:1178–1187. doi: 10.1038/sj.emboj.7600113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hanada K., Budzowska M., Modesti M., Maas A., Wyman C., Essers J., Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuraoka I., Kobertz W.R., Ariza R.R., Biggerstaff M., Essigmann J.M., Wood R.D. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J. Biol. Chem. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- 58.Pichierri P., Averbeck D., Rosselli F. DNA cross-link-dependent RAD50/MRE11/NBS1 subnuclear assembly requires the Fanconi anemia C protein. Hum. Mol. Genet. 2002;11:2531–2546. doi: 10.1093/hmg/11.21.2531. [DOI] [PubMed] [Google Scholar]
- 59.Fekairi S., Scaglione S., Chahwan C., Taylor E.R., Tissier A., Coulon S., Dong M.Q., Ruse C., Yates J.R., 3rd, Russell P., et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Svendsen J.M., Smogorzewska A., Sowa M.E., O'Connell B.C., Gygi S.P., Elledge S.J., Harper J.W. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Munoz I.M., Hain K., Declais A.C., Gardiner M., Toh G.W., Sanchez-Pulido L., Heuckmann J.M., Toth R., Macartney T., Eppink B., et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 62.Roques C., Coulombe Y., Delannoy M., Vignard J., Grossi S., Brodeur I., Rodrigue A., Gautier J., Stasiak A.Z., Stasiak A., et al. MRE11-RAD50-NBS1 is a critical regulator of FANCD2 stability and function during DNA double-strand break repair. EMBO J. 2009;28:2400–2413. doi: 10.1038/emboj.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Knipscheer P., Raschle M., Smogorzewska A., Enoiu M., Ho T.V., Scharer O.D., Elledge S.J., Walter J.C. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kimm L.R., deLeeuw R.J., Savage K.J., Rosenwald A., Campo E., Delabie J., Ott G., Muller-Hermelink H.K., Jaffe E.S., Rimsza L.M., et al. Frequent occurrence of deletions in primary mediastinal B-cell lymphoma. Genes Chromosomes Cancer. 2007;46:1090–1097. doi: 10.1002/gcc.20495. [DOI] [PubMed] [Google Scholar]
- 65.Mathew C.G. Fanconi anaemia genes and susceptibility to cancer. Oncogene. 2006;25:5875–5884. doi: 10.1038/sj.onc.1209878. [DOI] [PubMed] [Google Scholar]
- 66.Patel K.J., Joenje H. Fanconi anemia and DNA replication repair. DNA Repair (Amst.) 2007;6:885–890. doi: 10.1016/j.dnarep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 67.Hicks J.K., Chute C.L., Paulsen M.T., Ragland R.L., Howlett N.G., Gueranger Q., Glover T.W., Canman C.E. Differential roles for DNA polymerases eta, zeta, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol. Cell Biol. 2009;30:1217–1230. doi: 10.1128/MCB.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 69.Bruun D., Folias A., Akkari Y., Cox Y., Olson S., Moses R. siRNA depletion of BRCA1, but not BRCA2, causes increased genome instability in Fanconi anemia cells. DNA Repair (Amst.) 2003;2:1007–1013. doi: 10.1016/s1568-7864(03)00112-5. [DOI] [PubMed] [Google Scholar]
- 70.Kim J.M., Kee Y., Gurtan A., D'Andrea A.D. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood. 2008;111:5215–5222. doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.