SUMMARY
Tissue microenvironment is an important determinant of carcinogenesis. We demonstrate that ionizing radiation, a known carcinogen, affects cancer frequency and characteristics by acting on the microenvironment. Using a mammary chimera model in which an irradiated host is transplanted with oncogenic Trp53 null epithelium, we show accelerated development of aggressive tumors whose molecular signatures were distinct from non-irradiated hosts. Molecular and genetic approaches show that TGFβ mediated tumor acceleration; molecular signatures implicated TGFβ and genetically reducing TGFβ abrogated the effect on latency. Surprisingly, tumors from irradiated hosts were predominantly estrogen receptor negative. This effect was TGFβ independent and linked to mammary stem cell activity. Thus the irradiated microenvironment affects latency and clinically relevant features of cancer through distinct and unexpected mechanisms.
INTRODUCTION
Currently, very little is known about how early changes in the microenvironment contribute to breast cancer. Ionizing radiation is one of a few demonstrable human breast carcinogens (Land et al., 1980). The prevailing view is that radiation induces cancer through DNA damage (NAS/NRC, 2006). However, this viewpoint is an oversimplification that is inconsistent with many experimental studies showing that ionizing radiation evokes acute and persistent, short and long range, effects (Kaplan et al., 1956; Ehrhart et al., 1997; Amundson et al., 1999a; Mancuso et al., 2008). We and others have postulated that radiation’s carcinogenic potential is perpetuated via so-called non-targeted radiation effects like altered signaling and microenvironment changes (Barcellos-Hoff et al., 2005; Durante and Cucinotta, 2008; Wright, 2010). We established a radiation chimera model in which the mammary gland is cleared of endogenous epithelium before the mouse is irradiated and subsequently transplanted with unirradiated, non-malignant epithelial cells (Barcellos-Hoff and Ravani, 2000). Mice irradiated with a high dose (400 cGy) that were transplanted up to two weeks later with unirradiated, immortalized mammary epithelial cells develop large, aggressive tumors even though normal outgrowths form in non-irradiated hosts.
The challenge remains to demonstrate that non-targeted radiation effects contribute to carcinogenesis following doses relevant to human populations. In the present studies, we use the radiation chimera to assess the frequency, rate and/or characteristics of carcinogenesis of a donor epithelium primed to undergo neoplastic transformation by genetic loss of p53. Carcinogenesis in Tp53 null tissue is similar to human breast cancer in that tumors exhibit genomic instability, differential expression of estrogen receptor α (ER) and heterogeneous histology (Jerry et al., 2000; Medina et al., 2002). Over the course of a more than a year, most (~70%) Trp53 null mammary epithelial transplants in wildtype mouse mammary stroma progress in situ from ductal outgrowths to ductal carcinoma in situ to invasive breast carcinomas (Medina et al., 2002). To test whether specific signals induced by radiation in the microenvironment contribute to its carcinogenic action, we used the Trp53 null mammary chimera in which only the host was exposed to low radiation doses (10–100 cGy) to determine the effect on the latency and type of cancer.
Results
Host irradiation affects development of spontaneous Trp53 null breast cancer
The radiation chimera model consists of surgically clearing the mammary epithelium from the inguinal glands of 3-week old BALB/c mice, irradiating or sham irradiating these mice at 10–12 weeks of age, and transplanting 3 days later with syngeneic mammary fragments (Figure 1A). Based on our prior study using 400 cGy, we first asked whether host irradiation was sufficient to promote cancer of orthotopically transplanted wildtype mammary epithelium. Mice were monitored by palpation for 60 weeks yet no palpable tumors arose from wildtype epithelium in either sham or irradiated hosts. These data indicated that neither transplantation itself (Figure 1B) nor host irradiation alone is sufficient to induce neoplastic transformation in wildtype epithelium.
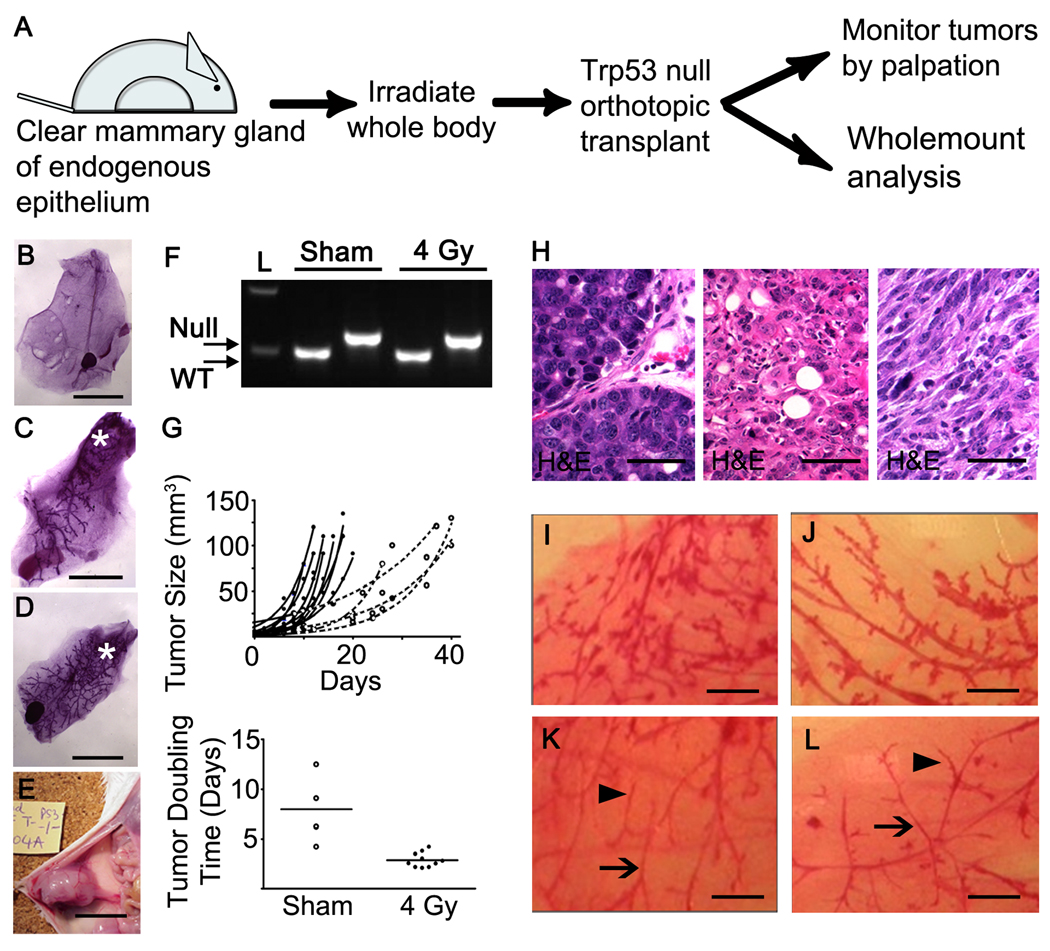
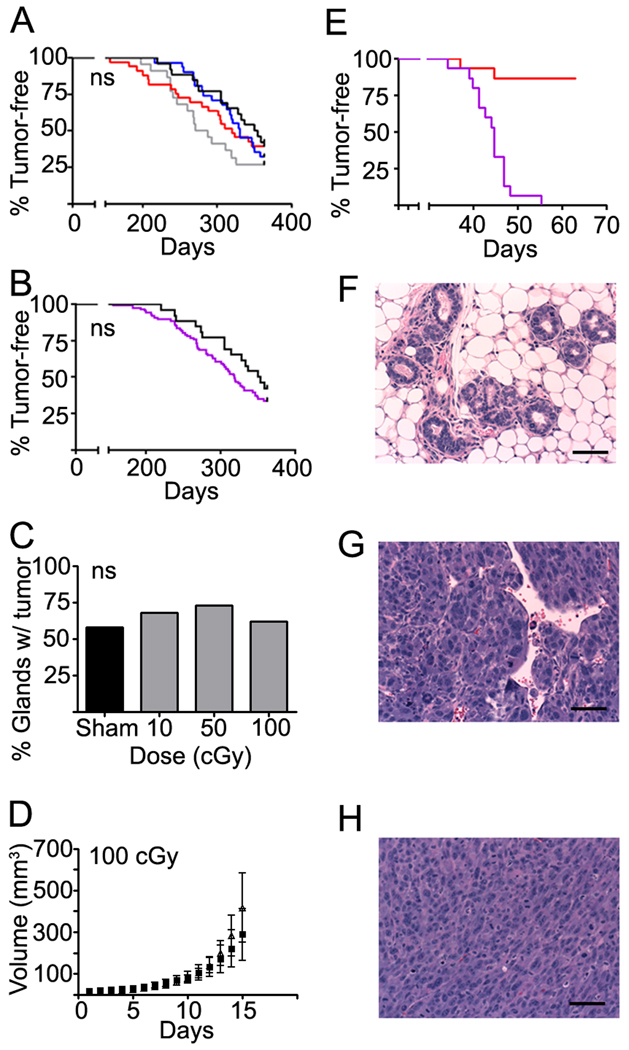
Figure 1. Host irradiation affects tumor features.
