Abstract
Olfactory receptor (OR) loci frequently cluster and are present on most human chromosomes. They are members of the seven transmembrane receptor (7-TM) superfamily and, as such, are part of one of the largest mammalian multigene families, with an estimated copy number of up to 1000 ORs per haploid genome. As their name implies, ORs are known to be involved in the perception of odors and possibly also in other, nonolfaction-related, functions. Here, we report the characterization of ORs that are part of the MHC-linked OR clusters in human and mouse (partial sequence only). These clusters are of particular interest because of their possible involvement in olfaction-driven mate selection. In total, we describe 50 novel OR loci (36 human, 14 murine), making the human MHC-linked cluster the largest sequenced OR cluster in any organism so far. Comparative and phylogenetic analyses confirm the cluster to be MHC-linked but divergent in both species and allow the identification of at least one ortholog that will be useful for future regulatory and functional studies. Quantitative feature analysis shows clear evidence of duplications of blocks of OR genes and reveals the entire cluster to have a genomic environment that is very different from its neighboring regions. Based on in silico transcript analysis, we also present evidence of extensive long-distance splicing in the 5′-untranslated regions and, for the first time, of alternative splicing within the single coding exon of ORs. Taken together with our previous finding that ORs are also polymorphic, the presented data indicate that the expression, function, and evolution of these interesting genes might be more complex than previously thought.
[The sequence data described in this paper have been submitted to the EMBL nucleotide data library under accession nos. Z84475, Z98744, Z98745, AL021807, AL021808, AL022723, AL022727, AL031893, AL035402, AL035542, AL050328, AL050339, AL078630, AL096770, AL121944, AL133160, and AL133267.]
Olfactory receptor genes (ORs) were first identified in rat olfactory epithelium as small, intronless genes with easily identifiable consensus motifs in conserved domains of the predicted seven transmembrane (7-TM) structure (Buck and Axel 1991). This work stimulated much interest in understanding the molecular basis of olfaction, leading to a large number of ORs being identified. ORs are best known for their involvement in the perception of odors, which is accomplished through OR expression in two anatomically and functionally different organs within the nose: the main olfactory epithelium (MOE) and the vomeronasal organ (VNO). In general, ORs expressed in the MOE are believed to recognize environmental odors (conscious odor perception), whereas ORs expressed in the VNO are believed to recognize odors such as pheromones (subconscious odor perception). However, two recent studies suggest that most of the VNO-type 1 ORs (V1Rs) are nonfunctional pseudogenes in humans (Giorgi et al. 2000; Rodriguez et al. 2000). Nonolfaction-associated OR function such as cell-cell recognition in embryogenesis has also been suggested (Dreyer 1998). For recent reviews on the molecular and cellular biology of ORs, see the special Science issue of October 27, 1999, on olfaction (Science vol. 286; Mombaerts 1999a,b). Public databases currently hold over 600 OR and OR-like genes and pseudogenes, from invertebrates such as Caenorhabditis elegans and Drosophila melanogaster to complex vertebrates, including more than 200 from Homo sapiens. However, the analysis of these ORs has somehow been hampered because only partial sequences are available for most of them (at least for the mammalian ORs). This shortcoming is largely the result of the quick but imperfect approach to identify new ORs by degenerate PCR, and it will soon be corrected as more genomic sequences become available.
A recent genome-wide survey revealed that MOE-type ORs are present on most human chromosomes (Rouquier et al. 1998). The fact that ORs occur in clusters rather than being randomly distributed was recognized early on (Ben-Arie et al. 1994), and a combination of repeated single gene and block duplications was proposed as the underlying mechanism (Lancet and Ben-Arie 1993; Glusman et al. 1996, 2000; Sullivan et al. 1996; Trask et al. 1998a,b). The existence of major histocompatibility complex (MHC)-linked ORs on human chromosome 6 was first discovered in 1995 (Fan et al. 1995). Using a cDNA selection approach, several cDNAs were identified (including FAT11) and mapped telomeric of the MHC. Together with the recently published sequence of the classical MHC (The MHC Sequencing Consortium 1999), the OR cluster reported here forms over 4.5 Mb of contiguous genomic sequence. The region including the OR cluster has previously been shown to be in strong linkage disequilibrium with the MHC (although possibly not in all haplotypes) and has been proposed to be part of the extended MHC (Malfroy et al. 1997; Stephens et al. 1999). This raises the possibility that these ORs are not only physically linked to the MHC but also may have some functional association (e.g., mate selection) with genes of the complex (Ehlers et al. 2000; Ziegler et al. 2000a). Therefore, we have sequenced the human MHC-linked OR cluster and discuss here our findings in comparison with our preliminary data from the orthologous murine OR cluster.
RESULTS AND DISCUSSION
Gene Organization and Genomic Environment
Taking the previously established region of conserved synteny between human and mouse (Yoshino et al. 1997) into account, we have divided the human MHC-linked OR cluster into two subclusters: the MHC-linked major OR cluster (562 kb between positions 105 and 667 kb in Fig. 1) and the MHC-linked minor OR cluster (between HFE and RFP). As illustrated in Figure 1, the ∼3-Mb-long region between HFE and RFP is not yet completely finished (∼90% finished, four out of 30 clones still unfinished), but all of the OR loci have been finished and are included in the analysis presented here. Throughout this study, we follow the OR naming convention previously proposed by us (Ehlers et al. 2000; Ziegler et al. 2000a).
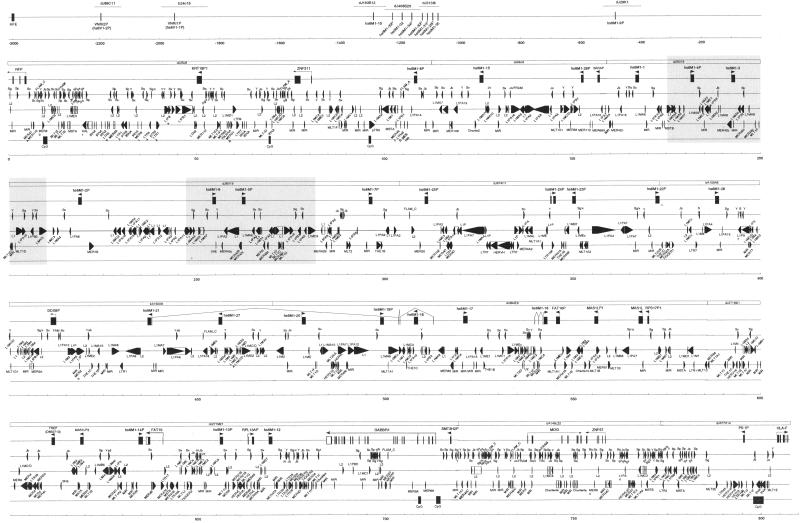
Figure 1.
