Abstract
A 30-fold redundant human bacterial artificial chromosome (BAC) library with a large average insert size (178 kb) has been constructed to provide the intermediate substrate for the international genome sequencing effort. The DNA was obtained from a single anonymous volunteer, whose identity was protected through a double-blind donor selection protocol. DNA fragments were generated by partial digestion with EcoRI (library segments 1–4: 24-fold) and MboI (segment 5: sixfold) and cloned into the pBACe3.6 and pTARBAC1 vectors, respectively. The quality of the library was assessed by extensive analysis of 169 clones for rearrangements and artifacts. Eighteen BACs (11%) revealed minor insert rearrangements, and none was chimeric. This BAC library, designated as “RPCI-11,” has been used widely as the central resource for insert-end sequencing, clone fingerprinting, high-throughput sequence analysis and as a source of mapped clones for diagnostic and functional studies.
The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AQ936150–AQ936491.]
The main goal of the publicly funded human genome project is to completely determine the human genomic DNA sequence. Five large centers in the United States and the United Kingdom (the G5 group) along with three smaller centers in France, Germany, and Japan (the G8 group) are the major contributors to the sequencing effort. The initial draft version of the human DNA sequence was completed on June 26, 2000, and a high-quality version will become accessible by 2003. The human genome project presents unique ethical and political requirements with respect to the source DNA for library construction, because never before has an individual's genetic blueprint been deciphered completely. One or more volunteers were required to donate their DNA for the sequencing effort. Donor recruitment must comply with regulations (Botkin and Gut 1996; Marshall 1996) to protect the individual's interests and requires informed consent. In addition, it is preferable to obtain the first human genome sequence with the focus on the composition of genes across the prototypical human genome rather than exploring the diversity of genes across the human population. With only a few donors contributing to the prototype of the human genome, it is likely that the prototype will not be equally derived from all ethnic or social groups. To avoid a willful bias with respect to representatives from one group or another, a double-blind donor selection protocol was desirable and was formulated in compliance with the stated policies of the funding agencies (see http://www.nhgri.nih.gov:80/Grant_info/Funding/Statements/RFA/human_subjects.html).
Large-insert genomic DNA libraries in bacteria, such as bacterial artificial chromosome (BAC; Shizuya et al. 1992) and P1-derived artificial chromosome (PAC; Ioannou et al. 1994) libraries, provide a way to divide the complexity of the human genome into a composite of large DNA segments of reduced complexity. Ideally, BAC libraries should completely represent the genome without cloning artifacts or rearrangements and should be provided in an addressable format with clones physically separated. Libraries arrayed in microtiter dishes provide the opportunity for many researchers around the world to accumulate and use information on particular clones (Green and Olson 1990; Nizetic et al. 1991; Evans et al. 1992; Cohen et al. 1993; Marra et al. 1997; Zhao et al. 2000), thus permitting resource sharing through central repositories. BAC libraries are used as a source of substrates for shotgun sequencing projects, to create a database of end sequences (Mahairas et al. 1999; Zhao 2000; Zhao et al. 2000) and restriction fingerprints for building overlapping clone sets (contigs; Marra et al. 1997, 1999). BACs also provide scaffolding information for mapping sequence contigs to localized genomic regions by using a direct genomic shotgun sequencing approach (Adams et al. 2000; Hoskins et al. 2000). The BAC library (RPCI-11) described in this manuscript represents one of the first libraries constructed in compliance with the policies of the U.S. funding agencies for DNA sequence resources and has been used to a larger extent than any other library. A detailed characterization of the RPCI-11 BAC library has not been reported, although its use in clone end sequencing, fingerprinting, and complete chromosome sequencing has been described (Dunham et al. 1999; Mahairas et al. 1999; Hattori et al. 2000; Soderlund et al. 2000; Zhao et al. 2000; McPherson et al. 2001).
To definitively determine that clones contain a single cloned fragment and thus are nonchimeric, both ends of the BAC inserts were independently mapped by screening against other clones within a very redundant clone contig spanning 1.5 Mb of human chromosome 14. The rationale is that if the BAC is chimeric, the ends will be from different genomic regions. To examine the level of rearrangements within clones, the contig was interrogated with ∼300 markers for inconsistent map results. In addition, all the BACs in the contig also were fingerprinted using high-resolution restriction fragment pattern analysis. This article establishes that chimeric clones containing multiple unrelated genomic DNA segments are essentially absent (at or below the 1% level) and that minor rearrangements can occur in ∼10% of the clones under normal growth conditions.
RESULTS
Donor Selection
Several years ago, the National Center for Human Genome Research (NCHGR, now the National Human Genome Research Institute, NHGRI) and the U.S. Department of Energy (DOE) formulated their policy on the inclusion of human volunteers for the Human Genome Project (HGP; Marshall 1996; http://www.nhgri.nih.gov:80/Grant_info/Funding/Statements/RFA/human_subjects.html). A central element of the policy was the need to protect the private information of the participants through a double-blind procedure such that the donors remain anonymous and their privacy will be protected to the maximum extent. Although it is possible to prepare BAC libraries from a mixture of DNA samples derived from many volunteers, this was not perceived desirable at the time. ixing many samples would lead to problems resolving the DNA sequence for difficult genomic regions. However, DNA sequencing of a single volunteer might be unethical and might prove to be politically unwise. Use of a single volunteer might raise the interest and curiosity of the public and the press to discover the identity of the donor and thus complicate efforts to protect the person's identity and privacy. Moreover, a procedure using a limited number of donors would have raised questions about perceived preferences with respect to the ethnicity, social group, or gender of the single donor. In view of these considerations, the policy was formulated to sequence the human genome from a composite of ∼10 BAC clone resources each contributing ∼10% of the donor's DNA to the final genome sequence at the completion of the HGP. The RPCI-11 BAC library was the first large insert clone collection to be prepared under the NCHGR/DOE policy.
Briefly, donors were recruited after a request for volunteers was advertised in a local newspaper, The Buffalo News, on Sunday, March 23, 1997. The first 10 male and 10 female volunteers replying by phone were invited to make an appointment with the genetic counselors at the Roswell Park Cancer Institute (RPCI) to give informed consent and provide a 50-mL blood sample. The informed consent form did not include any identifying information except for the signature of the blood donor. The blood samples were simply identified by arbitrary numbers and by gender. From 20 samples, one male and one female sample were selected at random within the cloning laboratory. The samples were used for establishing EBV-transformed cells and for extracting high molecular weight DNA to be embedded in agarose. The genetic counselors were not involved in the final sample selection and were not made aware of the outcome. The consent forms have been placed in sealed envelopes and stored in a locked cabinet only accessible to the genetic counselors at RPCI. The male DNA sample was used to construct the first library (RPCI-11), and the female sample was used to prepare the second library (RPCI-13, not described). Unfortunately, the attempt to prepare EBV-transformed cells for the RPCI-11 donor failed. As a consequence of the double-blind donor selection procedure, it was impossible to obtain a second sample from the same male donor for a second attempt to establish transformed cells.
BAC Library Construction and Characteristics
The new human BAC library was constructed using optimized cloning procedures (Osoegawa et al. 1998).