(A) Schematic of the experimental protocol. Wholemounts of (B) cleared mammary gland; (C) 6 week Trp53 null outgrowth; (D) 10 week Trp53 null outgrowth; (E) tumor bearing Trp53 null outgrowth. (F) Examples of tumor genotype defined by PCR of wildtype and null allele. (G) Tumor growth rate as a function of host irradiation. Tumors that arose in irradiated hosts grew significantly faster compared to those in the sham group (top panel). Tumor doubling time was approximately 2 days in the irradiated host group compared to 8 days in the sham group (bottom panel). (H) Histopathology of Trp53 mouse mammary tumors: left, adenocarcinoma, middle, squamous cell carcinoma, and right, spindle cell carcinoma; scale bar=100 µm. Wholemounts from Trp53 null epithelium transplanted to mice that were sham-irradiated (I) or irradiated with (J) 100 cGy, (K) 200 cGy, or (L) 400 cGy before transplantation. Doses of 200 cGy and above exhibit reduced branching, thinner ducts (arrows), and lack of alveolar buds (arrow heads), indicative of ovarian insufficiency. Scale bar=1 mm.
In contrast to wildtype epithelium, most Trp53 null transplants developed palpable tumors. The percentage of successful transplants into cleared fat pads was 81±2% S.D. in control hosts (n=55) and 77±10% in irradiated hosts (n=54) in 4 consecutive experiments. Syngeneic Trp53 null mammary outgrowths in wildtype hosts are morphologically normal at weeks 6 and 10 post transplantation (Figure 1C,D). Tumors developed with a similar mean latency in sham (61 weeks ± 7.4 S.D.) and irradiated (63±5.5 S.D.) hosts (Figure 1E), which were confirmed to contain the p53 null allele (Figure 1F). The growth rate of tumors that arose in hosts irradiated with 400 cGy was increased in comparison to sham-irradiated hosts (Figure 1F). As described previously, Trp53 null mammary tumors were diverse in terms of histology, proliferation, lineage markers and ER (Figure 1H). Tumor histological types included poorly differentiated, solid adenocarcinomas with little stroma, spindle-cell morphology, and squamous cell carcinomas.
Unexpectedly, the frequency of Trp53 null tumors in irradiated hosts was reduced by 21% (p<0.01) compared to sham-irradiated hosts. Since women who receive an ovarian dose of >500 cGy have a greatly reduced risk of breast cancer (Inskip et al., 2009) and ovariectomy decreases cancer development by Trp53 null mammary transplants (Medina et al., 2003), we considered the possibility that radiation exposure compromised ovarian function. To test this idea, Trp53 8 week ductal outgrowth were examined. Outgrowths from 400 cGy irradiated mice were noted to have thinner mammary gland ducts and significantly (p<0.001) fewer branches (0.31±0.1/unit length) compared to controls (0.56±0.1). This defect in branching morphogenesis persisted one year after transplantation into hosts irradiated with 200 cGy or more but Trp53 null mammary outgrowths in hosts irradiated with 100 cGy or less were histologically indistinguishable from those of sham-irradiated mice (Figure 1H–K).
To avoid confounding by ovarian effects, and to better represent relevant human exposures, we focused subsequent radiation-chimera experiments on doses of 10–100 cGy (Figure 2). The rate at which tumors developed in transplants increased in irradiated hosts compared to sham-irradiated hosts (Figure 2A). When all radiation dose groups were pooled and compared to the sham-irradiated control group, host irradiation unequivocally accelerated tumorigenesis (Figure 2B). The first tumors were detected at about 170 days post transplantation in both irradiated and non-irradiated hosts but by 300 days, 100% of transplants in hosts irradiated with either 10 or 100 cGy had developed tumors compared to 54% of transplants in unirradiated hosts. Median tumor latency was significantly reduced by 72 day for 10 cGy, 82 days for 100 cGy, and 63 days for all doses pooled compared to sham irradiated mice. At 365 days after transplantation, all outgrowths in irradiated hosts (n=45) had developed tumors compared to 69% (n= 20/29) in sham-irradiated mice (Figure 2C, p<0.05, Chi-square test). Furthermore, as was observed in hosts irradiated with 400 cGy, tumor growth rate increased with increasing host radiation exposure (Figure 2D). Thus, low doses of ionizing radiation altered the course of carcinogenesis, even when radiation was administered in the absence of the epithelium and exposure preceded detectable cancer by many months.
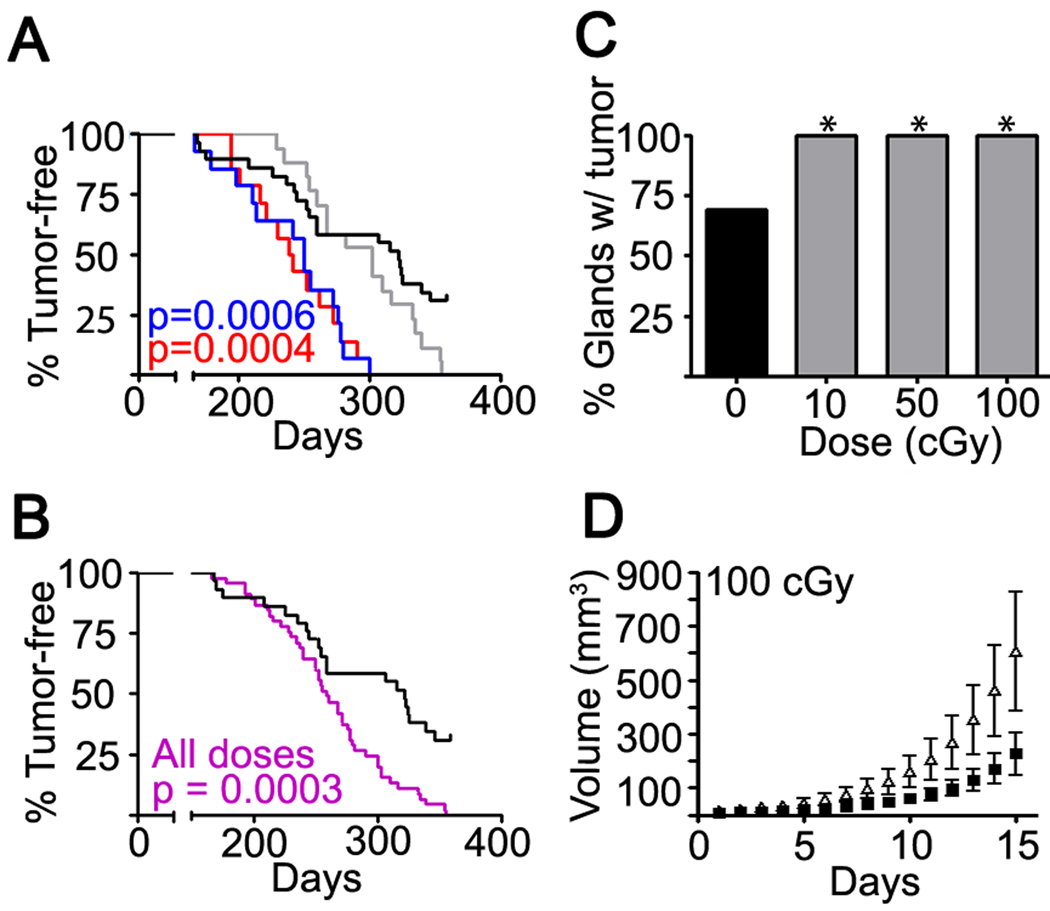
Figure 2. Low dose irradiation promotes tumor development.
(A) Analyses of the time-to-tumor occurrence of tumors in sham (black) and hosts irradiated with 10 (blue), 50 (grey), or 100 (red) cGy. Significance was calculated by the log-rank test. (B) Tumor occurrence in transplants pooled from all radiation doses groups (purple, n=45) compared to sham irradiated controls (black, n=29) was accelerated (p<0.0005, log-rank test). (C) Tumor frequency at experiment termination in each dose group (sham, 20/29; 10, 14/14; 50, 17/17 ; 100 cGy, 14/14; p<0.05, Chi-square test). (D) Trp53 null tumor growth rate was increased in hosts previously irradiated with 100 cGy (open symbols) compared to sham (closed symbols) hosts (bars, SEM). Host irradiation at lower doses showed a similar trend but with wider variance. See also Table S1.
Molecular features of breast cancer are altered by host irradiation
Breast cancer in women is a heterogeneous disease in terms of histology, marker expression and prognosis (Parise et al., 2009), as are breast tumors that develop in Trp53 knockout mice (Jerry et al., 2000). We next considered the possibility that acceleration in irradiated hosts was because the specific tumor type was affected. We classified Trp53 null tumors arising in unirradiated hosts and low dose irradiated hosts by histological type (n=81). Most tumors from unirradiated mice were adenocarcinomas (43%) or spindle cell carcinomas (33%), the remaining tumors were myoepitheliomas or squamous carcinoma (Table S1). Tumor type was not significantly associated with host irradiation status or latency per se.