Genomic organization of the MHC-linked olfactory receptor (OR) genes on chromosome 6. In accordance with the agreed sequence orientation for the human genome, the orientation shown here is from telomere (left) to centromere (right). Except for the top segment, each sequence segment consists of a scale bar, a bar for CpG islands, a bar for short interspersed repeats, a bar for long interspersed repeats, a bar for Alu repeats, a bar for exons, and a bar for the sequenced clone tile path. Classification of all repeats is according to the RepeatMasker program (see Methods). Transcriptional orientations are shown by arrows under the gene names and EST-confirmed splicing in the 5′-UTRs of hs6M1-16 and hs6M1-21 is indicated by interconnecting the corresponding exons. Gene positions in the 3-Mb top segment are approximate. The duplication of a 35-kb segment containing two olfactory receptor genes is boxed grey.
Figure 1 summarizes the genomic organization of the region between RFP and HLA-F that, as defined above, contains the major OR cluster and turns out to be just over 800 kb in size. It reveals the cluster to consist of 25 MOE-type OR loci, of which 12 (hs6M1 -1, -3, -6, -12, -15, -16, -17, -18, -20, -21, -27, -28) have complete open reading frames and, therefore, are predicted to be functional. The remaining 13 loci (hs6M1 -2P, -4P, -5P, -7P, -8P, -13P, -14P, -19P, -22P, -23P, -24P, -25P, -26P) are predicted to be pseudogenes (P) on the basis of disabling stop codons, rearrangements, and/or frameshift mutations. The ratio of genes versus pseudogenes is likely to be different in different individuals owing to OR polymorphism. For instance, we showed previously that the stop codon rendering hs6M1-4P, a pseudogene here, is not present in six out of 10 cell lines tested for OR gene polymorphism (Ehlers et al. 2000; Ziegler et al. 2000a). A similar scenario of gene versus pseudogene status was also established for hs6M1-17, hs6M1-19P, and hs6M1-29P (Ehlers et al. 2000; data not shown). Taken together, these data indicate that the gene versus pseudogene ratio for the MHC-linked major OR cluster is closer to 1 rather than the previously reported average of about 0.3 (Rouquier et al. 1998). In addition, the region also contains a number of other genes, including MOG (Pham-Dinh et al. 1993), GABBR1 (Kaupmann et al. 1997), FAT10 (Liu et al. 1999), ZNF57, the human counterpart of Zfp57 (Okazaki et al. 1994), a novel Mas-like G-protein-coupled receptor (MAS1L), a novel zinc finger protein (ZNF311), and 11 pseudogenes. Interestingly, GABBR1 and MAS1L are also members of the 7-TM superfamily. Dot-matrix analysis of the entire 807-kb region reveals one major duplication event of about 35 kb involving ORs hs6M1-4P/hs6M1-3 and hs6M1-5P/hs6M1-6 (Fig. 1, grey blocks). Comparisons of further coding regions show that more duplications and/or gene conversions are likely to have occurred—examples are hs6M1-12, -13P and -16, which are on average 90% identical (DNA level) to each other (Ziegler et al. 2000b). A detailed phylogenetic analysis of MHC-linked OR genes is described below.
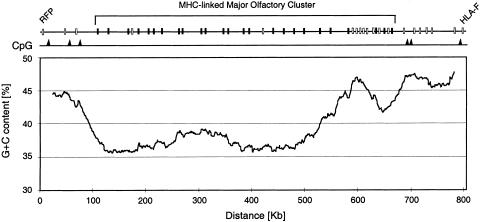
Figure 2 shows a G + C content plot for the MHC-linked major OR cluster and its immediate flanking regions. This analysis defines the entire cluster as a low G + C (average 37.83%) isochore (L-family), including a local G + C increase around position 600 kb owing to the insertion of five non-OR loci (see also Fig. 1). CpG analysis reveals that there are no CpG islands within the cluster but several within the flanking regions. These results are consistent with the observations made on the chromosome 17 cluster. Although the cluster on chromosome 17 also resides within an L-family isochore, it contains four CpG islands but they are not coupled to any of the OR genes (Glusman et al. 2000). The 11 OR loci of the MHC-linked minor cluster (distal of RFP) are only shown at their approximate positions. Among them are two VNO-type 1 pseudogenes, VNRI1P and VNRI2P (also known as hs6V1-1P and hs6V1-2P). Of the remaining nine MOE-type loci, three (hs6M1-10, -32, -35) are predicted to be functional and six (hs6M1-9P, -29P, -30P, -31P, -33P, -34P) are predicted to be pseudogenes (although apparently not in all individuals; see above), giving a gene-to-pseudogene ratio of 0.5 compared with about 1 for the major cluster. In all, we have identified 36 novel human OR loci, resulting in a density of 1 OR per 23 kb for the MHC-linked major cluster. In comparison, the OR cluster on chromosome 17p13.3 is of similar size (412 kb) and of similar density (1 OR per 24 kb) as the MHC-linked major cluster, but it has a higher gene-to-pseudogene ratio (1.83) and no intervening non-OR (pseudo)genes (Glusman et al. 2000). Dense clustering of functionally related genes has also been observed in other gene families and is thought to be advantageous for coordinate regulation (Gumucio et al. 1988; Zimmer et al. 1992; Wright et al. 1995). Quantitative feature analysis (data not shown) shows the major OR cluster to reside within an L-isochore with a distinct preponderance of L1 repeats, confirming the possibility of an L1-mediated duplication mechanism. For instance, the boundary sequences of the block duplication in Figure 1 (grey boxes) are all L1 repeats. A similar L1-mediated mechanism has been shown to be responsible for the duplication of the γ-globin locus (Fitch et al. 1991).
Figure 2.
G + C content plot of the MHC-linked major olfactory receptor (OR) cluster and immediate flanking regions. The mean G + C% (smoothed per 50-kb interval) is plotted per 1 kb at the midpoint of the interval starting at 25 kb. (Black boxes) OR loci, (white boxes) non-OR loci, (black triangles) positions of CpG islands. The average G + C content of the cluster is 37.83% (see also Table 2), defining it as a low G + C (L-family) isochore (Bernardi 1993).
In Silico Transcript Analysis
There is increasing direct and indirect evidence that OR expression is not limited to the MOE. OR-like sequences have been found in a number of other tissues, such as testis (Parmentier et al. 1992), colon, kidney, liver (Dreyer 1998), and heart (Drutel et al. 1995), suggesting a role for ORs outside the olfactory system. Based on in silico transcript analysis of the OR cluster described here, we can confirm and add to this evidence. Screening of publicly available expressed sequence tag (EST) databases produced hits as summarized in Table 1. The overall low hit rate is not surprising, as there are no public EST data available from MOE tissue. Only five out of the 36 MHC-linked ORs show any matches to ESTs >90% similarity. However, these matches confirm that some ORs are transcribed in non-MOE tissue such as lung, kidney, colon, prostate, testis, and germ cell tumors and, therefore, may be involved in nonolfaction-associated function.
Table 1.