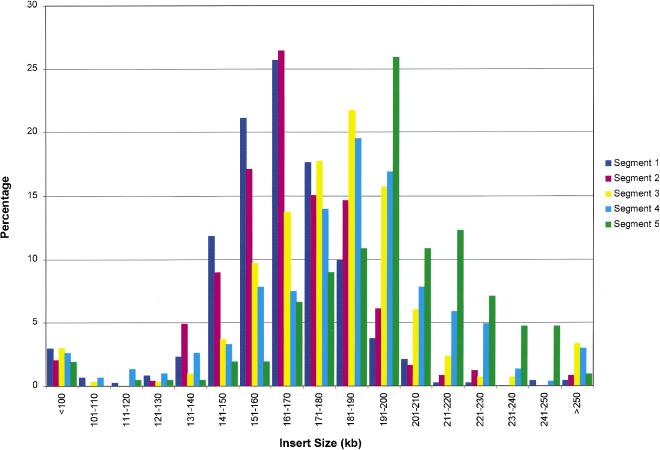
The goal was to generate a sufficient number of clones to provide at least 20-fold redundant representation of the human genome, thus ensuring nearly complete presence of all clonable sequences. The first set of ∼440,000 clones was prepared from human DNA fragmented by partial digestion using EcoRI and EcoRI methylase. A total of three partial digestions and size fractionations were performed, and five eluted DNA fractions were used for construction of the EcoRI library portion (Table 1A). Reliance on a combination of these enzymes might result in a biased genome representation because of the putative presence of preferential digestion or methylation sites or because of simple sequence regions with an aberrant incidence of the restriction sites. Therefore, an additional 106,000 BAC clones were prepared from the same donor DNA by partial digestion with MboI. Two DNA fractions derived from two partial digestions and size fractionations were used for construction of the MboI library part (Table 1A). All clones were arrayed into 384-well dishes by using a colony-picking robot, and the arrayed library was organized for logistical reasons into five segments, comprising a total of 1440 microtiter dishes, as summarized in Table 1B. The pBACe3.6 vector (Frengen et al. 1999) was used to construct the EcoRI library section (segments 1–4) and the pTARBAC1 vector (Zeng et al. 2001) was used for the MboI library (segment 5). The pTARBAC1 vector is derived from pBACe3.6 by insertion of the yeast centromeric element, CEN6, and the yeast-selectable marker His3. BAC clones in the pTARBAC1 vector can be deleted through digestion with restriction enzymes, which do not cut into the vector sequence. The linearized deleted BACs have a complete vector sequence on a single fragment and include the adjacent vector-insert junctions. Such deletion clones can be repaired to full size through homologous recombination with human genomic DNA during cotransformation into yeast spheroplasts, an approach that has been designated as transformation associated recombination (TAR) cloning (Larionov et al. 1996). High-density replica filters were prepared to screen the library as described previously (Osoegawa et al. 2000). One hundred forty-three nonrecombinant clones in the entire library containing the intact vector were identified and distinguished from insert containing clones by colony hybridization by using the vector as a probe. Nonrecombinant clones contain the high copy number pUC replicon, lacking in the recombinant clones, thus resulting in a much higher hybridization signal of vector sequences. While analyzing BAC clones by using pulsed-field gel electrophoresis to determine the average insert size, we observed noninsert clones containing a small deleted vector fragment consistent with sucrose resistance. The ratio of noninsert clones was estimated to be 1.7%, 0.5%, 1.0%, 1.0%, and 0.0% in each segment, respectively (Table 1). The average insert size for the BACs is 163, 168, 181, 183, and 195 kb with a standard deviation of 28.2, 25.3, 43.6, 39.8, and 30.5 for segments 1–5, respectively (Table 1, Fig. 1). The increase in average insert size for the later segments reflects improvements in cloning skills while generating the library. The distribution of insert sizes is shown in Figure 1. The entire library contains 29.4 human genome equivalents, assuming the size of the human genome size to be 3.3 gigabase.
Figure 1.
Size distribution of the RPCI-11 library. A total of 483, 246, 299, 308, and 212 clones from each of segments 1–5 was picked randomly and analyzed with a CHEF apparatus after NotI digestion. The horizontal axis refers to the size range of insert DNA, and the vertical axis indicates percentage of clones corresponding to each size range. Purple, pink, yellow, blue, and green bars correspond to segment 1, 2, 3, 4, and 5, respectively.
α-Satellite Clones
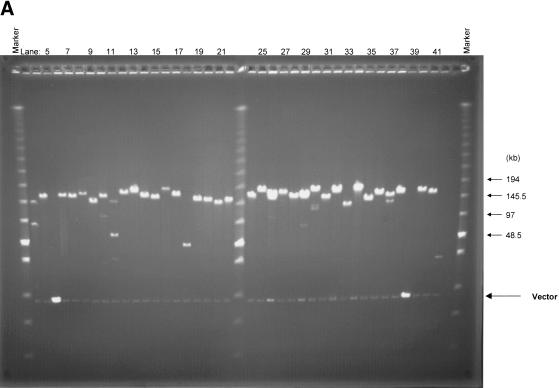
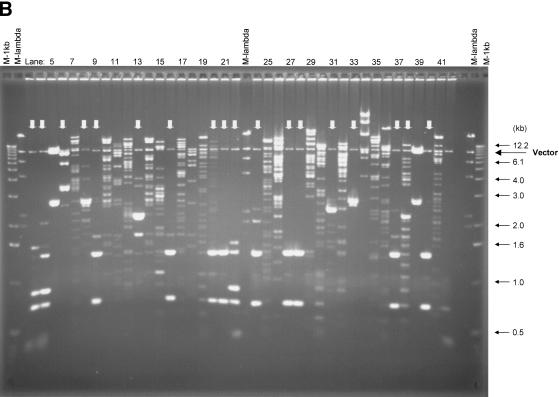
Approximately 10% of the human genome consists of large repeat blocks of α-satellite sequences centered around the centromeres (Tyler-Smith and Willard 1993). These repeat blocks include the 340-bp EcoRI dimeric satellite sequence and the 680-bp EcoRI tetrameric repeat (Baldini et al. 1989, 1990), whereas on average EcoRI sites occur at a much lower incidence in the rest of the genome at 3–4-kb intervals. Because EcoRI was used to fragment the human genome in preparation for the bulk of the BAC library, it is conceivable that nonoptimal fragmentation of the genome might have resulted in over- or underrepresentation of alphoid elements. To examine the presence of these tandem repeat elements, we screened the first 48 microtiter dishes (18,432 clones) of the library (segment 1) by colony hybridization using a probe containing alphoid repeats. Only 1.1% (209) of the BAC clones were positive, indicating that these sequences are underrepresented. Thirty-seven α-satellite-containing clones were analyzed by fingerprinting and pulsed-field gel electrophoresis after digestion with EcoRI or NotI (Fig. 2). Surprisingly, all but six of the BAC clones had a distinct unique insert size as revealed after NotI digestion and pulsed-field gel electrophoresis (Fig. 2A). The absence of a smear of multiple fragments and the absence of minor NotI bands indicate relative stability of the α-satellite repeat blocks in BACs. None of the BAC clones contained the 340-bp EcoRI repeat elements.
Figure 2.
Analysis of BAC clones containing α-satellite sequence. Thirty-seven α-satellite positive BAC clones were isolated and digested with NotI or EcoRI. (A) The NotI-digested DNA was separated using CHEF with the Low Range PFG Marker loaded on both sides, and the sizes of the markers are indicated with arrows. (B) The EcoRI-digested DNA was fractionated with standard agarose gel electrophoresis with the 1-kb DNA ladder (outside) and the λ DNA/HindIII fragments (inside) loaded on both sides. Vector bands that are indicated by arrows are observed at 8.8 kb in both panels. False-positive clones that contain intact vector are shown in lanes 5 and 39 in both panels. Stronger intensity of the vector bands is due to the high copy number of plasmid derived from pUC19-stuffer fragment. A variety of EcoRI repetitive blocks are observed in B and indicated with vertical arrows.
Eighteen clones contained the EcoRI repeat regions with periodicities of 680 bp and larger, observed as high-intensity bands after digestion with EcoRI (indicated with an arrow in Fig. 2B). The remaining 19 clones showed random EcoRI digestion patterns indicating the absence of large stretches of α-satellite sequence with an EcoRI site in these BACs. To analyze the stability of the cloned alphoid repeat blocks, we streaked all clones to single colonies, and 10 subclones for each of the original 37 clones were analyzed by fingerprinting. For 15 of the 37 clones, all of the 10 subclones revealed identical patterns by dual analysis of EcoRI fingerprinting and NotI restriction analysis (not shown). Rearrangements occurred in 10%–50% of the subcolonies for 22 of the 37 clones, shown either in the fingerprints or after pulsed-field analysis. These results indicate that the relative stability is high for the BAC clones containing highly repetitive elements in this library.