To further explore how tumors arising in irradiated hosts are distinct from those that occur in non-irradiated hosts, we used Affymetrix gene chips to profile total RNA from individual adenocarcinoma or spindle cell tumors that arose in non-irradiated mice (n=9) and irradiated mice (n=23). Raw data was background normalized and unsupervised hierarchical clustering (UHC) was performed using a 1 S.D. filter cut-off of gene expression change of at least 2-fold that yielded 2,547 probes. UHC did not readily separate tumors on the basis of host irradiation status (Figure 3A). To explicitly compare tumors from irradiated hosts and non-irradiated hosts (reference group), we performed a supervised analysis of genes with a p-value of 0.05 and a minimum 2-fold change, using significance of analysis of microarray (SAM) methodology and permutation analysis under a leave-one-out bootstrap scheme (Tusher et al., 2001). This strategy resulted in 24 genes, which we referred to as the irradiated host core signature (24-IHC), enriched in tumors that developed in irradiated hosts. Using the 24-IHC gene expression list, UHC segregated tumors of non-irradiated hosts from those of irradiated hosts (Figure 3B). Ingenuity pathway analysis (IPA) resulted in two major networks (Figure S1A,B). The first containing 12/21 identified genes described a network characterizing cell morphology and amino acid metabolism; the second containing 10/21 identified genes, associated with cellular movement, cellular growth and proliferation, and cancer. Quantigene validation of expression differences confirmed 22 genes of the 24-IHC; this subset still segregated tumors from irradiated or non-irradiated hosts.
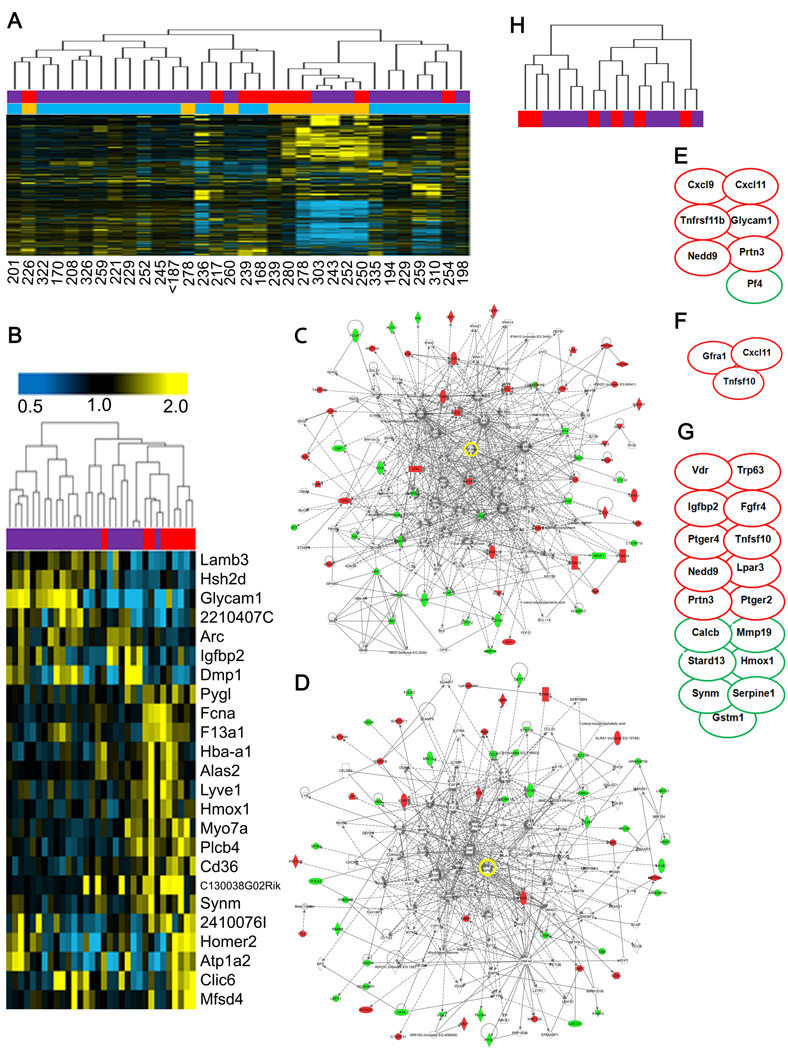
Figure 3. Tumors from irradiated hosts exhibit distinct gene expression.
(A) UHC of Trp53 null mouse tumors based on SD of 1.0 from sham (red) or irradiated (purple) hosts that were either spindle cell carcinoma (gold) or adenocarcinoma (turquoise). Latency for each tumor is listed below the column. (B) Supervised hierarchical clustering of permutation analysis using SAM with a threshold of 2-fold change identified 24 genes that classified tumors that arose in irradiated (purple) hosts versus sham-irradiated (red). The genes of the irradiated host core (24-IHC) are listed at the right. IPA networks of gene interactions among the 24-IHC include cell-to-cell signaling and interaction, cellular development, hematopoiesis, and cellular assembly and organization. (C,D) IPA network of the top two gene networks generated from the 156-IHC. Note that TGFβ is a central node in both networks (yellow circle). IPA of 156-IHC also revealed enrichment for genes involved (E) leukocyte chemo-attraction and binding (p=0.007), (F) monocyte maturation (p=0.006), and (G) proliferation of tumor cell lines (p=0.0007). Red ovals, induced; green ovals, suppressed. (H) Dendrogram of tumor expression profiles based on the 24–IHC genes indicates that unsupervised hierarchical clustering did not segregate tumors from sham-irradiated (red) versus irradiated (purple) Tgfb1 +/− mice. See also Figure S1 and Table S2.
To define the global biology of tumors arising in irradiated hosts, a gene list was generated using a 1.5 fold-change threshold (Table S2), which also segregated tumors from irradiated or non-irradiated hosts (Figure S1C). IPA using these 156 genes invoked cell-cell interaction, cancer, hematological system development, and DNA replication, recombination, and repair (Figure 3C and D). IPA analysis of the 156-IHC also revealed enrichment for genes involved leukocyte chemo-attraction and binding (Figure 3E, p=0.007), monocyte maturation (Figure 3F, p=0.006), and proliferation of tumor cell lines (Figure 3G p=0.0007).
The top-ranked networks contained a node occupied by the cytokine TGFβ1, which although not transcriptionally regulated, is known to play a central role in the response of tissues to radiation. Consistent with this, we found that the 156 gene list significantly overlapped (p=<0.01) gene lists describing mouse mammary tumors driven by cooperation between Wnt and TGFβ (Labbe et al., 2007). Previous work from our group has shown that TGFβ is persistently activated in the irradiated mouse mammary gland (Barcellos-Hoff et al., 1994; Ehrhart et al., 1997). Thus we hypothesized the TGFβ mediates tumor promotion of Trp53 null transplants in irradiated wild type mice.
TGFβ mediates persistent tissue radiation responses
To determine the extent to which radiation changes in gene expression can be attributed to TGFβ, we next conducted comprehensive analysis of the contribution of TGFβ signaling in irradiated mammary gland by expression profiling Tgfb1 heterozygote and wildtype mammary glands at 1 and 4 weeks after whole body exposure to 10 cGy, the lowest dose used in the tumor experiment. Microarray analysis showed that radiation regulated 178 identified genes (p=0.05 and 1.25-fold differences) similarly in Tgfb1 wildtype and heterozygote mammary gland (Figure 4A; Table S3), which constitutes those genes that are independent of TGFβ dose. The top down-regulated genes in both irradiated genotypes suggested that epithelial cell differentiation was affected. Down regulation of amphiregulin (Areg), inhibin beta b (Inhibb), Wnt5a and suppressor of cytokine signaling 3 (Socs3) also suggest decreased differentiation. Up-regulated genes included Adamts18, which is a disintegrin-like and metallopeptidase with thrombospondin type 1 motif indicative of extracellular matrix remodeling, heat shock protein 8 (Hspa8) reflecting persistent stress, and chemokine (C-X-C motif) receptor 4 (Cxcr4) associated with expanding vascular networks. Consistent with these, IPA networks invoked antigen presentation, cell-to-cell signaling and interaction, hematological system development and function.
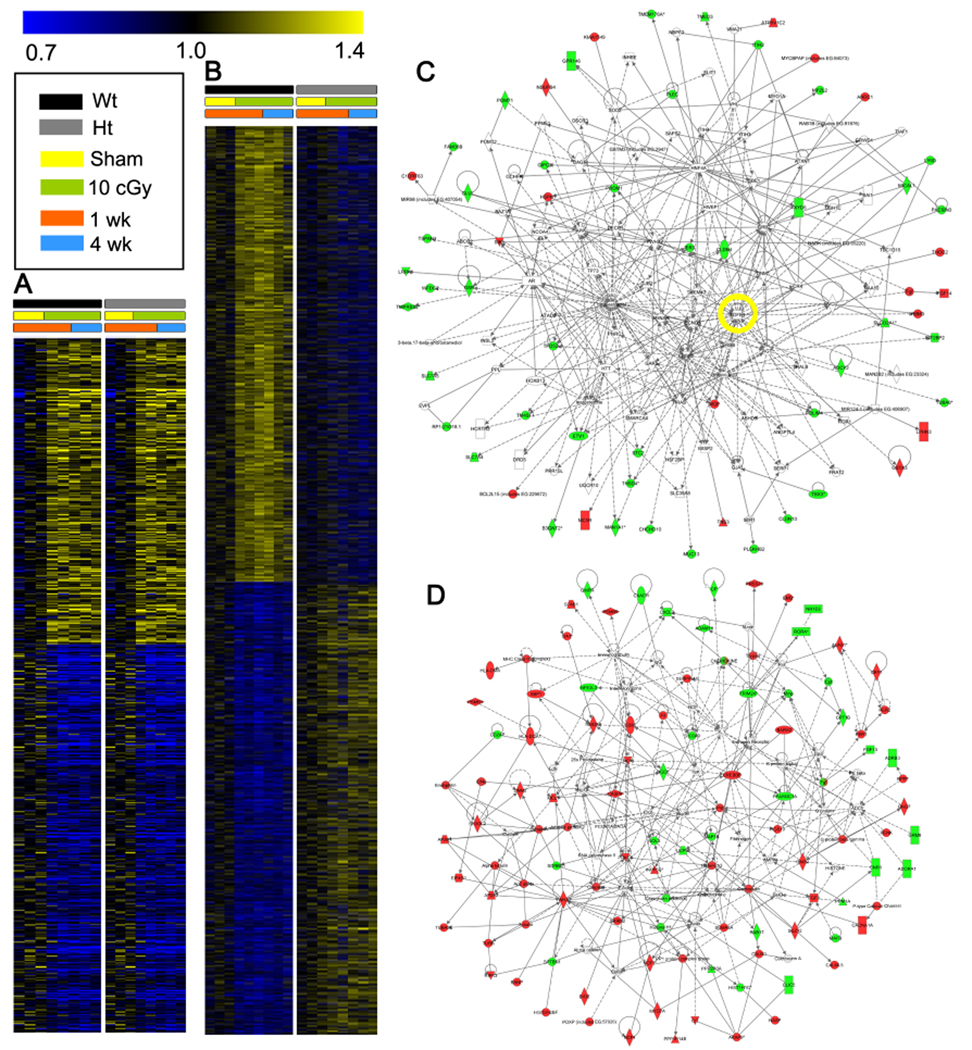
Figure 4. A single low radiation dose elicits persistent changes in gene expression that are highly modulated by TGFβ.