List of Expressed Sequence Tags (ESTs) Matching MHC-Linked OR Genes
| OR | Genomic AC no. | EST AC no. | Tissue | EST length | Position in EST | Position in genomic clone | Identity (%) |
|---|---|---|---|---|---|---|---|
| hs6M1-14P | AL031983 | AW071655 | Germ cell tumors | 457 | 1–457 | 84185–83729 | 100 |
| AI912965 | Kidney | 534 | 1–534 | 84185–83652 | 100 | ||
| AI763023 | Kidney | 527 | 1–527 | 84167–83641 | 99 | ||
| AI304583 | Colon | 435 | 1–435 | 84167–83549 | 100 | ||
| AI813634 | Lung | 580 | 1–580 | 84306–84885 | 100 | ||
| AI476350 | Lung, testis, B-cell | 491 | 1–491 | 84306–84795 | 99 | ||
| hs6M1-16 | AL035542 | AI023490 | Testis | 477 | 4–366 | 41365–41727 | 99 |
| 367–477 | 41981–42091 | 100 | |||||
| AA382326 | Testis | 352 | 1–11 | 45171–45161 | 100 | ||
| 12–63 | 43327–43276 | 100 | |||||
| 64–319 | 42236–41981 | 97 | |||||
| 320–352 | 41727–41695 | 91 | |||||
| hs6M1-21 | AL096770 | AA936177 | Lung, testis, B-cell | 387 | 3–169 | 32709–32543 | 100 |
| 170–245 | 81125–81050 | 100 | |||||
| 246–283 | 80707–80670 | 100 | |||||
| 284–387 | 71870–71767 | 100 | |||||
| hs6M1-24P | AL050339 | AA922169 | Lung, testis, B-cell | 385 | 3–157 | 24461–24615 | 100 |
| 158–385 | 27485–27712 | 99 | |||||
| hs6M1-2 | AL133267 | N68399 | Fetal liver, spleen | 428 | 1–324 | 21789–21466 | 99 |
| Z98744 | 325–425 | 57723–57624 | 99 |
All 36 MHC-linked ORs were searched with BLASTN against the human EST database and matching ESTs were aligned with the genomic DNA to determine any splice sites. Where splicing events were identified, the corresponding match positions and identities are given for each exon separately. Some ESTs had been derived from pooled libraries, hence the listing of multiple tissues in such cases.
Alignment of these ESTs to the genomic sequence reveals unusual splicing in the 5′-UTRs of several ORs. For instance, the alignment for hs6M1-21 reveals three 5′-UTR exons and indicates that the primary transcript starts at least 74 kb upstream (position 512 kb in Fig. 1) of the hs6M1-21 ATG start codon (position 438 kb in Fig. 1). The predicted transcript spans four other OR loci, two of which are in the same (hs6M1-18, −27) and two in the opposite (hs6M1-19P, −20) transcriptional orientation. It is quite conceivable that such long transcripts may play a role in the coordinate expression of clustered ORs, for example, via alternative splicing and/or antisense regulation.
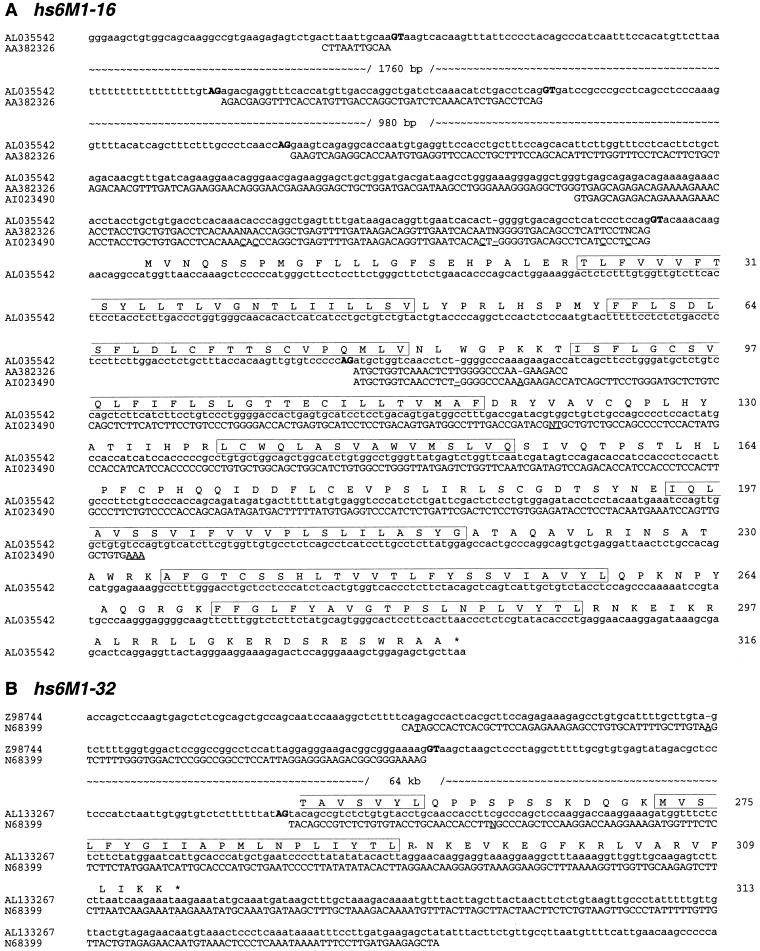
In the case of hs6M1-16 (position 542 kb in Fig. 1), the alignment with two ESTs (both from testis) also reveals 3 exons in the 5′-UTR but only up to 3 kb upstream of the predicted ATG start codon. Interestingly, both ESTs splice around the expected start codon to the third methionine (amino acid position 79) within the single coding exon of hs6M1-16, producing a predicted protein lacking the first 78 amino acids and, therefore, the first two transmembrane domains (Fig. 3A). A similar scenario is also observed for hs6M1-32 (Fig. 3B). In this case, the first half of the EST matches to a presumed noncoding sequence in PAC 193B13 (Z98744), and the second half matches to PAC 408B20 (AL133267) and splices into amino acid position 254 of hs6M1-32,. This results in a potential 5′-UTR of at least 64 kb and, using the first in-frame methionine, a predicted gene product of only 41 amino acids. To our knowledge, these two examples are the first evidence of alternative splicing within the single coding exon of any OR. Alternative splicing or alternative use of ATG start codons may also explain some of the differences observed between mouse and human ORs. hs6M1-14P, for example, is considered a pseudogene because it misses the first 78 amino acids compared to its murine ortholog, mm17M1-6 (see below). Yet, it is the only OR matching a comparatively large number of ESTs all between 99% and 100% similarity and from nonolfaction-associated tissues (Table 1). Although the position of sequence divergence coincides perfectly with the presence of an acceptor splice site, several ESTs span the position, indicating that this splice site is not used—at least not in the tissues from which the ESTs were derived (data not shown). Our interpretation of the data is that, similar to hs6M1-16, hs6M1-14P could make use of an alternative ATG start codon, most likely the one corresponding to the methionine mentioned above for hs6M1-16, resulting again in the omission of the first two transmembrane domains. In fact, the described alternative splicing or use of alternative ATG start codons may be quite common, because the methionine corresponding to amino acid position 79 in Figure 3A is conserved in 62% of the MHC-linked MOE-type ORs presented here. Of these, ten (hs6M1-2P, -7P, -8P, -9P, -15, -16, -21, -22P, -24P, -35) have apparently functional acceptor splice sites that would allow expression from this methionine as for hs6M1-16. The splicing would effectively avoid the frameshift mutations in hs6M1-7P and hs6M1-22P, making these two pseudogenes potentially expressable as proteins. In all examples discussed here, the AGGT splice consensus motif has been preserved and the corresponding splice phases are matching.