Marker Screening Results
The library, consisting of five segments, has 29.4 human genome equivalents as calculated from the average insert size and the total number of clones (Table 1). To examine the genomic representation for specific sequences, we screened the library via colony hybridization with 45 different probes. Overlapping oligonucleotide probes (“overgos”; McPherson 1999) were designed from an arbitrary set of chromosome 5, 19, and 21 markers. The hybridization positive clones were confirmed using restriction enzyme fingerprinting and Southern hybridization. The screening results are summarized in Table 2. A total of 1076 clones from the EcoRI library and 272 clones from the MboI library were identified using the 45 unique probes. These results indicate the average genome redundancy of the library to be 23.9 per marker with a standard deviation of 6.56 for EcoRI library section and 6.0 per marker with a standard deviation of 3.04 for MboI section. The combined genomic representation (29.9) from EcoRI and MboI library segments is in close agreement with the expected genome equivalents (29.4).
Table 2.
Screening the Libraries Using Various Single Locus Markers
| Chromosome 5 | Chromosome 19 | ||||||
|---|---|---|---|---|---|---|---|
| Markers | Locus | No. of positives | Markers | Locus | No. of positives | ||
| Segment 1–4 | Segment 5 | Segment 1–4 | Segment 5 | ||||
| D5S417 | 5.5 cM | 27 | 11 | D19S221 | 35.5 cM | 22 | 2 |
| D5S406 | 10.7 cM | 26 | 9 | D19S411 | 31 | 10 | |
| D5S635 | 13.8 cM | 26 | 7 | D19S425 | 58.7 cM | 21 | 9 |
| D5S676 | 15.6 cM | 28 | 7 | D19S220 | 61.4 cM | 18 | 2 |
| D5S1986 | 44.5 cM | 37 | 3 | D19S422 | 62.5 cM | 14 | 8 |
| D5S426 | 51.6 cM | 25 | 5 | D19S219 | 69.9 cM | 21 | 3 |
| D5S634 | 59.9 cM | 34 | 5 | D19S412 | 69.9 cM | 22 | 5 |
| D5S2076 | 62.5 cM | 21 | 5 | D19S596 | 77.6 cM | 16 | 6 |
| D5S407 | 65.0 cM | 24 | 13 | D19S214 | 106.1 cM | 21 | 8 |
| D5S491 | 67.2 cM | 25 | 8 | ||||
| D5S2028 | 69.0 cM | 31 | 5 | Chromosome 21 | |||
| D5S2029 | 92.5 cM | 25 | 10 | Markers | No. of positives | ||
| D5S456 | 109.3 cM | 24 | 2 | D21S1904 | 0.0 cM | 23 | 7 |
| D5S505 | 111.6 cM | 24 | 2 | D21S1911 | 0.1 cM | 28 | 3 |
| D5S2065 | 121.7 cM | 20 | 10 | D21S262 | 32.6 cM | 26 | 6 |
| D5S657 | 130.1 cM | 27 | 4 | D21S1252 | 38.7 cM | 13 | 2 |
| D5S2017 | 144.8 cM | 23 | 7 | D21S1893 | 48.1 cM | 27 | 10 |
| D5S2090 | 149.9 cM | 31 | 5 | D21S1260 | 51.6 cM | 22 | 3 |
| D5S673 | 155.5 cM | 16 | 2 | ||||
| D5S487 | 157.6 cM | 18 | 8 | Average number | |||
| D5S412 | 161.0 cM | 29 | 5 | 23.9 | 6.0 | ||
| D5S403 | 162.2 cM | 15 | 13 | ||||
| D5S2066 | 164.9 cM | 17 | 4 | Standard deviation | |||
| D5S2032 | 168.5 cM | 46 | 9 | 6.56 | 3.04 | ||
| D5S672 | 85.5 cM | 24 | 3 | ||||
| D5S413 | 150.0 cM | 15 | 2 | ||||
| D5S496 | 172.6 cM | 24 | 6 | ||||
| D5S2030 | 190.9 cM | 12 | 7 | ||||
| D5S1987 | 21.9 cM | 31 | 5 | ||||
| D5S1991 | 25.9 cM | 26 | 6 | ||||
A total of 45 markers (30 from chromosome 5, 9 from chromosome 19, and 6 from chromosome 21) have been used to screen the RPCI-11 library. The average genome representation of EcoRI and MboI segments was determined to be 23.9 and 6.0 based on the screening results. Standard deviation from the average number of positive clones for EcoRI and MboI segments were estimated to be 6.56 and 3.04, respectively. Overall genome redundancy has been estimated at 23.1-fold for EcoRI library part and 6.3-fold for MboI based on the average insert size and the number of clones. The locus column indicates the human chromosome location from the top of each chromosome linkage group.
A High-Resolution BAC/STS-Content Map
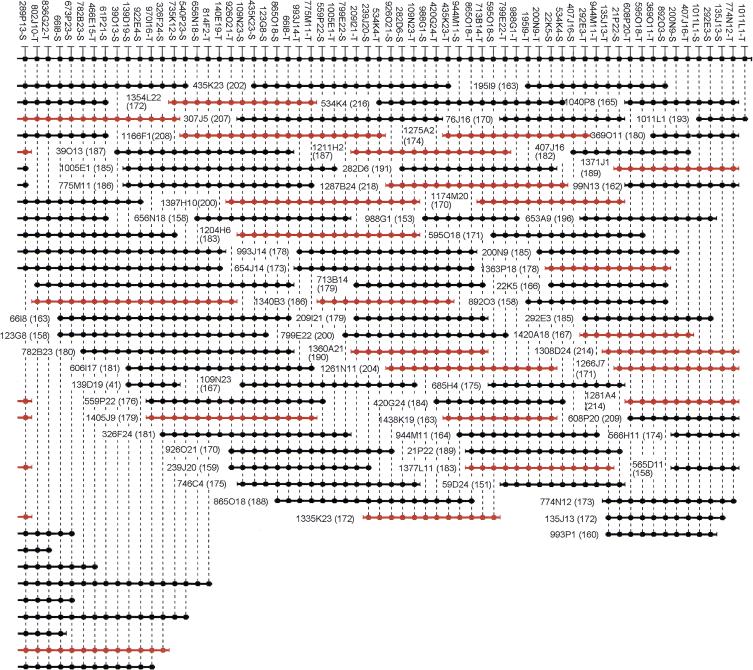
Ideally, BAC clones should represent random, unbiased cloning of the human genome and should retain the insert fragment without cloning artifacts, such as chimeras and rearrangements. To explore the genomic fidelity and integrity of the BACs, we arbitrarily chose a 1.5-Mb region from chromosome 14q24.3 for a detailed characterization of all clones and examination of the fidelity of the cloning process. The region has been used previously to characterize the RPCI-1, -3, -4, -5, and -6 human PAC libraries (C. Wu, B. Zhao, C. Chen, J.J. Catanese, P. Ioannou and P.J. de Jong, unpubl.). A high-resolution PAC/STS-content map provided the markers to isolate all corresponding BAC clones and additional new markers were generated after determination of all the insert-end sequences. All BAC end sequences have been deposited in GenBank under accession nos. AQ936150–AQ936491. The new BAC contig contains 121 clones and 48 clones derived from the EcoRI and MboI library sections, respectively. For high-resolution comparison of the BACs, 121 STS markers from BAC ends and 168 pre-existing STS markers, mostly derived from PAC clone ends, were mapped to the BAC clones in the contig. Because most of the markers were designed from BAC and PAC ends, it was possible to determine a definitive linear order for all markers (Fig. 3) by using SEGMAP, an STS-content mapping program (Green and Green 1991). Note that the 168 pre-existing markers are not shown in Figure 3.