(A) Heat map based on PTM (p<0.05 and threshold of 1.25-fold) for radiation-induced genes common to mammary gland from Tgfb1 wildtype (black) and heterozygote (grey) littermates at 1 (orange) and/or 4 (blue) weeks after sham irradiation (yellow) or 10 cGy (green) exposure. (B) Heat map based on PTM (p<0.05) and threshold of 1.25-fold change for genes that are down regulated (blue) or up regulated (red) in mammary gland from irradiated wildtype (black) but not Tgfb1 heterozygote (grey) littermates at 1 (orange) or 4 (blue) weeks after sham (yellow) or 10 cGy (green) radiation exposure. (C) IPA networks of the genes up-regulated by radiation in both genotypes invoked cellular growth and proliferation, reproductive system development and function, and organismal development. Note TGFβ is a node (yellow circle). (D) IPA network of the genes induced by radiation only in wildtype hosts included functions involved in hematological disease, metabolic disease, and connective tissue development and function. See also Tables S3, S4, and S5.
In contrast, more than twice as many genes (n=488) were regulated by radiation in a TGFβ-dose dependent manner (Figure 4B). TGFβ transcriptional targets, including Tgfbi, Col1a1, and Gadd45b were increased in the wildtype but not Tgfb1 heterozygote gland, consistent with prior studies showing that radiation induces TGFβ activation (Barcellos-Hoff, 1993). IPA of genes regulated only in wildtype mice (Table S4) and TGFβ-dependent, radiation regulated genes (Table S5) identified cell-to-cell signaling, cell signaling and development, and cancer as the top wildtype networks. These analyses support the premise that a low radiation dose elicits persistent changes in gene expression (Amundson et al., 1999b), one third of which are independent and two thirds of which are dependent upon TGFβ dose. In contrast, antigen presentation, cellular assembly and organization, and cell cycle were identified in the expression profiles of irradiated Tgfb1 +/− mammary glands compared to unirradiated tissue, which implicates TGFβ signaling as a critical determinant of the pattern of radiation response. We noted that 29 genes regulated by radiation in mammary gland overlapped the IHC-156 list from tumors arising in irradiated hosts.
TGFβ mediates tumor latency in irradiated hosts
Although the specific epithelial actions of TGFβ suggest that it functions as a tumor suppressor early in cancer (Cui et al.), its roles on development of epithelial cancers in the context of irradiated tissue are unknown. To investigate whether host TGFβ contributes to the radiation effect on Trp53 null carcinogenesis, the radiation-chimera experiment was repeated using syngeneic Tgfb1 +/− hosts. Strikingly, Tgfb1 +/− host irradiation did not affect the frequency, latency, or growth rate of Trp53 null carcinomas (Figure 5A–D), nor molecular characteristics (Table S6), providing strong genetic proof that a critical threshold of TGFβ is an essential facet of radiation induced tumorigenesis and acceleration.
Figure 5. TGFβ promotes carcinogenesis in irradiated hosts.
(A) Kaplan-Meier analyses of the time-to-tumor occurrence in Tgfb1 heterozygote hosts irradiated with sham (black), 10 (blue), 50 (grey), and 100 (red) cGy. Host irradiation did not decrease tumor latency. Significance was calculated by the log-rank test. (B) Tumor occurrence in transplants into Tgfb1 heterozygote hosts pooled from all radiation doses groups (purple, n=86) compared to sham irradiated controls (black, n=26). (C) Tumor incidence of Trp53 null outgrowths does not significantly increase in irradiated Tgfb1 heterozygote hosts compared to sham hosts at 365 days post transplantation. Sham, n=15/26; 10 cGy, n=21/31; 50 cGy, n=16/22; and 100 cGy, n=20/33 (ns, not significant). (D) Tumor growth rate was not affected by host irradiation (bars, SEM). (E) TGFβ treatment significantly (p<0.0001) increased mammary tumor incidence (black) compared to control parental CDβGeo cells (grey) transplanted to cleared mammary glands. (F) Most CDβGeo cells give rise to ductal outgrowths, as shown in a representative tissue section (H&E, bar=50 µm). (G) A few CDβGeo injections give rise to nodular tumors (H&E, bar=50 µm). (H) CDβGeo cells exposed to prolonged TGFβ in vitro rapidly generate solid tumors (H&E, bar=50 µm). See also Table S6.
Given that genetically reducing host TGFβ rescued tumor promotion caused by host irradiation, we asked whether the 24-IHC derived from tumors of irradiated wild type hosts could segregate tumors that arose in non-irradiated Tgfb1 +/− mice (n=6) from those that arose in irradiated hosts (n=10). Neither the 24-IHC nor a similar SAM bootstrap analysis could segregate tumors from non-irradiated versus irradiated Tgfb1 +/− hosts (Figure 3H). As radiation did not accelerate carcinogenesis in the Tgfb1 +/− hosts, these tumors can be considered a validation set of the distinct biology of the microenvironment that accelerates carcinogenesis.
Given that reducing the host TGFβ abolished the radiation effect on tumor latency, we next sought to test whether chronic TGFβ could alter malignant progression. To do so we used a derivative of COMMA-1D cells, CDβGeo, which produce ductal and alveolar structures when transplanted in cleared fat pads (Deugnier et al., 2006) and exposed them to 14 days of continuous TGFβ treatment (5ng/ml) in vitro. These and the parental cells were then injected (500,000 cells/gland) into contralateral inguinal cleared mammary fat pads of WT BALB/c host mice (n=15). As shown previously (Barcellos-Hoff and Ravani, 2000), un-treated parental cells injected into cleared mammary glands mostly gave rise to ductal outgrowths (Figure 5E) and a few nodular tumors (2/15; Figure 5F). In contrast, TGFβ treated CDβGeo cells rapidly formed solid tumors (Figure 5G) with a mean latency of 44 days, such that by 9 weeks all fat pads had tumors compared to 13% of those injected with parental cells (Figure 5H). Together these data support chronic TGFβ activity as the mechanism by which host irradiation accelerates Trp53 null mammary carcinogenesis.
Host Irradiation Mediates Tumor ER Status Independently of TGFβ dosage
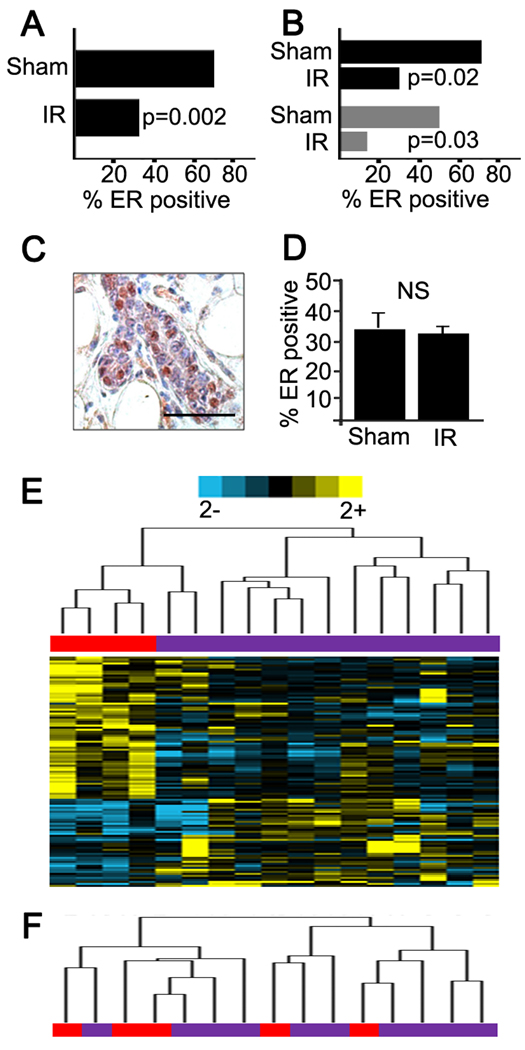
The presence of estrogen receptor is perhaps the most important clinical marker in breast cancer and is associated with distinct risk factors, pathological features, and clinical behavior (Jensen and Jordan, 2003). We determined the ER status of Trp53 null tumors using the Allred scoring system (Harvey et al., 1999). Host irradiation significantly increased development of ER-negative tumors (p=0.002, Fisher’s exact test). Sixty-five percent of tumors that arose in sham-irradiated hosts were ER-positive (28/45, both genotypes) compared to only 35% of tumors (33/93, pooled radiation doses, both genotypes) in irradiated hosts (Figure 6A). This effect of host irradiation to increase ER-negative tumors was observed in both genotypes irradiated with 10 cGy (p<0.05; 5B) and therefore was not associated with the effect of radiation on latency per se.