Figure 3.
Alignment of ESTs to the genomic sequences of (A) hs6M1-16, (B) hs6M1-32. AG/GT splice sites are highlighted in bold. Long intron sequences are not shown, but their sizes are indicated. The numbers on the right of the alignments refer to the conceptual amino acid positions of the unspliced protein. Positions of sequence disagreement are underlined and predicted transmembrane domains are boxed. Dashes were introduced where required to maximize the alignment. For more details, see Table 1.
Our in silico transcript analysis suggests that some ORs (including ORs currently classified as pseudogenes) may be expressed in a truncated, yet functional, form. Alternative splicing, although not over distances as long as reported here, and the expression of OR pseudogenes have been reported previously (Asai et al. 1996; Crowe et al. 1996; Walensky et al. 1998). Furthermore, the deletion of the first two transmembrane domains (as in the case of hs6M1-16) has been shown not to affect the functional expression of other members of the 7-TM G-protein-coupled receptor gene family (Ling et al. 1999). In this context, it should be noted that alternative splicing is very common. A recent EST-based study showed alternative splicing to take place in 35% of genes in the TIGR human gene index (Mironov et al. 1999). Most of the splicing events occurred within the 5′-UTRs, which was interpreted as evidence for alternative regulation mechanisms. Concerning the MHC-linked ORs, experimental evidence is now needed to determine (1) whether these splice events serve to regulate OR expression, (2) whether they correlate with nonolfaction-associated function, and (3) whether they contribute toward the generation of alternative OR gene products with novel ligand-binding properties. The in silico analysis presented here is a first step in this direction.
Human–Mouse Comparison
Conserved function correlates well with conserved synteny, which makes comparative genomic analyses so informative (Koop and Hood 1994; Baxendale et al. 1995; Ansari-Lari et al. 1998). Comparisons between human and mouse are particularly informative because the two species have diverged enough to distinguish potential coding sequences from noncoding sequences, but not too much for many regulatory sequences to be still identifiable (Hardison et al. 1997). For these and many other reasons, we are interested in analyzing the MHC-linked OR cluster in mouse alongside the human OR cluster. The MHC linkage of the mouse OR cluster on mouse chromosome 17 (also known as Tu42 and Leh89 gene clusters) was established previously (Amadou et al. 1995; Szpirer et al. 1997), and the cloning of the entire cluster is almost complete (Amadou et al. 1999). Here, we report our results from sequencing the first two clones (BACs 573K1 and 332P19) of this contig.
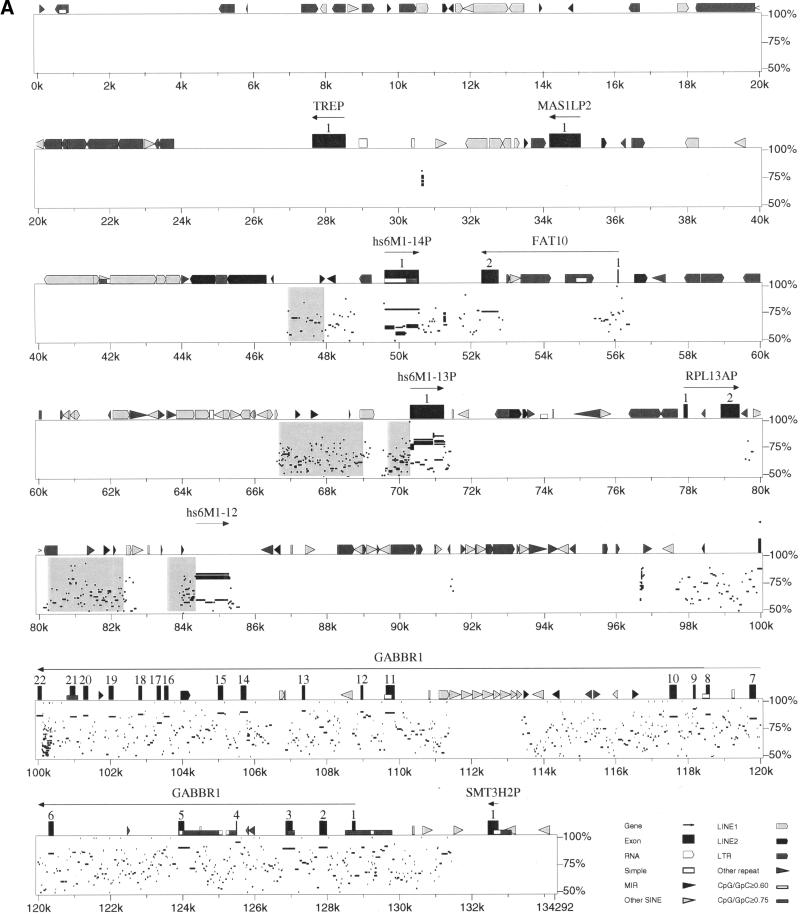
Figure 4A shows a comparison of human PAC 271M21 with mouse BAC 573K1 in a Percent Identity Plot (PIP) (Hardison et al. 1997). Segments of 50%–100% identity between the two sequences are plotted using the coordinates of the subject sequence, in this case the human sequence. Features in the subject sequence such as exons, repeats, and CpG islands are also plotted for orientation. The plot shows clearly that the two sequences are highly related, although the four non-OR pseudogenes (TREP, MAS1LP2, RPL13AP, and SMT3H2P) are not present in the mouse sequence. For instance, all 22 exons of the gamma-amino-butyric acid receptor B1 (GABBR1) are ∼80% identical (DNA level), whereas the introns show recognizable but partial similarity only, and part of intron 10 (position 111–113.5 kb) is not conserved at all owing to human-specific repeat expansion. The three OR loci (hs6M1-12, -13P and -14P) clearly have related genes (>75% DNA identity) in the mouse clone and the presence of multiple stacked homology bars (compared with single bars for GABBR1 and FAT10) indicates additional mouse-specific OR duplications. This becomes more obvious when re-plotting the PIP using the mouse sequence as the subject sequence (data not shown). Figure 4B gives a schematic summary of both analyses. Three loci (GABBR1, FAT10, and hs6M1-14P), including one OR, are identified as true orthologs based on positional and sequence conservation. Although the remaining ORs (hs6M1-12 and -13P in human and mm17M1-1, -2, -3, -4, -5P in mouse) are clearly closely related by sequence, their exact relationship is less obvious because of species-specific duplications (see also phylogenetic analysis below). The remaining species-specific pseudogenes (MAS1LP2, RP13AP and SMT3H2P in human and the Vhl-LP gene fragment in mouse) must all have arisen by insertion or deletion after the two species diverged. Another interesting feature of the PIP analysis is the identification of conserved sequence blocks (Fig. 4A, boxed grey) upstream of all three OR loci, indicating the presence of potential regulatory elements. The conservation of such blocks is consistent with our findings, discussed above, that the 5′-UTRs of ORs can extend over considerable distances upstream of the ATG start codons and may include several splicing events. Experimental work to identify the true 5′-ends of all ORs and to test such potential regulatory elements in functional promoter assays is now in progress.
Figure 4.