Figure 3.
A 1.5-Mb BAC contig map localized on chromosome 14q24.3. A total of 169 BAC clones were identified from the RPCI-11 BAC library. The contig has been assembled according to the hybridization results using SEGMAP. The deduced markers are depicted with a black circle along the top, and each short horizontal line with black circles represents a BAC clone. Combination of the plate number and well position represents the clone name from the RPCI-11 library. For example, 1035K4 is found in plate 1035 at well position K4. The markers derived from the T7 or SP6 vector end are condensed as −T or −S after the clone name. The size of each clone is indicated in kilobases in parentheses. Markers have arbitrarily been assigned even spacing for diagrammatic purposes, and the length of the horizontal lines does not accurately represent the insert size. Only 121 markers derived from BAC ends are shown in this figure. Note that seven markers (between 289P13-S and 466E15-T) are overlapping in two contiguous figures. BACs derived from MboI library section (segment 5) are shown in red.
Chimeric Clone Levels
Chimeric clones are one of the problems encountered in constructing an accurate physical map and in determining the genomic sequence. A priori, it is presumed that chimeric BACs are generated under different conditions from chimeric YAC clones. Although chimeric YACs can originate from a coligation linking unrelated genomic fragments, most chimeric YACs result from homologous recombination between incomplete YACs immediately after transformation into yeast spheroplasts (Green et al. 1991; Haldi et al. 1994; Wada et al. 1994).
Recombination and transformation of multiple large DNA fragments are expected to occur at much lower levels in Escherichia coli as compared with yeast, hence leaving coligation as the most plausible mechanism for creating chimeric BACs. Moreover, recombinant BAC clones are created by ligation with genomic DNA fragments that have been purified using the improved double size fractionation procedure with pulsed-field gel electrophoresis (Osoegawa et al. 1998). At the molecular level, it thus is most likely that two unrelated large fragments coligate and then connect with a vector fragment. However, such chimeras will have double the insert size and will transform E. coli at very low efficiency because of the strong size bias in E. coli transformation. It thus is postulated that chimeras—if found at all in the BAC system—will be rare and will be derived from one small and one large fragment. Such unbalanced chimeras will be difficult to detect by fluorescent in situ hybridization (FISH), which frequently is used to detect YAC chimeras. Thus, a different approach was implemented to identify chimeric clones by mapping of the opposite insert-end sequences for many clones, either through conventional wet-laboratory screening approaches or through BLAST searches against sequences in GenBank. All 169 BACs from the 1.5-Mb contig were analyzed to determine if both insert ends mapped to other clones from this validated contig. Overgo probes were designed from the BAC end sequences for the EcoRI library to generate new markers. For 58 clones, both end probes were unique and were used for the hybridization screening. All were found to map to overlapping clones thus proving that 58 of 58 clones consisted of a single contiguous genome segment. Independent of the marker screening, 338 insert-end sequences from the 169 BACs have been used to search GenBank with the BLASTN program (Altschul et al. 1990). GenBank contains the draft sequences for the region sequenced from our earlier PAC contig. Through the GenBank searches, an additional 109 clones were determined to be nonchimeric by using the BLAST searches either alone or in combination with the probe screening results. It was not possible to determine the status of the remaining two clones as either chimeric or nonchimeric because of the poor quality of end sequences. Thus, 167 of 167 clones with informative ends contained a single contiguous genome segment, indicating that chimerism within the BAC library is low (1%) or does not occur at all. The similar result was reported that only one chimeric clone was found of 113 clones during the course of characterization of mouse BAC/PAC libraries (Osoegawa et al. 2000).
Clone Rearrangements
Traditionally, comparison of fingerprinting patterns from overnight cultures versus serial cultures have been used to examine the stability of BAC and PAC inserts (Shizuya et al. 1992; Ioannou et al. 1994; Woo et al. 1994; Cai et al. 1995; Woon et al. 1998). This is a simple method used to determine if BAC and PAC clones are maintained without major rearrangements after many cell generations. However, BAC clones are not routinely used as sequencing templates after an excessive number of generations obtained through serial culturing. It thus appears more important to determine whether BAC clones have subtle rearrangements after more typical growth conditions. The goal is to show clonal integrity and fidelity of the BAC clones for use as sequencing templates. If a rearrangement occurred during the bacterial transformation or shortly thereafter, then independently obtained overlapping clones would end up with inconsistent genomic structures. Therefore, the 169 overlapping BACs from the 1.5-Mb contig were compared by pulsed-field gel electrophoresis, high-resolution STS-content mapping (Renault et al. 1997), and fingerprinting (Marra et al. 1997). Five parallel subcolonies from each BAC were analyzed by pulsed-field gel electrophoresis to exclude the occurrence of major rearrangements or clonal contamination. Although most (92.3%) of the clones revealed identical subcolonies, 13 clones of 169 showed multiple insert sizes indicating either clonal contamination or DNA rearrangements. Contaminating clones are not expected to map to the same contig area and can be identified because they are predicted to be negative for all the contig markers. Unlike the previously discussed α-satellite clones, all major size inconsistencies between the subcolonies resulted from clonal contamination. For high-resolution comparison of the BACs, 121 STS markers from BAC ends and 168 STS markers derived mainly from PAC clone ends were mapped to the BAC clones in the contig (Fig. 3). A clone deletion may be indicated by negative screening results from hybridization or PCR for one or more internal markers, whereas all the flanking markers correctly identify the particular clone. The average marker spacing is 5.2 kb based on an approximate 1.5-Mb contig size. No inconsistent results were found for any of the clones by using all of the 289 markers in colony hybridization and PCR analyses. This indicates the absence of small deletions spanning more than one marker interval. However, it was not possible to exclude potential deletions occurring between markers. To increase the sensitivity of the screening for rearranged clones, fingerprinting analysis (Marra et al. 1997) has been applied to detect small rearrangements within the resolving power of the agarose gel electrophoresis and computer software. The fingerprinting analysis also may identify chimeric clones as inconsistent fragment patterns (Osoegawa et al. 2000). Fingerprinting was performed in duplicate using the EcoRI restriction fragment patterns from two single-colony isolates for each clone. Clonal instability may be detected as heterogeneity in the duplicate fingerprints. Rearrangements also may be detected as inconsistent fragment patterns by comparing different clones from the same contig. BAC clones from the MboI library part were prepared using MboI partially digested DNA and do not have any vector-derived EcoRI sites.
Consequently, the EcoRI fingerprints of overlapping clones from the MboI library segment always have at least one clone-specific fragment not found in any of the overlapping clones. This fragment measures in excess of 10.6 kb, consisting of the complete vector and variable-size insert-end sequences. BACs from the EcoRI library part always have an identical vector EcoRI fragment of 8.8 kb, smaller than 10.6 kb because different vectors were used. Sixteen small rearrangements were detected within the contig as heterogeneity between duplicate subcolonies (Fig. 4). These rearrangements are defined by single fragment differences between the restriction fingerprints of the related subcolonies, with fragment sizes (or fragment size differences) between 200 bp and 10 kb. Two possible fingerprint inconsistencies were found within clones by comparing them with all their corresponding overlapping clones. These clone-specific fragments were each found in single clones within a 30-fold redundant contig, hence suggesting that they do not represent polymorphic differences within the diploid genome of the donor. In summary, small rearrangements were observed in 18 of 169 clones as a result of alterations to a single genomic fragment during or after the cloning process.