Figure 6. The frequency of ER negative Trp53 null tumors is increased by host irradiation.
(A) The frequency ER-negative tumors was significantly greater (p<0.002) in irradiated hosts compared to sham hosts. (B) The frequency of ER-negative Trp53 null tumors arising of hosts irradiated with 10 cGy was significantly increased in either host genotype (black, p<0.05; grey, Tgfb1 +/−, p<0.05). (C) ER immunohistochemistry in 5 week old outgrowths of Trp53 null mammary outgrowths; scale bar=100 µm. (D) The frequency ER-positive cells in outgrowths was not affected by host irradiation (sham hosts, 34±6% SEM, n=3 vs irradiated hosts, 31±2% SEM, n=9). (E) The ER-115 profile clusters ER-negative tumors that arose in irradiated (purple) from sham-irradiated (red) hosts. (F) Dendrogram showing the ER-115 does not cluster ER-positive tumors. See also Table S7.
To confirm the distinct biology associated with ER status, we localized progesterone receptor (PR) in a subset of 20 tumors. Most (8/10) ER-positive tumors were PR-positive, while few (4/10) ER-negative tumors were PR-positive. We considered the possibility that the frequency of ER-positive cells Trp53 null outgrowths was affected by host irradiation but the frequency of ER positive cells was unaffected in irradiated compared to control hosts (Figure 6 C,D).
What determines the prevalence of ER negative cancer is not well-understood (Allred et al., 2008). ER negative breast cancer is most frequent in young women and certain racial groups, particularly African-American women (Parise et al., 2009). The observation that irradiated hosts were significantly more likely to give rise to ER-negative and PR-negative tumors implicates radiation-induced heterotypic signaling in determining critical clinical features of breast cancer. We then asked how different the expression profiles of ER-negative tumors were in sham and irradiated hosts as a means to infer whether they develop via similar paths. SAM-tandem-bootstrap identified 115 genes (Table S7) that cluster ER-negative tumors from irradiated versus sham-irradiated hosts (Figure 6E), but not ER-positive tumors (Figure 6F).
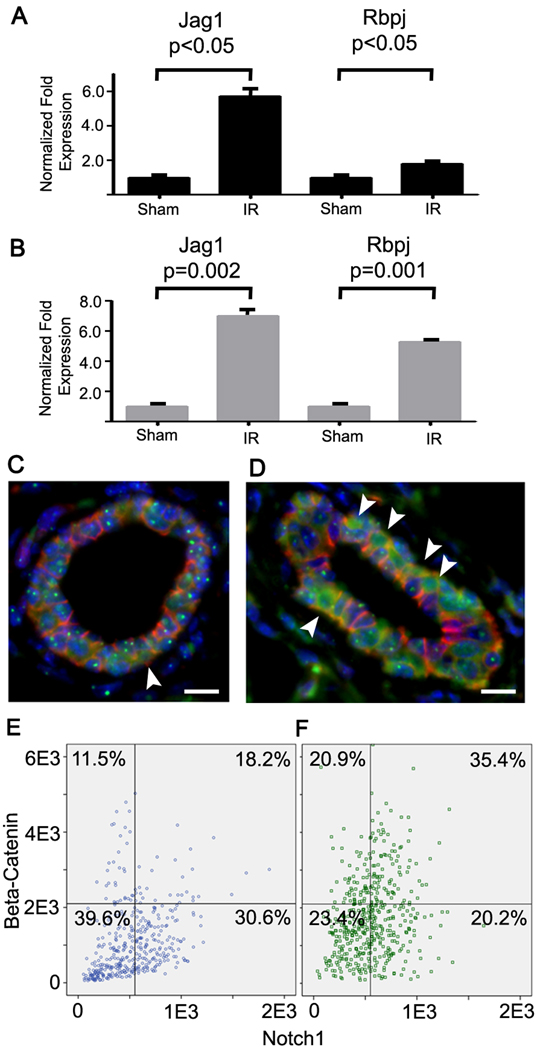
It is has been proposed that breast cancer heterogeneity is determined in part by the cell of origin and its position within the epithelial lineage hierarchy of normal organs (Sell and Pierce, 1994). A corollary is that the tumors retain fundamental programming that remains evident in the biology, behavior, and signature of the cancer subtype. Indeed the expression profiles of isolated mammary stem cells (MaSC), thought to give rise to luminal progenitor (LP) cells that in turn generate mature luminal (ML) cells segregate breast cancers with specific markers and prognosis (Lim et al., 2010). Mouse Trp53 null tumors are similar to claudin-low breast cancer (Prat et al., 2010) and both are enriched in the MaSC signature (Lim et al., 2010). Moreover, neoplastic transformation in this model is thought to be enhanced by increased stem cell self-renewal (Cicalese et al., 2009), which is mediated by Notch signaling (Tao et al., 2010). Notch is preferentially activated in the normal ductal luminal epithelium and promotes commitment of MaSC in vivo (Bouras et al., 2008). We noted significant core enrichment for the Notch pathway in irradiated tissues of both genotypes at 4 weeks when compared to corresponding sham controls. Activation of this pathway was confirmed in an independent experiment using qRT-PCR of Jag1 and Rbpj, which are a key effector and transducer of Notch signaling respectively. Both genes are significantly induced in Tgfb1 wildtype and heterozygote tissues following irradiation with 10 cGy. We then used high content image analysis to localize epithelial Notch based on β-catenin immunoreactivity (Figure 7C, D). We found that nuclear co-localization of both proteins was significantly increased by radiation (Figure E,F). These data suggested that radiation could affect stem cell activity by inducing key regulators of mammary self-renewal and lineage commitment.
Figure 7. Radiation induces Notch and β-catenin activity.
(A) Notch ligand, Jag1, is increased at 1 wk and a transducer of Notch signaling, Rpbj, is increased at 4 wk after irradiation as measured by qRT-PCR (bars, SEM). (B) Notch ligand, Jag1, and a transducer of Notch signaling, Rpbj, are increased at 4 wk in irradiated Tgfb1 heterozygote mammary tissue as measured by qRT-PCR (bars, SEM). (C, D) Dual immunostaining of Notch (green) and β-catenin (red) in mammary epithelium in which nuclei are stained with DAPI (blue; Bar=25 µm). Arrowheads indicate cells that have high nuclear Notch immunoreactivity and β-catenin that are increased in irradiated tissues (D) compared to sham-irradiated tissue (C). (E,F) Multiscale in situ sorting of nuclear Notch and β-catenin immunoreactivity shows that radiation (F; n=486 cells) significantly increased the frequency of Notch positive cells (p<0.05, Fisher’s exact test) and dual stained cells (p<0.001, Fisher’s exact test) compared to sham-irradiated tissues (E; n=424 cells).
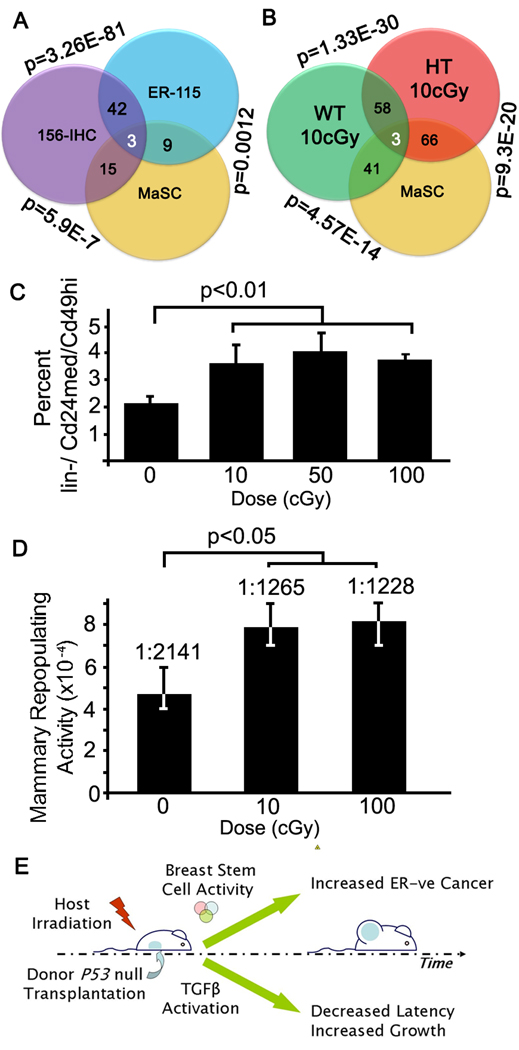
Since the mammary stem cell is ER-negative, as are tumors that are enriched in the MaSC signature, we asked whether the MaSC profile relates to IHC-156 and ER-115 profiles (Figure 8A). Genes up-regulated in the IHC-156 showed a highly significant (p=5.4 E-05) enrichment using ConceptGen for genes up regulated in the MaSc profile, as was the ER-115 signature (p=0.01). These data suggest that tumors arising in the irradiated host have a strong MaSC profile. Similarly, MaSC genes were significantly enriched after irradiation in mammary gland (Figure 8B). Together these data suggested the hypothesis that low dose host irradiation might affect the mammary lineage hierarchy by altering self-renewal in mammary stem cells.