(A) Percent identity plot (PIP) of the human-mouse comparison for the centromeric boundary of the MHC-linked olfactory receptor (OR) gene cluster. The two sequences used are accession no. AL031983 for the human sequence and accession no. AL078630 for the mouse sequence. The human sequence was used as the subject sequence and is annotated along the top line. Regions between 50% and 100% conservation to mouse are plotted under the corresponding human positions. The grey shaded boxes mark conserved regions possibly involved in the regulation of the corresponding OR loci. (B) Schematic summary of the human-mouse comparative analysis. OR loci are shown as black boxes and non-OR loci as white boxes. Orthologous gene loci are connected by dotted lines. 'cen' and 'tel' define directions towards centromere and telomere, respectively.
Our comparative analysis suggests at least one orthologous MHC-linked OR and established a high level of conserved synteny between the two OR clusters of human and mouse. In the two mouse clones (BACs 573K1 and 332P19) sequenced thus far a total of 14 OR loci have been identified of which at least 10 (mm17M1-1, -2, -3, -4, -6, -10, -11, -12, -13, -14) are predicted to be expressed. In addition to mm17M1-5P, which has multiple frameshift mutations, ORs mm17M1-7P, -8P, -9P are defined here as pseudogenes owing to a A > G transition at position 1, changing the initiation of translation from a methionine to a valine. The same mutation has been shown before to prevent normal initiation in other human genes (Fojo et al. 1989; Breimer et al. 1994), but it is still possible that these ORs are initiated by the second in-frame methionine at position 33 (see above). In any case, extrapolation from the above numbers indicates that the total number of expressed OR loci in the mouse cluster is higher than in humans, as has been suggested before for the entire murine contingent of OR genes (Mombaerts 1999b).
Phylogeny
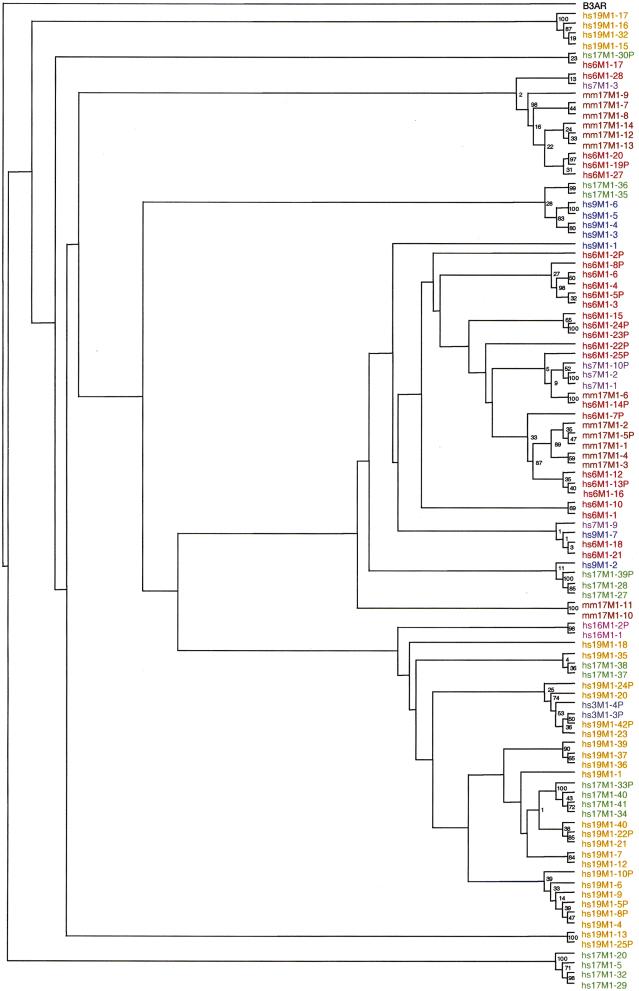
To establish the relationship of the MHC-linked ORs to each other and to ORs from other clusters and species, we performed a phylogenetic analysis. Publicly available ORs were compiled into a BLAST searchable protein database. This database cross-references all original accession numbers, previous gene names, etc., and is available from us (see Methods).
Figure 5 shows a phylogenetic tree of the MHC-linked ORs reported here and representatives from other human and mouse OR clusters. Apart from some notable exceptions, most ORs group on branches corresponding to their respective chromosomal clusters, indicating that local duplication is the main mechanism of OR gene pool expansion. However, local duplications cannot account for all ORs, and there are several examples of ORs that are more closely related to ORs found in other clusters than in their own. hs6M1-17, -18, -19P, -20, -21, -27 and -28, for instance, appear to be the most diverged of the human MHC-linked ORs although hs6M1-19P, -20, -27 and -28 still cluster with MHC-linked ORs from mouse (mm17M1-7, -8, -9, -12, -13 and -14). Regarding the comparison to mouse, the tree confirms orthology between hs6M1-14P and mm17M1-6 (100% bootstrap confidence) and paralogy between hs6M1-12, -13 and mm17M1-1, -2, -3, -4, -5P (87% bootstrap confidence). The only mouse ORs that do not cluster with any other mouse or human MHC-linked ORs are mm17M1-10 and mm17M1-11. They were either inserted into the MHC cluster after divergence of the two species or the human counterparts were deleted.
Figure 5.
Phylogenetic tree of olfactory receptor (OR) genes from the MHC-linked clusters in human and mouse and representatives from other human clusters. The most conserved block of 98 amino acids (including TM2 and TM3) was aligned in 99 ORs, analyzed by the maximum parsimony method and confirmed by 1000 bootstrap replicates (values shown only for most recent divergences). The alignment and all the OR sequences used here are available from our ftp site (see Methods for details). For the alignment of OR pseudogenes (suffixed P), a total of nine dashes were introduced where required to correct for frameshift mutations. The human β-3 adrenergic receptor (B3AR), accession no. P13945, was used as an outgroup.
Conclusions
Apart from our demonstration that the human MHC-linked OR cluster is among the largest in the human genome and shows limited but significant homology to its counterpart in the genome of the mouse, the most intriguing aspect of this study is the EST-based finding of long distance and alternative splicing within the 5′-UTR and coding regions of some OR genes. If experimentally verified, it seems likely that this feature will be connected to regulatory control properties and diverse functions. It remains to be seen whether common control mechanisms govern the expression of OR genes in different species, and different tissues. Our study provides the foundation of such analyses for the MHC-linked OR genes.
METHODS
Mapping, Sequencing and Analysis
A sequence-ready contig of the 800-kb region between HLA-F and RFP was generated by integration of several published contigs (kindly provided by A. Volz, J. Gruen, and D. Ruddy) with clones from the chromosome 6 mapping effort at the Sanger Centre (Lauer et al. 1997; Mungall et al. 1997; Volz et al. 1997; Ahn and Gruen 1999). The contig is part of a 7.5-Mb contig (including the extended MHC) that will be described elsewhere. The corresponding mouse contig was also described previously and was extended by fingerprint analysis of additional clones (Yoshino et al. 1998; Amadou et al. 1999).