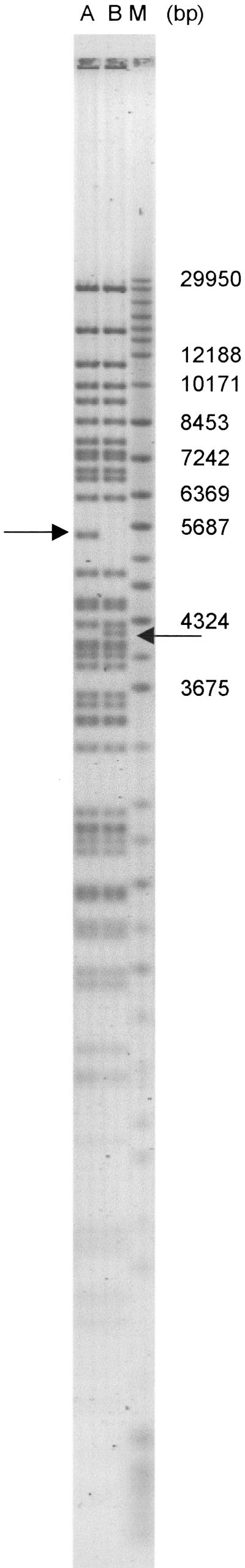
Figure 4.
This picture shows clonal heterogeneity that is derived from the same clone. (M) Marker lane that contains a mixture of two commercially available markers as described in Methods. The DNAs shown in lanes A and B were isolated from different single colonies that are derived from the same clone. (Arrows) Inconsistent EcoRI fragments between the clones. Some of the marker sizes are indicated. The fingerprints show rearrangements that took place during cell duplication.
Randomness of EcoRI and MboI Partial Digestion
The optimal library size for complete representation of the human genome depends on statistical considerations but also on the randomness of the BAC cloning process. The randomness of cloning can be affected by two factors: the restriction cutting of the genome, and possible sequence-content bias in creating a viable bacterial colony. To test the randomness of the original restriction digest, we compared and analyzed all insert-end sequences for the 169 clones in the 1.5-Mb contig for evidence of repeated use of the same cut sites within independent clones. Specifically, it would be of interest to reveal sites preferentially cleaved. Such sites would more likely turn up at the end of cloned fragments and could possibly cause gaps in contig maps. One way to analyze preferential cutting is to search for independent cloning events sharing the same restriction site at the insert end at an incidence in excess of statistical expectation. The likelihood of independent clones with a shared insert end can be predicted by the Poisson equation [P (X = k) = e−μμk/k!]. In this case, the Poisson variable (μ) is the number of BAC ends per EcoRI or MboI site. The incidence of the identical BAC ends is represented by k where k = 2 for duplicate, k = 3 for triplicate and k = 4 for quadruplicate sharing of the same BAC insert-end sequence. The average GC base composition of the human genome is 42% (Shapira 1976) and, from this, average sizes of EcoRI and MboI restriction fragments are calculated at 3.2 kb and 270 bp, respectively. Therefore, a 1.5-Mb contig with 121 and 48 BACs from the EcoRI and MboI library sections contains 469 and 5556 possible EcoRI and MboI cutting sites, respectively. This translates into 0.258 BAC ends per EcoRI site and 0.0086 BAC ends per MboI site (μ = 0.2580 and μ = 0.0086). In addition, a survey of the high-throughput genomic (HTG) sequence database (Ouellette and Boguski 1997) as of December 26, 2000 revealed that there are 65 small gaps in the genomic sequence between marker 1035K4-S and 1011L1-T (Fig. 3). The total length of the sequence between these markers was 1,629,088 bp, the GC base composition is 43.7%, and 403 EcoRI sites and 4553 MboI sites were found in the sequence. This translates to 0.300 BAC ends per EcoRI site and 0.0105 BAC ends per MboI site, if all the EcoRI and MboI sites were found. Table 3 shows the results for shared clone ends in the EcoRI and MboI library segments as compared with the expected values. As a result, the incidence of identical BAC ends is 1.4–1.5-fold, 2.2–2.9-fold and 5–7.7-fold higher for EcoRI for duplicate, triplicate, and quadruplicate events and 4–4.9-fold for MboI for duplicate than the anticipated numbers. Two clones (1234F15 and 1174G20) from different plates in the MboI library have identical sequences at both ends and thus are likely derived from a single transformation event resulting in two colonies through cell duplication. Therefore, a 4–4.9-fold higher than expected rate does not reflect preferential MboI cutting sites. Four additional clones (123I8, 140F20, 1266P7, and 1266P8) were identified as members in this contig. These clones are identical with 123G8, 140E19, and 1266J7, respectively.
Table 3.
Numbers of Incidences of Clones with Shared Insert Ends within the 1.5 Mb BAC Contig
| BACs sharing the same end | EcoRI library section | MboI library section | ||||
|---|---|---|---|---|---|---|
| A | B | C | A | B | C | |
| 2 | 24.1 | 26.9 | 37 | 0.41 | 0.5 | 0 (2) |
| 3 | 2.1 | 2.7 | 6 | 1.1 × 10−3 | 1.7 × 10−3 | 0 |
| 4 | 0.13 | 0.2 | 1 | 0 | 0 | 0 |
The expected numbers of events that two, three, or four clones share the same end within the 1.5 Mb contig were calculated using the formula 2 × expected frequency (P) × number of expected restriction sites. “A” columns show the numbers calculated based on a 42% GC content for the entire genome. “B” columns show the number of incidences based on the actual numbers of EcoRI and MboI restriction sites within the BAC contig region as found in GenBank. “C” columns show the empirical numbers found by comparing the end-sequences of the BAC clones within the contig. For the MboI library only two occurrences were found of duplicate sharing of ends. Because these two instances were from the same two clones, they reflect a clonal duplication prior to colony picking and not an independent sharing of the same insert end.
Because these clones were contained in the same plates, they likely resulted from an arraying error by the picking robot. The ratio of the duplicated clones by cell duplication and the robotic error would be estimated to be 2.9% (five of 173 clones). In addition to possible cloning bias due to preferential cutting of some restriction sites, it is conceivable that the sucrose positive selection system has resulted in some bias. For instance, it is possible that some of the human insert sequences have fortuitous E. coli promoter-like sequences positioned close to the open reading frame of the SacB gene in the vector. This might result in expression of the SacB gene with a resulting sucrose sensitivity and nonviability of the clones. In fact, ∼20% additional true recombinant clones can be generated if sucrose selection is avoided (data not shown). Sucrose selection nevertheless is preferred because it significantly reduces the background of nonrecombinant clones. It is presumed that the bias caused by the sucrose selection is more or less random and does not make genomic regions unclonable. No instances of contig gaps due to a nonrandom distribution of fortuitous bacterial promoter elements in human DNA are known.