Figure 8. Radiation affects the mammary stem cell pool.
(A) The overlap between the MaSC signature (Lim et al., 2010), IHC-156 and ER-115 is indicated within the Venn diagram and the p-value for enrichment determined with ConceptGen is shown outside the regions of interest. (B) Venn diagram showing the overlap between the MaSC signature and the genes regulated by radiation in the Tgfb1 wildtype (WT) and heterozygote (HT) mammary gland as described for A. (C) Radiation significantly (p<0.01) increased the proportion of lin-/Cd24med/Cd49hi cells determined by FACS analysis of mammary epithelial cells isolated from tissue of mice irradiated 6 weeks before compared to sham-irradiated mice (bars, SEM). Dose was not associated with the degree of response. (D) The mammary repopulating capacity of cells from mice irradiated as in C is significantly increased (p<0.05) as determined by limiting dilution estimation (+95% C.I.). (E) Schematic of distinct mechanisms by which host irradiation affects tumor latency and type.
To test this idea, mice were irradiated with graded low doses at 3 weeks of age and cells isolated from fully mature glands were analyzed by FACs using Cd24med/Cd49hi mammary repopulation markers (Shackleton et al., 2006). Similar cell numbers were recovered from irradiated mouse mammary gland, which is expected for these very low doses and the extended recovery period. The proportion of lin-/ Cd24med/Cd49hi cells in irradiated mice was significantly increased (p<0.05) compared to sham-irradiated mice (Figure 8C). Note the absence of dose dependence, indicating that this effect is not mediated by cell kill per se. Functional analysis of repopulating potential is the gold standard to assess mammary stem cells (Purton and Scadden, 2007). Thus we isolated mammary cells from 8 week old mice that were sham-irradiated or irradiated with 10 cGy at 3 weeks. Mammary repopulating activity increased ~1.7 fold (p<0.05) in irradiated mice compared to sham-irradiated mice, again without evidence of dose dependence (Figure 8D).
Thus, low doses of ionizing radiation induce a tumor promoting microenvironment by two distinct mechanisms (Figure 8E). One mechanism is the induction of TGFβ activity that acts to accelerate tumorigenesis. The other mechanism, which is not affected by host Tgfβ1 haploinsufficiency and hence appears to be unaffected by TGFβ dosage, radiation induction of Notch pathway and mammary stem cell activity, correlates with the increased frequency of ER-negative tumors.
Discussion
Even though engineered mouse models have shown that microenvironment is critical in determining whether cancer ensues from a specific oncogenic event (Bhowmick et al., 2004; de Visser et al., 2006; Kuperwasser et al., 2004), few studies have examined whether carcinogens modify stroma to actively participate in multistep carcinogenesis. Here, we employ the radiation chimera model to provide compelling evidence that a known human carcinogen, ionizing radiation, promote breast cancer through effects on the microenvironment. Several features of carcinogenesis in irradiated hosts parallel those documented in irradiated women: early onset, a more aggressive phenotype and worse prognosis defined by markers. We identified TGFβ in gene expression profiles of irradiated tissue and tumors arising in irradiated hosts, and used a genetic knockdown model to confirm that radiation-induced host TGFβ accelerated carcinogenesis. We also used this combined molecular and genetic approach to show that the effect of radiation on tumor ER status was independent of TGFβ host status and genetically separable from the effect on latency. Rather, radiation induced Notch pathway activation and deregulation of mammary stem cell activity was correlated with ER status of tumors, a mechanism in which radiation alters tissue composition, thus affecting development of specific breast cancer types.
Although it is common in risk modeling to extrapolate from high to low radiation doses, our data demonstrate that radiation effects on cell interactions and host physiology is different at high versus low doses. High dose (4Gy) host irradiation inhibited Trp3 null tumor development, even though mammary ductal outgrowth occurred efficiently. We observed that branching morphogenesis was reduced in mice irradiated with high doses, consistent with ovarian hormone deficiency. Similarly, young women whose cancer treatment induces premature ovarian failure (Inskip et al., 2009) and older women who undergo radiotherapy have reduced risk of breast cancer because ovarian hormones regulate mammary proliferation (Morin Doody et al., 2000).
In contrast, even though many months elapsed between host irradiation and tumor appearance, low radiation doses accelerated cancer and increased tumor growth rate, suggesting a paradigm in which radiation promotes carcinogenesis by altered heterotypic cell interactions. Distinct from rapid molecular responses to DNA damage, signals from irradiated cells can induce a range of events both in distant unirradiated cells and in the progeny of irradiated cells. These phenomena are encompassed under a class of actions now called non-targeted effects postulated to impact radiation carcinogenesis (Barcellos-Hoff et al., 2005; Wright, 2010). The few studies to explicitly test this hypothesis have used high doses that may alter host physiology (Barcellos-Hoff and Ravani, 2000; Kaplan et al., 1956). The lowest dose reported herein is near that at which humans show increased cancer risk.
Prior studies using expression profiles have argued that the biology following low dose radiation differs from that following high doses, but it has proven difficult to use these differences to identify key drivers of processes that affect cancer risk. We identified a gene signature that clustered tumors arising in irradiated hosts from those that arose in naïve hosts. Network analysis of the IHC-156 revealed TGFβ hubs and enrichment of TGFβ mediated genes. Our earlier functional studies showed that radiation-induced TGFβ activation in vivo mediates extracellular matrix remodeling, cell fate decisions, ATM kinase control of the DNA damage response, and EMT (reviewed in (Andarawewa et al., 2007)). Expression analysis of irradiated Tgfb1 heterozygote and wild type mammary gland further underscored the considerable influence of TGFβ in the tissue response to radiation and motivated the radiation chimera experiment using Tgfb1 heterozygote hosts. The radiation chimera model unequivocally demonstrates that radiation-induced host TGFβ mediates promotion in this model, even though the transplanted epithelium is competent to both produce and respond to TGFβ. Consistent with this, we found that mammary epithelial cells chronically exposed to TGFβ in vitro readily progress to tumors in vivo.
The radiation chimera not only showed accelerated carcinogenesis, but altered the expression profiles of tumors that arose from unirradiated epithelium many months after host exposure. Broeks and colleagues reported that gene expression profiles of breast cancers in women treated with radiation for Hodgkin’s lymphoma cluster separately from tumors from unirradiated women diagnosed at the same age and were consistent with a more aggressive tumor type (Broeks et al., 2010). More to the point, young women treated with radiation for childhood cancer not only have a significantly increased risk of breast cancer at an early age, but a much greater likelihood to have ER-negative cancer compared to age matched controls (Castiglioni et al., 2007). Importantly, even though the epithelium is not irradiated in the radiation chimera model, unlike the breasts of women exposed to radiation by diverse medical sources, it recapitulates all of the clinically relevant features of radiation-preceded breast cancer. Thus it not only provides a relevant model of radiation carcinogenesis but also unequivocally shows that the stroma is a pathologically relevant target of radiation.
We found that the frequency of ER-negative mammary tumors increased in irradiated hosts, which was independent of host Tgfb1 haploinsufficiency, was associated with radiation-induced Notch pathway activation and stem cell activity. ER-negative cancers are thought to arise from the early, undifferentiated cells of the mammary gland, either mammary stem cells or luminal progenitor cells (Lim et al., 2010). We hypothesize that radiation exerts significant effects on mammary epithelial hierarchy because the expression profiles of tumors arising in irradiated hosts as well as the irradiated mammary gland significantly overlapped the MaSC profile, recently described by Visvader and colleagues (Lim et al., 2010). Lifetime breast cancer risk is correlated with factors that drive stem cell proliferation (Savarese et al., 2007). We explicitly tested this idea using cell surface markers and functional repopulating capacity in cells isolated from irradiated mice, which showed that radiation significantly increased the mammary repopulating activity, and could thereby increase the number of target cells that could initiate cancer. Taken together these data lead to the hypothesis that aberrant heterotypic interactions induced by radiation early in life may set the stage for stem cell expansion and increase the risk of developing ER-negative breast cancer, as observed in the radiation-chimera and in women treated with radiation for childhood cancers (Castiglioni et al., 2007).
It has become increasingly evident that cell function and dysfunction during cancer development are highly intertwined with the microenvironment (Barcellos-Hoff and Medina, 2005; Bissell et al., 2002; Gonda et al., 2009). Our studies suggest that radiation have very early and persistent effects on the tissue microenvironment that are critical to its carcinogenic potential. Although radiation therapy for cancer is effective, it comes at the price of increased cancer risk that is a life-long burden for cancer patients, particularly those diagnosed during childhood. Radiotherapy for childhood cancers in which breast is exposed dramatically increases breast cancer at an early age (Castiglioni et al., 2007). Our study raises the possibility that cancer risk could be decreased by targeting host biology mediated by TGFβ after radiation exposure.