A minimum tile path of overlapping clones was selected from both contigs, and each clone was randomly subcloned into M13mp18 and pUC18 (Bankier et al. 1987). Clone-specific details, such as library source and overlap sizes, are given in the corresponding EMBL submission headers. The DNA sequence was determined using the enzymatic dideoxy chain termination sequencing chemistry (Sanger et al. 1977) and automated ABI 373/377/3700 DNA sequencers (Applied Biosystems). The generated reads were quality clipped, screened for cloning and sequencing vectors, and assembled as previously described (The Sanger Centre 1998).
The sequences reported here have been submitted under the following clone names and accession numbers to the EMBL nucleotide databank.
Human: 25J6: Z84476; 88J8: AL035402; 80I19: AL022727; 974I11: AL050339; 150A6: AL096770; 994E9: AL035542; 145L22: AL050328; 271M21: AL031983; 377H14: AL022723; 86C11: AL021807; 24o18: AL021808; 193B12: Z98744; 408B20: AL133267; 313I6: AL121944; 29K1: Z98745.
Mouse: 573K1: AL078630; 332P19:AL133160.
Please note that, for all analyses described here, the sequence of the following accession numbers was inverted to reflect their true genomic orientation (p-telomere to centromere): Z84476, AL050339, AL096770, AL035542, AL031983, AL022723, AL078630.
The sequences were analyzed using the Sanger Centre's analysis strategy (http://www.sanger.ac.uk/HGP/Humana/). The genomic environment analysis was performed using the RepeatMasker program (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker/) to identify repeats in each sequence and parsing the output with a perl script to produce an Excel readable table of the repeat composition. ESTs were identified by BLASTN (Altschul et al. 1990) searching the human EST database at http://www.ncbi.nlm.nih.gov/ and were aligned manually. The PIP of the human and mouse sequences was generated with the advanced PIPmaker program at http://globin.cse.psu.edu/cgi-bin/pipmaker?advanced (Hardison et al. 1997).
The phylogenetic analysis of the human and murine MHC-linked ORs was performed by two different methods (neighbor-joining and maximum parsimony) using the PHYLO_WIN package (Galtier et al. 1996). Alignments were made with CLUSTALW (Thompson et al. 1997) program and some minor manual adjustments. The final alignment is available at ftp.sanger.ac.uk/pub/rmy/Younger_et_al.pdf. Based on distance estimates derived from the Dayhoff Percent Accepted Mutations (PAM250) substitution matrix (Dayhoff et al. 1978), the maximum parsimony (Fitch 1971), and the neighbor-joining (Saitou and Nei 1987) methods were used for tree construction. Both methods produced essentially identical trees confirmed by 1000 bootstrap replicates. Trees were drawn using the TreeView program (Page 1995).
OR Database
Public DNA and protein databases were searched for OR genes that were compiled into a nonredundant BLAST searchable (FASTA format) protein database of 331 ORs, following the naming convention previously proposed by us (Ehlers et al. 2000; Ziegler et al. 2000a). The database cross-references any previous gene names, original accession numbers and, where available, protein identification (PID) numbers and is available from our ftp site (ftp.sanger.ac.uk/pub/rmy/ROLFdb).
Figure 4b.
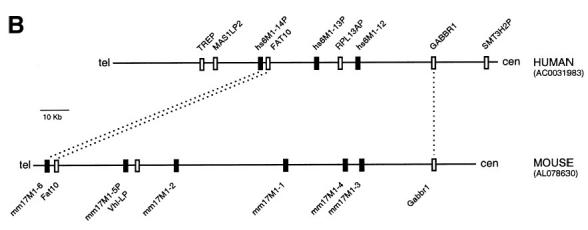
(Continued.) (B) Schematic summary of the human-mouse comparative analysis. OR loci are shown as black boxes and non-OR loci as white boxes. Orthologous gene loci are connected by dotted lines. 'cen' and 'tel' define directions towards centromere and telomere, respectively.
Acknowledgments
We thank all past and present members of the Chromosome 6 Project group (http://www.sanger.ac.uk/HGP/Chr6/), in particular C. Edwards, K. Evans, S. Humphray, M. Mashreghi-Mohammadi, L. Matthews, S. Phillips, V. Rand, S. Sims, S. Smith, A. Tracey, B. Tubby, H. Whitaker, A. Wild, L. Wilming, S. Williams, and J. Rogers. S.B., G.B., R.H., S.M., and A.J.M. were funded by the Wellcome Trust. A.E., S.F., J.T., A.V., and A.Z. were supported by a grant from the Volkswagen-Stiftung. J.T. was funded by a Wellcome Trust program grant. C.A. and K.F.L. were supported by the Howard Hughes Medical Institute, and C.A. also by the IPSEN Foundation. R.M.Y. was supported by a studentship from the UK Medical Research Council (MRC). A.Z. and S.B. also acknowledge the receipt of a Wellcome Trust travel grant.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL beck@sanger.ac.uk; FAX 44 (0) 1223-494919.
Article published on-line before print: Genome Res., 10.1101/gr.160301.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.160301.
REFERENCES
- Ahn J, Gruen J. The genomic organization of the histone clusters on human 6p21.3. Mamm Genome. 1999;10:768–770. doi: 10.1007/s003359901089. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amadou C, Ribouchon M, Mattei M, Jenkins N, Gilbert D, Copeland N, Avoustin P, Pontarotti P. Localization of new genes and markers to the distal part of the human major histocompatibility complex region and comparison with the mouse: New insights into the evolution of mammalian genomes. Genomics. 1995;26:9–20. doi: 10.1016/0888-7543(95)80077-y. [DOI] [PubMed] [Google Scholar]
- Amadou C, Kumanovics A, Jones EP, Lambracht-Washington D, Yoshino M, Fischer Lindahl K. The mouse major histocompatibility complex: Some assembly required. Immunol Rev. 1999;167:211–221. doi: 10.1111/j.1600-065x.1999.tb01394.x. [DOI] [PubMed] [Google Scholar]
- Ansari-Lari MA, Oeltjen JC, Schwartz S, Zhang Z, Muzny DM, Lu J, Gorrell JH, Chinault AC, Belmont JW, Miller W, et al. Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res. 1998;8:29–40. [PubMed] [Google Scholar]
- Asai H, Kasai H, Matsuda Y, Yamazaki N, Nagawa F, Sakano H, Tsuboi A. Genomic structure and transcription of a murine odorant receptor gene: Differential initiation of transcription in the olfactory and testicular cells. Biochem Biophys Res Commun. 1996;221:240–247. doi: 10.1006/bbrc.1996.0580. [DOI] [PubMed] [Google Scholar]
- Bankier AT, Weston KM, Barrell BG. Random cloning and sequencing by the M13/dideoxynucleotide chain termination method. Methods Enzymol. 1987;155:51–93. doi: 10.1016/0076-6879(87)55009-1. [DOI] [PubMed] [Google Scholar]