DISCUSSION
During recent years, bacterial artificial chromosome libraries have become a central reagent for physical mapping and sequencing of complex genomes. Although six to 10 large insert libraries including PACs, BACs and cosmids have been used for the publicly funded human genome project, a survey of the HTG sequence database (Ouellette and Boguski 1997) as of July 8, 2000 revealed that ∼80% of the large insert clones are from the RPCI-11 BAC library. Among the reasons for the disproportionate usage are the consistent high-average insert sizes as well as the timely and widespread dissemination of copies of the RPCI-11 collection. The consistency in insert sizes and library quality is a direct consequence of the improved cloning procedures we introduced (Osoegawa et al. 1998). This and other aspects of library quality are consistent with the characterization described in this article. The preferential use of one library over others has important consequences. It permits the accumulation of overlapping data sets for the same clones and thus provides a real opportunity for the development of a reference library of the human genome. For instance, only two human BAC libraries, the CalTech D library (http://www.tree.caltech.edu/lib_status.html) and RPCI-11 were used for sequencing the insert ends to provide sequence information on the BAC clones (Mahairas et al. 1999; Zhao et al. 2000). BAC end sequencing was introduced as a tool for virtual library screening to search for BACs overlapping with large sequence contigs and to use these BACs as sources to expand the contiguous sequence. Because of the option to use insert-end sequences to connect existing sequence data with clones, the term STC for sequence tag connectors was introduced (Venter et al. 1996). The database of BAC end sequences also provides a way for selecting unrelated clones not yet present in the fraction of the genome already being sequenced. Such unrelated clones thus could serve as nucleation points for new sequence contigs. Finally, the two sequenced ends of large insert clones provide a mechanism to determine relative map location for sequence contigs derived from whole-genome shotgun sequencing (WGS). This application, designated as genome scaffolding (Adams et al. 2000; Hoskins et al. 2000), requires the availability of a reference library with BAC end sequence information accumulated for many clones. Relative BAC mapping information deduced from direct or virtual screening can be combined with map information based on fingerprinted BAC clones, to either expand the contigs or provide a means to check data sets for mutual compatibility. Although such data can be obtained on a case-by-case basis for clones picked out by screening, it is more economical and consistent to obtain fingerprints for most of the clones in the BAC library under precisely defined conditions and preferably within the same laboratory. This has, in fact, been performed only for 270,000 clones of the RPCI-11 BAC library at Washington University (McPherson et al. 2001; http://genome.wustl.edu/gsc/human/human_database.shtml). Additional mapping information has been accumulated for the same reference library through marker screening and chromosome walking procedures (Dunham et al. 1999; Hattori et al. 2000). For instance, several research efforts have aimed at isolating a subset of characterized BAC clones, which are mapped to well-defined, regularly spaced locations along the human genome. Such clone sets will be applied as diagnostic clone collections for characterizing chromosomal rearrangements through in situ hybridization procedures for cancer applications (Strausberg et al. 1997; Cheung et al. 2001; The Cancer Genome Anatomy Project [CGAP, http://www.ncbi.nlm.nih.gov/ncicgap/]) or to define inborn deletion syndromes (Shapira 1998; Developmental Genome Anatomy Project [DGAP, http://dgap.harvard.edu]). The mapped clones can be used not only as hybridization probes but also as hybridization templates on glass slide arrays for comparative genomic hybridization (CGH; Pinkel et al. 1998). In addition, BACs can be used as a source of “reproducible” PCR fragments by using primers to highly repeated dispersed sequence elements such as the Alu repeat sequence. Such PCR fragments are a rich source of haplotype-specific polymorphisms, and procedures have been developed to determine the haplotype information for large genomic regions by arraying the BAC-derived Alu-PCR fragments on high-density glass arrays (Cheung et al. 1998). In addition, the fraction of clones in the pTARBAC vector (segment 5 only) may provide a means to reisolate the same genomic segments from other human haplotypes and possibly from other primates through transformation-associated recombination cloning in yeast (Zeng et al. 2001). The abundant use of the same BAC library for many data sets has ensured that this clone collection will not only serve as a transient tool for the sequencing of the human genome, but will continue its use as a reference set of well-characterized clones for functional research in cell-based expression studies, for creating phenotypes in mice through human BAC transgenes (Antoch et al. 1997; Yang et al. 1997, 1999; Probst et al. 1998) and for use in diagnostic applications (Schmitt et al. 1998; Marinescu et al. 1999; Orsetti et al. 1999). Precise sequence information was available for 18,688 RPCI-11 BAC clones on July 8, 2000, which represent nearly 80% of the human genome and were used for shotgun sequencing projects. The availability of this sequenced set of BAC clones permits the design of gene modification strategies to support research questions that can be addressed through BAC transfection assays and BAC transgenic animals.
METHODS
Library Construction
Leukocytes were isolated from a single male anonymous donor. Agarose blocks containing high molecular weight DNA were prepared as described previously (Osoegawa et al. 1999). The pBACe3.6 (Frengen et al. 1999) and pTARBAC1 vectors (Zeng et al. 2001) were used to clone EcoRI- and MboI- partially digested DNA. High molecular weight DNA was size-fractionated with a CHEF apparatus (BioRad) and the size-fractionated DNA was eluted by an electroelution procedure (Osoegawa et al. 1998). The ligation product was transformed into electrocompetent E. coli DH10B cells (ElectroMAX DH10B; Life Technologies). The detailed protocol for construction of a BAC library was described previously (Osoegawa et al. 1998, 1999). High-density replica filters were prepared as described previously (Osoegawa et al. 2000).
Insert Size Analysis
A total of 483 clones from segment 1, 246 from segment 2, 299 from segment 3, 308 from segment 4, and 212 from segment 5 were picked by taking different size fractionations and transformations into consideration (Table 1A). These clones were incubated in LB medium containing 20 μg/mL chloramphenicol. BAC DNA was purified using an automated plasmid isolation machine (AutoGen 740; Integrated Separation Systems). DNA was analyzed after digestion with NotI (New England Biolabs) by using a CHEF or a FIGE (BioRad) as described previously (Osoegawa et al. 1998). The insert sizes were determined using an Alpha Innotech IS1000 digital imager. Low Range PFG Marker (New England Biolabs) containing a mixture of λ DNA-HindIII fragments and λ concatemers was used as marker DNA for size determination.
α-Satellite Clones
α-Satellite DNA was amplified using primer 1: 5′-GGTTCAACT CTGTGAG-3′, and primer 2: 5′-CACTCTTTTTGTAGAATC TG-3′;. The PCR was performed using 10 ng human genomic DNA as a template with 0.15 μM of each primer, 250 μM dNTPs, 1 × PCR buffer (Boehringer-Mannheim), and 0.4 units Taq DNA polymerase (Boehringer-Mannheim) in a 10-μL reaction volume. The amplification was performed at 94°C for 1 min and 94°C for 15 sec, 58°C for 15 sec, 72°C for 20 sec, 5 cycles, then followed with 94°C for 15 sec, 56°C for 15 sec, 72°C for 20 sec, 30 cycles, and 72°C for 1 min in the GeneAmp PCR system 9600 (Perkin-Elmer Cetus). The PCR product was purified from agarose gel electrophoresis and labeled with α-32P dCTP (3000 Ci/mmol; Amersham) by using the PCR incorporation procedure (Osoegawa et al. 2000). The hybridization was performed with low-density (six plates per membrane) and high-density (48 plates per membrane) filters. To accurately determine the position of the positive clones, we mixed a low concentration of 32P-labeled vector DNA in the hybridization to visualize every clone in the grid. The positive clones were analyzed with CHEF and conventional agarose gel electrophoresis after digestion with NotI and EcoRI, respectively. The Low Range PFG Marker was used as marker DNA for CHEF and the 1-kb DNA ladder (Life Technologies), and λ DNA-HindIII fragments (Life Technologies) were loaded on both sides of the gel for standard electrophoresis.
Screening the Library
A total of 45 STS markers specific for chromosomes 5, 19, and 21 were selected. Thirty markers were derived from chromosome 5, nine markers from chromosome 19, and six markers from chromosome 21. Overlapping oligonucleotide (overgo; McPherson 1999) probes have been designed based on the publicly available marker sequence. The hybridization procedure was described previously (Osoegawa et al. 2000).
Contig Construction and Fingerprinting Analysis
A 3.5-Mb PAC contig map has been constructed previously on chromosome 14q24.3 (C. Wu, B. Zhao, C. Chen, J.J. Catanese, P. Ioannou, and P.J. deJong, unpubl.). A region of 1.5 Mb that was defined by 205 STS markers from this PAC contig was selected to construct a BAC contig from the RPCI-11 library. Hybridization, overgo probe design, STS-content mapping, and fingerprinting have been described previously (Osoegawa et al. 2000). BACs for the 1.5-Mb region were identified by hybridization screening of high-density colony membranes by using a minimal set of nine PAC clones. The putatively positive clones then were rearrayed to prepare a sublibrary enriched for the region. To establish the STS-content map for all the clones in the BAC contig, we followed two approaches. BAC end sequences were used to design overgo probes, which were hybridized against the high-density replica filters from the sublibrary. In addition, 168 of the previous 205 STS markers were mapped to the BAC contig by using PCR. The composite of hybridization and PCR data was analyzed using the SEGMAP V. 3.49 software (Green and Green 1991) to establish the STS-content map. All the contig clones also were analyzed by fingerprinting (Marra et al. 1997). A mixture of Analytical Marker DNA, Wide Range Ladder (Promega), and Marker V (Roche Molecular Biochemicals) was loaded in marker lanes for the fingerprinting (Fig. 4).