Experimental Procedures
Mice
All animal experiments were performed at Lawrence Berkeley National Laboratory with institutional review and approval. BALB/c mice were purchased from Simonsen Laboratories (Gilroy, CA) and housed four per cage, fed with Lab Diet 5008 chow and water ad libidum. Trp53 null and Tgfb1 heterozygote BALB/c mice were bred in-house under similar conditions. For transplantation experiments, the epithelial rudiments in inguinal glands of 3-week-old mice were surgically removed. These host mice were irradiated whole body at 10–12 weeks of age to the indicated dose at a rate of 23 cGy/min using 60Co γ-radiation. Three days after irradiation, the cleared mammary glands of host mice were transplanted with a 1 mm3 fragment of non-irradiated Trp53 null BALB/c mammary gland harvested and pooled from 3 or more inguinal glands of 8–10 week old donor mice. Mice were monitored for 365 days.
An informative transplant was defined as that which had an epithelial outgrowth evident by tumor development or confirmation at sacrifice at 12 months. Time to tumor occurrence was plotted using Kaplan-Meier with significance determined by the log-rank test (Graphpad Prizm). Tumor growth curves in a treatment group were fitted to an exponential curve and averaged. Tumors were divided and frozen in liquid nitrogen, embedded in OCT and formalin fixed followed by paraffin embedding.
For tissue analysis, 10 week old Tgfb1 heterozygote and wildtype mice were injected with estrogen (1 µg) and progesterone (1 mg) dissolved in sesame oil 2 days before irradiation with 10 cGy. The lymph node was removed from inguinal mammary glands used to isolate RNA. For mammary stem cell activity, cells were isolated from 5–8 mice sham or irradiated 6 weeks before and processed for lin-/ Cd24med/Cd49hi FACS analysis as described (Shelton et al., 2010) in three experiments with technical triplicates. Mammary repopulation frequency was measured by limiting dilution as described (Illa-Bochaca et al., 2010) using 5 cell doses and 58 mice as recipients per treatment. The repopulating capacity in sham and irradiated mice was compared using L-Calc V1.1.1 (StemCell Technologies).
Immunohistochemistry
Sections were deparaffinized and rehydrated prior to antigen unmasking according to manufacturer’s instructions (Vector Labs, #H-3300), washed once with phosphate buffer saline and blocked with 0.5% casein and 0.1% Tween 20/PBS for 1 hr at room temperature. Primary antibody for ER C1355 (Millipore/Upstate, #06-935), PR (Neomarkers), αSMA (Sigma, # A2547), K6 (Covance, #PRB-169P), and K14 (Covance, #PRB-155P) were diluted in Superblock Blocking Buffer (Pierce, #37515) and refrigerated overnight. The slides were washed, followed by incubation with fluorochrome-conjugated secondary antibody, washed and counterstained with DAPI (2 µg/ml; Molecular Probes). Histopathological characteristics of the tumors were reviewed by two observers blinded to the experimental details of the mouse models. Tumors were classified and staining was analyzed by two pathologists (JSR-F & FCG) as previously described (McCarthy et al., 2007). ER scoring using the Allred scoring system (Harvey et al., 1999). Notch and β–catenin dual localization was assessed using multiscale in situ sorting (Fernandez-Gonzalez et al., 2009).
Expression Profiling
Total RNA quality and quantity was determined using Agilent 2100 Bioanalyzer and Nanodrop ND-1000. Affymetrix mouse Genechip MG-430 2.0 arrays were used according to manufacturer’s protocol. Background normalization was done using R software v2.10.1 with widgets specific to the Affymetrix platform. UHC was done using Gene Cluster v3.0 software and heatmaps were visualized using Java TreeView v1.1.4r3 software. No filter was used unless specified as a SD of 1.0 relative to the expression values of that gene across all samples. Adjusted data means of gene expression values was centered by medians. Gene clustering was done by an uncentered-correlation and array clustering was done by Spearman Rank correlation.
Affymetrix CEL files were normalized using Robust Multichip Average algorithm (Bolstad et al., 2003) in GeneSpring GX software (Agilent Technologies) and each probe was normalized to the median value of the unirradiated specimens for each genotype. Genes differentially regulated in mammary gland tissues were identified by feature selection algorithm Pavlidis template matching (Pavlidis and Noble, 2001) using a p-value <0.05 for pathway analysis. Heatmaps were incorporated in the multi-experiment viewer of the TIGR TM4 Analysis package (Saeed et al., 2003). Pathways were identified with Ingenuity Pathway Analysis, ConceptGen (http://conceptgen.ncibi.org/core/conceptGen/index.jsp), L2L (Newman and Weiner, 2005), or Gene Set Enrichment Analysis using MolDig v3 database. QuantiTect primers for murine Gapdh, Notch1, Jag1, Jag2, and Rbpj (Qiagen) were used with Qiagen's QuantiTect SYBR Green PCR Kit on a BioRad CFX96 Thermal Cycler according to manufacturer's protocols.
Accession Number
NCBI Gene Expression Omnibus database accession number for irradiated mammary glands and tumors is GSE18216.
Statistical Analysis
Statistical analysis was performed using Prism (GraphPad). Differences between treatment groups was determined using the Chi-square test or two tailed Student’s t-test for differences, which were considered statistically significant at P<0.05.
Supplementary Material
Acknowledgments
The authors thank Drs. P. Cowin, C. Allred and P. Williams for helpful discussion, Dr. J. Paupert and Y. Huang for assistance with slide scoring, W. Chou, B. Chu, R. Chou and B. Yang for technical assistance, and Y. Zhang of the NYU Cancer Institute Genomics Facility for generating the microarray data. This research was supported by a DOD-BCRP predoctoral fellowship to DHN, funds from Breakthrough Breast Cancer to FCG and JSR-F; and grants to MHBH from California Breast Cancer Research Program, the Department of Energy, Office of Biological and Environment Research Low Dose program, and the Breast Cancer and the Environment Research Centers grant number U01 ES/CA 012801 (MHBH) and 012800 (SZH) from the National Institute of Environmental Health Sciences and the National Cancer Institute of the National Institutes of Health.
Abbreviations
- TGFβ
Transforming growth factor β1
- ER
estrogen receptor α
- PR
progesterone receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: M.H.B.H. designed research; H.A.O.R., I.I.B, S.A.R., S. H., F.C.G., J.S.R.-F., A.D.B. and D.H.N. performed research; D.J.J., and D.M. contributed reagents; D.H.N., I.I.B., J.Z., J-H. M. and M.H.B.H. analyzed data; and D.H.N., D.M. and M.H.B.H wrote the paper.
References
- Allred DC, Wu Y, Mao S, Nagtegaal ID, Lee S, Perou CM, Mohsin SK, O'Connell P, Tsimelzon A, Medina D. Ductal Carcinoma In situ and the Emergence of Diversity during Breast Cancer Evolution. Clin Cancer Res. 2008;14:370–378. doi: 10.1158/1078-0432.CCR-07-1127. [DOI] [PubMed] [Google Scholar]
- Amundson SA, Bittner M, Chen Y, Trent J, Meltzer P, Fornace AJJ. Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene. 1999a;18:3666–3672. doi: 10.1038/sj.onc.1202676. [DOI] [PubMed] [Google Scholar]
- Amundson SA, Do KT, Fornace AJJ. Induction of stress genes by low doses of gamma rays. Radiat Res. 1999b;152:225–231. [PubMed] [Google Scholar]
- Andarawewa KL, Kirshner J, Mott JD, Barcellos-Hoff MH. TGFb: Roles in DNA damage responses. In: Jakowlew S, editor. Transforming Growth Factor-Beta in Cancer Therapy, Volume II Cancer Treatment and Therapy. Totowa: Humana Press; 2007. pp. 321–334. [Google Scholar]
- Barcellos-Hoff MH. Radiation-induced transforming growth factor b and subsequent extracellular matrix reorganization in murine mammary gland. Cancer Res. 1993;53:3880–3886. [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Derynck R, Tsang ML-S, Weatherbee JA. Transforming growth factor-b activation in irradiated murine mammary gland. J Clin Invest. 1994;93:892–899. doi: 10.1172/JCI117045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Medina D. New highlights on stroma-epithelial interactions in breast cancer. Breast Cancer Res. 2005;7:33–36. doi: 10.1186/bcr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Park C, Wright EG. Radiation and the microenvironment - tumorigenesis and therapy. Nat Rev Cancer. 2005;5:867–875. doi: 10.1038/nrc1735. [DOI] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Ravani SA. Irradiated mammary gland stroma promotes the expression of tumorigenic potential by unirradiated epithelial cells. Cancer Res. 2000;60:1254–1260. [PubMed] [Google Scholar]