- Baxendale S, Abdulla S, Elgar G, Buck D, Berks M, Micklem G, Bates G, Brenner S, Beck S, Lehrach H. Comparative sequence analysis of the Human and Pufferfish Huntington's Disease gene. Nat Genet. 1995;10:67–76. doi: 10.1038/ng0595-67. [DOI] [PubMed] [Google Scholar]
- Beck S, Abdulla S, Alderton RP, Glynne RJ, Gut IG, Hosking LK, Jackson A, Kelly A, Newell WR, Sanseau P, et al. Evolutionary dynamics of non-coding sequences within the class II region of the human MHC. J Mol Biol. 1996;255:1–13. doi: 10.1006/jmbi.1996.0001. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Lancet D, Taylor C, Khen M, Walker N, Ledbetter D, Carrozzo R, Sheer D, Lehrach H, North M. Olfactory receptor gene cluster on human chromosome 17: Possible duplication of an ancestral receptor repertoire. Hum Mol Genet. 1994;3:229–235. doi: 10.1093/hmg/3.2.229. [DOI] [PubMed] [Google Scholar]
- Bernardi G. The isochore organization of the human genome and its evolutionary history—a review. Gene. 1993;135:57–66. doi: 10.1016/0378-1119(93)90049-9. [DOI] [PubMed] [Google Scholar]
- Breimer LH, Winder AF, Jay B, Jay M. Initiation codon mutation of the tyrosinase gene as a cause of human albinism. Clin Chim Acta. 1994;227:17–22. doi: 10.1016/0009-8981(94)90131-7. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Crowe ML, Perry BN, Connerton IF. Olfactory receptor-encoding genes and pseudogenes are expressed in humans. Gene. 1996;169:247–249. doi: 10.1016/0378-1119(95)00849-7. [DOI] [PubMed] [Google Scholar]
- Dayhoff M, Schwartz RM, Orcutt BC. A model of evolutionary change in proteins. In: Dayhoff M, editor. Atlas of protein sequence and structure. 5, Suppl. 3. Silver Spring, MD: National Biomedical Research Foundation; 1978. pp. 345–352. [Google Scholar]
- Dreyer W. The area code revisited: Olfactory receptors and other related transmembrane receptors may function as the last digits in a cell surface code for assembling embryos. Proc Natl Acad Sci. 1998;95:9072–9077. doi: 10.1073/pnas.95.16.9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drutel G, Arrang J-M, Diaz J, Wisnewsky C, Schwartz K, Schwartz J-C. Cloning of OL1, a putative olfactory receptor and its expression in the developing rat heart. Receptors Channels. 1995;3:33–40. [PubMed] [Google Scholar]
- Ehlers A, Beck S, Forbes S, Trowsdale J, Uchanska-Ziegler B, Volz A, Younger R, Ziegler A. MHC-Linked olfactory receptor loci exhibit polymorphism and contribute to extended HLA/OR-haplotypes. Genome Res. 2000;10:1968–1978. doi: 10.1101/gr.10.12.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Liu Y-C, Parimoo S, Weissman S. Olfactory receptor-like genes are located in the human major histocompatibility complex. Genomics. 1995;27:119–123. doi: 10.1006/geno.1995.1013. [DOI] [PubMed] [Google Scholar]
- Fitch DH, Bailey WJ, Tagle DA, Goodman M, Sieu L, Slightom JL. Duplication of the γ-globin gene mediated by L1 long interspersed repetitive elements in an early ancestor of simian primates. Proc Natl Acad Sci. 1991;88:7396–7400. doi: 10.1073/pnas.88.16.7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst Zool. 1971;20:406–416. [Google Scholar]
- Fojo SS, de Gennes JL, Chapman J, Parrott C, Lohse P, Kwan SS, Truffert J, Brewer HB., Jr An initiation codon mutation in the apoC-II gene (apoC-II Paris) of a patient with a deficiency of apolipoprotein C-II. J Biol Chem. 1989;264:20839–20842. [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. CABIOS. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Giorgi D, Friedman C, Trask BJ, Rouquier S. Characterization of nonfunctional V1R-like pheromone receptor sequences in human. Genome Res. 2000;10:1979–1985. doi: 10.1101/gr.10.12.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman G, Clifton S, Roe B, Lancet D. Sequence analysis in the olfactory receptor gene cluster on human chromosome 17: Recombinatorial events affecting receptor diversity. Genomics. 1996;37:147–160. doi: 10.1006/geno.1996.0536. [DOI] [PubMed] [Google Scholar]
- Glusman G, Sosinsky A, Ben-Asher E, Avidan N, Sonkin D, Bahar A, Rosenthal A, Clifton S, Roe B, Ferraz C, et al. Sequence, structure and evolution of a complete human olfactory receptor cluster. Genomics. 2000;63:227–245. doi: 10.1006/geno.1999.6030. [DOI] [PubMed] [Google Scholar]
- Gumucio DL, Wiebauer K, Caldwell RM, Samuelson LC, Meisler MH. Concerted evolution of human amylase genes. Mol Cell Biol. 1988;8:1197–1205. doi: 10.1128/mcb.8.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC, Oeltjen J, Miller W. Long human-mouse sequence alignments reveal novel regulatory elements: A reason to sequence the mouse genome. Genome Res. 1997;7:959–966. doi: 10.1101/gr.7.10.959. [DOI] [PubMed] [Google Scholar]
- Kaupmann K, Huggel K, Heid J, Flor PJ, Bischoff S, Mickel SJ, McMaster G, Angst C, Bittiger H, Froestl W, et al. Expression cloning of GABA(B) receptors uncovers similarity to metabotropic glutamate receptors. Nature. 1997;386:239–246. doi: 10.1038/386239a0. [DOI] [PubMed] [Google Scholar]
- Koop BF, Hood L. Striking sequence similarity over almost 100 kilobases of human and mouse T-cell receptor DNA. Nat Genet. 1994;7:48–53. doi: 10.1038/ng0594-48. [DOI] [PubMed] [Google Scholar]
- Lancet D, Ben-Arie N. Olfactory receptors. Curr Biol. 1993;3:668–674. doi: 10.1016/0960-9822(93)90064-u. [DOI] [PubMed] [Google Scholar]
- Lauer P, Meyer N C, Prass C E, Starnes SM, Wolff RK, Gnirke A. Clone-contig and STS maps of the hereditary hemochromatosis region on human chromosome 6p21.3-p22. Genome Res. 1997;7:457–470. doi: 10.1101/gr.7.5.457. [DOI] [PubMed] [Google Scholar]
- Ling K, Wang P, Zhao J, Wu Y-L, Cheng Z-J, Wu G-X, Hu W, Ma L, Pei G. Five-transmembrane domains appear sufficient for a G protein-coupled receptor: Functional five-transmembrane domain chemokine receptors. Proc Natl Acad Sci. 1999;96:7922–7927. doi: 10.1073/pnas.96.14.7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Pan J, Zhang C, Fan W, Collinge M, Bender JR, Weissman SM. A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2. Proc Natl Acad Sci. 1999;96:4313–4318. doi: 10.1073/pnas.96.8.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfroy L, Roth MP, Carrington M, Borot N, Volz A, Ziegler A, Coppin H. Heterogeneity in rates of recombination in the 6-Mb region telomeric to the human major histocompatibility complex. Genomics. 1997;43:226–231. doi: 10.1006/geno.1997.4800. [DOI] [PubMed] [Google Scholar]