Table 1A.
The RPCI-11 Human Male BAC Library
| Partial digestion | Fraction identity | Plate no. | Average insert size (kb) |
|---|---|---|---|
| EcoRI (segments 1–4) | M1B001E F6 | 1–280 | 163 |
| M1B004E F5 | 281–791 | 174 | |
| M1B003E F5 | 792–1068 | 184 | |
| M1B001E F5 | 1069–1107 | 197 | |
| M1B004E F6 | 1108–1152 | 154 | |
| MboI (segment 5) | M1B003Mb F4 | 1153–1329 | 192 |
| M1B002Mb F4 | 1330–1440 | 192 |
DNA source: Anonymous male donor.
Cloning vector: pBACe3.6 (segments 1–4) and pTARBAC1 (segment 5).
The average insert size of each fraction was determined by analyzing 120 clones using CHEF or FIGE apparatus after digestion with NotI restriction enzyme prior to the colony picking stage. The increase in average insert size for the later transformations reflects improvements in cloning skills while generating the library. Colonies from each fraction were picked and arrayed into 384-well plates by the order indicated in Table 1A. The libraries were organized into five segments as indicated in Table 1B.
Table 1B.
| Segment | Cloning enzyme | Total clones | Plate no. | Non-insert clones (%) | Insert size (kb) | Genomic redundancy |
|---|---|---|---|---|---|---|
| 1 | EcoRI | 108,499 | 1–288 | 1.7 | 163 | 5.4 |
| 2 | EcoRI | 109,496 | 289–576 | 0.5 | 168 | 5.6 |
| 3 | EcoRI | 109,657 | 577–864 | 1.0 | 181 | 6.0 |
| 4 | EcoRI | 109,382 | 865–1152 | 1.0 | 183 | 6.1 |
| 5 | MboI | 106,763 | 1153–1440 | 0.0 | 195 | 6.3 |
| Total | 543,797 | 1440 | 0.8 | 178 | 29.4 |
The “Total clones” column represents total wells after empty wells were subtracted [288 × 384–(empty wells)] in each segment. Average insert sizes in this table were estimated by analyzing 483 clones from segment one, 246 from segment two, 299 from segment three, 308 from segment four, and 212 from segment five. These clones were randomly picked from each fraction to correspond to their ratio within each segment, for example segment three comprises 75% of its clones from fraction M1B004E F5 and 25% from fraction M1B003E F5. Non-insert clones were observed as a single band that is smaller than the normal vector size on the gel. No non-recombinants containing the deleted vector fragment have been found among 212 clones analyzed from segment five. The genomic redundancy was estimated using 3300 Mb as the genome size.
Acknowledgments
We thank Norma J. Nowak and Jeffry Conroy for useful advice characterizing the library and Susan Rhodes for critical reading of the manuscript. We are grateful to Minako Tateno, Amy Beck, Alfred Cairo, Gregory P. Caldwell, Qingdan Chen, Melanie Hierl, David W. Kois, Yukiko Osoegawa, Beth A. Palka, Chung Li Shu, Barbara Swiatkiewcz, Gery Vessere, and Baohui Zhao for their technical advice and assistance. K.O. has been supported with a Postdoctoral Fellowship for Research Abroad from the Japan Society for the Promotion of Science (JSPS). This work was funded by grants from the National Institutes of Health (1RO1HL55700), the National Human Genome Research Institute (1RO1RG01165), and the U.S. Department of Energy (DE-FG02-94ER61883). Current availability of the RPCI-11 library and high-density replica filters can be found at http://www.chori.org/bacpac.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pdejong@mail.cho.org; FAX (510) 450-7924.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.169601.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini A, Smith DI, Rocchi M, Miller OJ, Miller DA. A human alphoid DNA clone from the EcoRI dimeric family: Genomic and internal organization and chromosomal assignment. Genomics. 1989;5:822–828. doi: 10.1016/0888-7543(89)90124-9. [DOI] [PubMed] [Google Scholar]
- Baldini A, Rocchi M, Archidiacono N, Miller OJ, Miller DA. A human α satellite DNA subset specific for chromosome 12. Am J Hum Genet. 1990;46:784–788. [PMC free article] [PubMed] [Google Scholar]
- Botkin JR, Gut IG. Whose genes are they and how can we identify them? Science. 1996;274:901. doi: 10.1126/science.274.5289.904a. [DOI] [PubMed] [Google Scholar]
- Cai L, Taylor JF, Wing RA, Gallagher DS, Woo S-S, Davis SK. Construction and characterization of a Bovine bacterial artificial chromosome library. Genomics. 1995;29:413–425. doi: 10.1006/geno.1995.9986. [DOI] [PubMed] [Google Scholar]
- Cheung VG, Gregg JP, Gogolin-Ewens KJ, Bandong J, Stanley CA, Baker L, Higgins MJ, Nowak NJ, Shows TB, Ewens WJ, et al. Linkage-disequilibrium mapping without genotyping. Nat Genet. 1998;18:225–230. doi: 10.1038/ng0398-225. [DOI] [PubMed] [Google Scholar]
- Cheung, V.G., Nowak, N., Jang, W., Kirsch, I.R., Zhao, S., Chen, X.-N., Kim, U.-J., Kuo, W.-L., Olivier, M., Conroy, J., et al. 2001. Integration of cytogenetic landmarks into the draft sequence of human genome. Nature (in press). [DOI] [PMC free article] [PubMed]
- Cohen D, Chumakov I, Welssenbach J. A first-generation physical map of the human genome. Nature. 1993;366:698–701. doi: 10.1038/366698a0. [DOI] [PubMed] [Google Scholar]
- Dunham I, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, Ainscough R, Almeida JP, Babbage A, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Evans GA, Snider K, Hermanson GG. Use of cosmids and arrayed clone libraries for genome analysis. Methods Enzymol. 1992;216:530–548. doi: 10.1016/0076-6879(92)16048-o. [DOI] [PubMed] [Google Scholar]
- Frengen E, Weichenhan D, Zhao B, Osoegawa K, van Geel M, de Jong PJ. A modular positive selection bacterial artificial chromosome vector with multiple cloning sites. Genomics. 1999;58:250–253. doi: 10.1006/geno.1998.5693. [DOI] [PubMed] [Google Scholar]
- Green ED, Olson MV. Systematic screening of yeast artificial-chromosome libraries by use of the polymerase chain reaction. Proc Natl Acad Sci. 1990;87:1213–1217. doi: 10.1073/pnas.87.3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ED, Green P. Sequence-tagged site (STS) content mapping of human chromosomes: Theoretical considerations and early experiences. PCR Methods Appl. 1991;1:77–90. doi: 10.1101/gr.1.2.77. [DOI] [PubMed] [Google Scholar]
- Green ED, Riethman HC, Dutchik JE, Olson MV. Detection and characterization of chimeric yeast artificial chromosome clones. Genomics. 1991;11:658–669. doi: 10.1016/0888-7543(91)90073-n. [DOI] [PubMed] [Google Scholar]
- Haldi M, Perrot V, Saumier M, Desai T, Cohen D, Cherif D, Ward D, Lander ES. Large human YACs constructed in a rad52 strain show a reduced rate of chimerism. Genomics. 1994;24:478–484. doi: 10.1006/geno.1994.1656. [DOI] [PubMed] [Google Scholar]
- Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, Park H-S, Toyoda A, Ishii K, Totoki Y, Choi D-K, et al. The DNA sequence of human chromosome 21. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- Hoskins RA, Nelson CR, Berman BP, Laverty TR, George RA, Ciesiolka L, Naeemuddin M, Arenson AD, Durbin J, David RG, et al. A BAC-based physical map of the major autosomes of Drosophila melanogaster. Science. 2000;287:2271–2274. doi: 10.1126/science.287.5461.2271. [DOI] [PubMed] [Google Scholar]
- Ioannou PA, Amemiya CT, Garnes J, Kroisel PM, Shizuya H, Chen C, Batzer MA, de Jong PJ. A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat Genet. 1994;6:84–89. doi: 10.1038/ng0194-84. [DOI] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Graves J, Chen X-N, Korenberg JR, Resnick MA. Specific cloning of human DNA as YACs by transformation-associated recombination. Proc Natl Acad Sci. 1996;93:491–496. doi: 10.1073/pnas.93.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahairas GG, Wallace JC, Smith K, Swartzell S, Holzman T, Keller A, Shaker R, Furlong J, Young J, Zhao S, et al. Sequence-tagged connectors: A sequence approach to mapping and scanning the human genome. Proc Natl Acad Sci. 1999;96:9739–9744. doi: 10.1073/pnas.96.17.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinescu RC, Johnson EI, Grady D, Chen XN, Overhauser J. FISH analysis of terminal deletions in patients diagnosed with cri-du-chat syndrome. Clin Genet. 1999;56:282–288. doi: 10.1034/j.1399-0004.1999.560405.x. [DOI] [PubMed] [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M, Kucaba T, Saekhon M, Hillier L, Martienssen R, Chinwalla A, Crockett J, Felele J, Grover H, Gund C, et al. A map for sequence analysis of the Arabidopsis thaliana genome. Nat Genet. 1999;22:265–275. doi: 10.1038/10327. [DOI] [PubMed] [Google Scholar]
- Marshall E. Whose genome is it, anyway? Science. 1996;273:1788–1789. doi: 10.1126/science.273.5283.1788. [DOI] [PubMed] [Google Scholar]
- McPherson JD. In: Genome analysis: A laboratory manual. Birren B, et al., editors. Vol. 4. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 207–213. [Google Scholar]
- McPherson, J.D., Marra, M., Hillier, L., Waterston R.H., Chinwalla, A., Wallis, J., Sekhon, M., Wylie, K., Mardis, E.R., Wilson, R.K. et al. 2001. A physical map of the human genome. Nature (in press).