- Bhowmick NA, Chytil A, Plieth D, Gorska AE, Dumont N, Shappell S, Washington MK, Neilson EG, Moses HL. TGF-{beta} signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303:848–851. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- Bissell MJ, Radisky DC, Rizki A, Weaver VM, Petersen OW. The organizing principle: microenvironmental influences in the normal and malignant breast. Differentiation. 2002;70:537–546. doi: 10.1046/j.1432-0436.2002.700907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat M-L, Oakes SR, Lindeman GJ, Visvader JE. Notch Signaling Regulates Mammary Stem Cell Function and Luminal Cell-Fate Commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Broeks A, Braaf LM, Wessels LF, Van de Vjver M, De Bruin ML, Stovall M, Russell NS, van Leeuwen FE, Van 't Veer LJ. Radiation-Associated Breast Tumors Display a Distinct Gene Expression Profile. International journal of radiation oncology, biology, physics. 2010;76:540–547. doi: 10.1016/j.ijrobp.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Castiglioni F, Terenziani M, Carcangiu ML, Miliano R, Aiello P, Bertola L, Triulzi T, Gasparini P, Camerini T, Sozzi G, et al. Radiation effects on development of HER2-positive breast carcinomas. Clin Cancer Res. 2007;13:46–51. doi: 10.1158/1078-0432.CCR-06-1490. [DOI] [PubMed] [Google Scholar]
- Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, Pelicci PG. The Tumor Suppressor p53 Regulates Polarity of Self-Renewing Divisions in Mammary Stem Cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- Cui W, Fowlis DJ, Bryson S, Duffie E, Ireland H, Balmain A, Akhurst RJ. TGFb1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell. 1996;86:531–542. doi: 10.1016/s0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- Deugnier M-A, Faraldo MM, Teulière J, Thiery JP, Medina D, Glukhova MA. Isolation of mouse mammary epithelial progenitor cells with basal characteristics from the Comma-D[beta] cell line. Developmental Biology. 2006;293:414–425. doi: 10.1016/j.ydbio.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Durante M, Cucinotta FA. Heavy ion carcinogenesis and human space exploration. Nat Rev Cancer. 2008;8:465–472. doi: 10.1038/nrc2391. [DOI] [PubMed] [Google Scholar]
- Ehrhart EJ, Carroll A, Segarini P, Tsang ML-S, Barcellos-Hoff MH. Latent transforming growth factor-b activation in situ: Quantitative and functional evidence following low dose irradiation. FASEB J. 1997;11:991–1002. doi: 10.1096/fasebj.11.12.9337152. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, Illa-Bochaca I, Welm BE, Fleisch MC, Werb Z, Ortiz-de-Solorzano C, Barcellos-Hoff MH. Mapping mammary gland architecture using multi-scale in situ analysis. Integr Biol. 2009;1:80–89. doi: 10.1039/b816933k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda TA, Tu S, Wang TC. Chronic inflammation, the tumor microenvironment and carcinogenesis. Cell Cycle. 2009;8:2005–2013. doi: 10.4161/cc.8.13.8985. [DOI] [PubMed] [Google Scholar]
- Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- Illa-Bochaca I, Fernandez-Gonzalez R, Shelton DN, Welm BE, Ortiz-de-Solorzano C, Barcellos-Hoff MH. Limiting-dilution transplantation assays in mammary stem cell studies. Methods Mol Biol. 2010;621:29–47. doi: 10.1007/978-1-60761-063-2_2. [DOI] [PubMed] [Google Scholar]
- Inskip PD, Robison LL, Stovall M, Smith SA, Hammond S, Mertens AC, Whitton JA, Diller L, Kenney L, Donaldson SS, et al. Radiation Dose and Breast Cancer Risk in the Childhood Cancer Survivor Study. J Clin Oncol. 2009;27:3901–3907. doi: 10.1200/JCO.2008.20.7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen EV, Jordan VC. The Estrogen Receptor: A Model for Molecular Medicine. Clin Cancer Res. 2003;9:1980–1989. [PubMed] [Google Scholar]
- Jerry DJ, Kittrell FS, Kuperwasser C, Laucirica R, Dickinson ES, Bonilla PJ, Butel JS, Medina D. A mammary-specific model demonstrates the role of the p53 tumor suppressor gene in tumor development. Oncogene. 2000;19:1052–1058. doi: 10.1038/sj.onc.1203270. [DOI] [PubMed] [Google Scholar]
- Kaplan HS, Carnes WH, Brown MB, Hirsch BB. Indirect Induction of Lymphomas in Irradiated Mice: I. Tumor Incidence and Morphology in Mice Bearing Nonirradiated Thymic Grafts Cancer Res. 1956;16:422–425. [PubMed] [Google Scholar]
- Kuperwasser C, Chavarria T, Wu M, Magrane G, Gray JW, Carey L, Richardson A, Weinberg RA. From The Cover: Reconstruction of functionally normal and malignant human breast tissues in mice. PNAS. 2004;101:4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe E, Lock L, Letamendia A, Gorska AE, Gryfe R, Gallinger S, Moses HL, Attisano L. Transcriptional Cooperation between the Transforming Growth Factor-{beta} and Wnt Pathways in Mammary and Intestinal Tumorigenesis. Cancer Res. 2007;67:75–84. doi: 10.1158/0008-5472.CAN-06-2559. [DOI] [PubMed] [Google Scholar]
- Land CE, Boice JD, Shore RE, Norman JE, Tokunaga M. Breast cancer risk from low-dose exposures to ionizing radiation: Results of parallel analysis of three exposed populations of women. J Natl Cancer Inst. 1980;65:353–376. [PubMed] [Google Scholar]
- Lim E, Wu D, Pal B, Bouras T, Asselin-Labat M-L, Vaillant F, Yagita H, Lindeman GJ, Smyth GK, Visvader JE. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Br Ca Res. 2010;12:R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso M, Pasquali E, Leonardi S, Tanori M, Rebessi S, Di Majo V, Pazzaglia S, Toni MP, Pimpinella M, Covelli V, Saran A. Oncogenic bystander radiation effects in Patched heterozygous mouse cerebellum. Proc Natl Acad Sci (USA) 2008;105:12445–12450. doi: 10.1073/pnas.0804186105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy A, Savage K, Gabriel A, Naceur C, Reis-Filho J, Ashworth A. A mouse model of basal-like breast carcinoma with metaplastic elements. J Pathol. 2007;211:389–398. doi: 10.1002/path.2124. [DOI] [PubMed] [Google Scholar]
- Medina D, Kittrell FS, Shepard A, Contreras A, Rosen JM, Lydon J. Hormone Dependence in Premalignant Mammary Progression. Cancer Res. 2003;63:1067–1072. [PubMed] [Google Scholar]
- Medina D, Kittrell FS, Shepard A, Stephens LC, Jiang C, Lu J, Allred DC, McCarthy M, Ullrich RL. Biological and genetic properties of the p53 null preneoplastic mammary epithelium. Faseb J. 2002;16:881–883. doi: 10.1096/fj.01-0885fje. [DOI] [PubMed] [Google Scholar]
- Morin Doody M, Lonstein JE, Stovall M, Hacker DG, Luckyanov N, Land CE. Breast cancer mortality after diagnostic radiography: findings from the U.S. Scoliosis Cohort Study. Spine. 2000;25:2052–2063. doi: 10.1097/00007632-200008150-00009. [DOI] [PubMed] [Google Scholar]
- NAS/NRC. Board on Radiation Effects Research (BEIRVII) Washington: National Academy Press; 2006. Health Risks from Exposure to Low Levels of Ionizing Radiation: Phase 2. [Google Scholar]
- Newman JC, Weiner AM. L2L: a simple tool for discovering the hidden significance in microarray expression data. Genome Biol. 2005;6:R81. doi: 10.1186/gb-2005-6-9-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parise CA, Bauer KR, Brown MM, Caggiano V. Breast Cancer Subtypes as Defined by the Estrogen Receptor (ER), Progesterone Receptor (PR), and the Human Epidermal Growth Factor Receptor 2 (HER2) among Women with Invasive Breast Cancer in California, 1998–2004. The Breast Journal. 2009;15:593–602. doi: 10.1111/j.1524-4741.2009.00822.x. [DOI] [PubMed] [Google Scholar]
- Pavlidis P, Noble W. Analysis of strain and regional variation in gene expression in mouse brain. Genome Biology. 2001;2 doi: 10.1186/gb-2001-2-10-research0042. research0042.0041 - research0042.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular charcterization of the claudin-low intrinsic subtype of breast cancer. Br Ca Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purton LE, Scadden DT. Limiting Factors in Murine Hematopoietic Stem Cell Assays. Cell Stem Cell. 2007;1:263–270. doi: 10.1016/j.stem.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Savarese T, Strohsnitter W, Low H, Liu Q, Baik I, Okulicz W, Chelmow D, Lagiou P, Quesenberry P, Noller K, Hsieh C-C. Correlation of umbilical cord blood hormones and growth factors with stem cell potential: implications for the prenatal origin of breast cancer hypothesis. Breast Cancer Research. 2007;9:R29. doi: 10.1186/bcr1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sell S, Pierce GB. Maturation arrest of stem cell differentiation is a common pathway for the cellular origin of teratocarcinomas and epithelial cancers. Lab Invest. 1994;70:6–22. [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shelton DN, Fernandez-Gonzalez R, Illa-Bochaca I, Ortiz-de-Solorzano C, Barcellos-Hoff MH, Welm BE. Use of stem cell markers in dissociated mammary populations. Methods Mol Biol. 2010;621:49–55. doi: 10.1007/978-1-60761-063-2_3. [DOI] [PubMed] [Google Scholar]
- Tao L, Roberts AL, Dunphy KA, Bigelow C, Yan H, Jerry DJ. Repression of Mammary Stem/Progenitor Cells by P53 is Mediated by Notch and Separable from Apoptotic Activity. STEM CELLS, N/A-N/A. 2010 doi: 10.1002/stem.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright EG. Manifestations and mechanisms of non-targeted effects of ionizing radiation. Mutat Res. 2010;687:28–33. doi: 10.1016/j.mrfmmm.2010.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.