- Mironov AA, Fickett JW, Gelfand MS. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P. Molecular biology of odorant receptors in vertebrates. Annu Rev Neurosci. 1999a;22:487–509. doi: 10.1146/annurev.neuro.22.1.487. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Odorant receptor genes in humans. Curr Opin Genet Dev. 1999b;9:315–320. doi: 10.1016/s0959-437x(99)80047-1. [DOI] [PubMed] [Google Scholar]
- Mungall AJ, Humphray SJ, Ranby SA, Edwards CA, Heathcott RW, Clee CM, Holloway E, Peck AI, Harrison P, Green LD, et al. From long range mapping to sequence-ready contigs on human chromosome 6. DNA Seq. 1997;8:151–154. doi: 10.3109/10425179709034066. [DOI] [PubMed] [Google Scholar]
- Okazaki S, Tanase S, Choudhury BK, Setoyama K, Miura R, Ogawa M, Setoyama C. A novel nuclear protein with Zinc fingers down-regulated during early mammalian cell differentiation. J Biol Chem. 1994;269:6900–6907. [PubMed] [Google Scholar]
- Page RD. TreeView: An application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Parmentier M, Libert F, Schurmans S, Schiffmann S, Lefort A, Eggerickx D, Ledent C, Mollereau C, Gerard C, Perret J, et al. Expression of members of the putative olfactory receptor gene family in mammalian germ cells. Nature. 1992;355:453–455. doi: 10.1038/355453a0. [DOI] [PubMed] [Google Scholar]
- Pham-Dinh D, Mattei MG, Nussbaum JL, Roussel G, Pontarotti P, Roeckel N, Mather IH, Artzt K, Fischer Lindahl K, Dautigny A. Myelin/oligodendrocyte glycoprotein is a member of a subset of the immunoglobulin superfamily encoded within the major histocompatibility complex. Proc Natl Acad Sci. 1993;90:7990–7994. doi: 10.1073/pnas.90.17.7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez I, Greer CA, Mok MY, Mombaerts P. A putative pheromone receptor gene expressed in human olfactory mucosa. Nat Genet. 2000;26:18–19. doi: 10.1038/79124. [DOI] [PubMed] [Google Scholar]
- Rouquier S, Taviaux S, Trask BJ, Brand-Arpon V, van den Engh G, Demaille J, Giorgi D. Distribution of olfactory receptor genes in the human genome. Nat Genet. 1998;18:243–250. doi: 10.1038/ng0398-243. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saittou M, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. J Mol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Stephens R, Horton R, Humpray S, Rowen L, Trowsdale J, Beck S. Gene organisation, sequence variation, and isochore structure at the centromeric boundary of the human MHC. J Mol Biol. 1999;291:789–799. doi: 10.1006/jmbi.1999.3004. [DOI] [PubMed] [Google Scholar]
- Sullivan SL, Adamson MC, Ressler KJ, Kozak CA, Buck LB. The chromosomal distribution of mouse odorant receptor genes. Proc Natl Acad Sci. 1996;93:884–888. doi: 10.1073/pnas.93.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpirer C, Szpirer J, Riviere M, Tazi R, Pontarotti P. Mapping of the Olf89 and Rfp genes to the rat genome: Comparison with the mouse and human and new insights into the evolution of the rodent genome. Cytogenet Cell Genet. 1997;78:137–139. doi: 10.1159/000134648. [DOI] [PubMed] [Google Scholar]
- The MHC Sequencing Consortium. Complete sequence and gene map of a human major histocompatibility complex. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- St. Louis The Sanger Centre and The Genome Sequencing Centre. Towards a complete human genome sequence. Genome Res. 1998;8:1097–1108. doi: 10.1101/gr.8.11.1097. [DOI] [PubMed] [Google Scholar]
- Thompson J, Gibson T, Plewniak F, Jeanmougin F, Higgins D. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask BJ, Massa H, Brand-Arpon V, Chan K, Friedman C, Nguyen OT, Eichler E, van den Engh G, Rouquier S, Shizuya H, et al. Large multi-chromosomal duplications encompass many members of the olfactory receptor gene family in the human genome. Hum Mol Genet. 1998a;7:2007–2020. doi: 10.1093/hmg/7.13.2007. [DOI] [PubMed] [Google Scholar]
- Trask BJ, Friedman C, Martin-Gallardo A, Rowen L, Akinbami C, Blankenship J, Collins C, Giorgi D, Iadonato S, Johnson F, et al. Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum Mol Genet. 1998b;7:13–26. doi: 10.1093/hmg/7.1.13. [DOI] [PubMed] [Google Scholar]
- Volz A, Davies A, Ragoussis I, Ziegler A. Dissection of the 5.5 Mbp region directly telomeric of HLA-B including a long range restriction map, YAC and PAC contigs. DNA Seq. 1997;8:181–188. doi: 10.3109/10425179709034071. [DOI] [PubMed] [Google Scholar]
- Walensky LD, Ruat M, Bakin RE, Blackshaw S, Ronnett GV, Snyder SH. Two novel odorant receptor families expressed in spermatids undergo 5′ splicing. J Biol Chem. 1998;273:9378–9387. doi: 10.1074/jbc.273.16.9378. [DOI] [PubMed] [Google Scholar]
- Wright KL, White LC, Kelly A, Beck S, Trowsdale J, Ting JP-Y. Coordinate regulation of the human TAP1 and LMP2 genes from a shared bi-directional promoter. J Exp Med. 1995;181:1459–1471. doi: 10.1084/jem.181.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino M, Xiao H, Jones EP, Kumanovics A, Amadou C, Fischer Lindahl K. Genomic evolution of the distal MHC class I region on mouse chromosome 17. Hereditas. 1997;127:141–148. doi: 10.1111/j.1601-5223.1997.00141.x. [DOI] [PubMed] [Google Scholar]
- Yoshino M, Xiao H, Amadou C, Jones EP, Fischer Lindahl K. BAC clones and STS markers near the distal breakpoint of the fourth t-inversion, In(17)4d, in the H2-M region on mouse Chromosome 17. Mamm Gen. 1998;9:186–192. doi: 10.1007/s003359900723. [DOI] [PubMed] [Google Scholar]
- Ziegler A, Ehlers A, Forbes S, Trowsdale J, Uchanska-Ziegler B, Volz A, Younger R, Beck S. Polymorphic olfactory receptor genes and HLA loci constitute extended haplotypes. In: Kasahara M, editor. Major histocompatibility complex: evolution, structure and function. Tokyo: Springer Verlag; 2000a. pp. 110–130. [Google Scholar]
- Ziegler A, Ehlers A, Forbes S, Trowsdale J, Volz A, Younger R, Beck S. Polymorphisms in olfactory receptor genes: A cautionary note. Hum Immunol. 2000b;61:1281–1284. doi: 10.1016/s0198-8859(00)00219-6. [DOI] [PubMed] [Google Scholar]
- Zimmer M, Medcalf RL, Fink TM, Mattmann C, Lichter P, Jenne DE. Three human elastase-like genes coordinately expressed in the myelomonocyte lineage are organized as a single genetic locus on 19pter. Proc Natl Acad Sci. 1992;89:8215–8219. doi: 10.1073/pnas.89.17.8215. [DOI] [PMC free article] [PubMed] [Google Scholar]