- Nizetic D, Zehetner G, Monaco AP, Gellen L, Young BD, Lehrach H. Construction, arraying, and high-density screening of large insert libraries of human chromosomes X and 21: Their potential use as reference libraries. Proc Natl Acad Sci. 1991;88:3233–3237. doi: 10.1073/pnas.88.8.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsetti B, Courjal F, Cuny M, Rodriguez C, Theillet C. 17q21-q25 aberrations in breast cancer: Combined allelotyping and CGH analysis reveals 5 regions of allelic imbalance among which two correspond to DNA amplification. Oncogene. 1999;18:6262–6270. doi: 10.1038/sj.onc.1203006. [DOI] [PubMed] [Google Scholar]
- Osoegawa K, Woon P-Y, Zhao B, Frengen E, Tateno M, Catanese JJ, de Jong PJ. An improved approach for construction of bacterial artificial chromosome libraries. Genomics. 1998;52:1–8. doi: 10.1006/geno.1998.5423. [DOI] [PubMed] [Google Scholar]
- Osoegawa K, de Jong PJ, Frengen E, Ioannou PA. In: Current protocols in human genetics. Dracopoli NC, et al., editors. New York: Wiley; 1999. pp. 5.15.1–5.15.33. [Google Scholar]
- Osoegawa K, Tateno M, Woon P-Y, Frengen E, Mammoser AG, Catanese JJ, Hayashizaki Y, de Jong PJ. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 2000;10:116–128. [PMC free article] [PubMed] [Google Scholar]
- Ouellette BFF, Boguski MS. Database divisions and homology search files: A guide for the perplexed. Genome Res. 1997;7:952–955. doi: 10.1101/gr.7.10.952. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kou W-L, Chen C, Zhai Y, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Probst FJ, Fridell RA, Raphael Y, Saunders TL, Wang A, Liang Y, Morell RJ, Touchman JW, Lyons RH, Noben-Trauth K, et al. Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science. 1998;280:1444–1447. doi: 10.1126/science.280.5368.1444. [DOI] [PubMed] [Google Scholar]
- Renault B, Hovnanian A, Bryce S, Chang JJ, Lau S, Sakuntabhai A, Monk S, Carter S, Ross CJ, Pang J, et al. A sequence-ready physical map of a region of 12q24.1. Genomics. 1997;45:271–278. doi: 10.1006/geno.1997.4888. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmitt H, Sasiadek M, Jagielski J, Blin N. Classical and molecular cytogenetic methods in diagnosis of a rare translocation t(3;21) Int J Mol Med. 1998;1:569–571. doi: 10.3892/ijmm.1.3.569. [DOI] [PubMed] [Google Scholar]
- Shapira HS. In: Handbook of biochemistry and molecular biology: Nucleic acids. Fasman GD, editor. Cleveland, OH: CRC Press; 1976. 2: 241–275. [Google Scholar]
- Shapira SK. An update on chromosome deletion and microdeletion syndromes. Curr Opin Pediatr. 1998;10:622–627. doi: 10.1097/00008480-199810060-00015. [DOI] [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim U-J, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C, Humphray S, Dunham A, French L. Contigs built with fingerprints, markers and FPC V4.7. Genome Res. 2000;10:1772–1787. doi: 10.1101/gr.gr-1375r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg RL, Dahl CA, Klausner RD. New opportunities for uncovering the molecular basis of cancer. Nat Genet. 1997;15:415–416. doi: 10.1038/ng0497supp-415. [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C, Willard HF. Mammalian chromosome structure. Curr Opin Genet Dev. 1993;3:390–397. doi: 10.1016/0959-437x(93)90110-b. [DOI] [PubMed] [Google Scholar]
- Venter JC, Smith HO, Hood L. A new strategy for genome sequencing. Nature. 1996;381:364–366. doi: 10.1038/381364a0. [DOI] [PubMed] [Google Scholar]
- Wada M, Abe K, Okumura K, Taguchi H, Kohno K, Imamoto F, Schlessinger D, Kuwano M. Chimeric YACs were generated at unreduced rates in conditions that suppress coligation. Nucleic Acids Res. 1994;22:1651–1654. doi: 10.1093/nar/22.9.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S-S, Jiang J, Gill BS, Paterson AH, Wing RA. Construction and characterization of a bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 1994;22:4922–4931. doi: 10.1093/nar/22.23.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woon P-Y, Osoegawa K, Kaisaki PJ, Zhao B, Catanese JJ, Gauguier D, Cox R, Levy ER, Lathrop GM, Monaco AP, et al. Construction and characterization of a 10-fold genome equivalent rat P1-derived artificial chromosomal library. Genomics. 1998;50:306–316. doi: 10.1006/geno.1998.5319. [DOI] [PubMed] [Google Scholar]
- Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotech. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- Yang XW, Wynder C, Doughty ML, Heintz N. BAC-mediated gene-dosage analysis reveals a role for Zipro1 (Ru49/Zfp38) in progenitor cell proliferation in cerebellum and skin. Nat Genet. 1999;22:327–335. doi: 10.1038/11896. [DOI] [PubMed] [Google Scholar]
- Zeng, C., Kouprina, N., Zhu, B., Cairo, A., Hoek, M., Cross, G., Osoegawa, K., Larinov, V., and de Jong, P.J. 2001. A new BAC/YAC shuttle vector for selective re-isolation of genomic regions through recombination in yeast. Genomics (in press). [DOI] [PubMed]
- Zhao S. Human BAC ends. Nucleic Acids Res. 2000;28:129–132. doi: 10.1093/nar/28.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Malek J, Mahairas G, Fu L, Nierman W, Venter JC, Adams MD. Human BAC ends quality assessment and sequence analyses. Genomics. 2000;63:321–332. doi: 10.1006/geno.1999.6082. [DOI] [PubMed] [Google Scholar]