Abstract
Different steps in gene expression are intimately linked. In Saccharomyces cerevisiae, the conserved TREX complex couples transcription to nuclear messenger RNA (mRNA) export. However, it is unknown how TREX is recruited to actively transcribed genes. Here, we show that the Prp19 splicing complex functions in transcription elongation. The Prp19 complex is recruited to transcribed genes, interacts with RNA polymerase II (RNAPII) and TREX, and is absolutely required for TREX occupancy at transcribed genes. Importantly, the Prp19 complex is necessary for full transcriptional activity. Taken together, we identify the Prp19 splicing complex as a novel transcription elongation factor that is essential for TREX occupancy at transcribed genes and that thus provides a novel link between transcription and messenger ribonucleoprotein (mRNP) formation.
Keywords: transcription, mRNA export, TREX, Syf1, Prp19 complex
Transcription of protein-coding genes by RNA Polymerase II (RNAPII) is facilitated by a multitude of transcription factors. Transcription initiation factors regulate transcription and recruit RNAPII to the promoter. When RNAPII starts to synthesize the messenger RNA (mRNA), initiation factors are exchanged for elongation factors (Pokholok et al. 2002). Transcription elongation factors are thought to increase the processivity of RNAPII by facilitating chromatin passage as well as mRNA processing (Hirose and Manley 2000; Orphanides and Reinberg 2000, 2002; Ahn et al. 2004; Mason and Struhl 2005; Perales and Bentley 2009). Consistently, it has been shown in recent years that transcription and downstream processes such as capping, splicing, polyadenylation, cleavage, packaging of the mRNA into a mature messenger ribonucleoprotein (mRNP), mRNP quality control, and mRNP export are intimately linked (Maniatis and Reed 2002; Reed 2003; Stutz and Izaurralde 2003; Iglesias and Stutz 2008; Komili and Silver 2008; Carmody and Wente 2009).
In Saccharomyces cerevisiae, the conserved TREX complex couples transcription to mRNP export (for review, see Reed and Cheng 2005; Kohler and Hurt 2007; Carmody and Wente 2009; Katahira and Yoneda 2009; Rondon et al. 2010). TREX consists of the heterotetrameric THO complex (comprised of the proteins Tho2, Hpr1, Mft1, and Thp2), the intranuclear mRNA export factors Sub2 and Yra1, the SR proteins Gbp2 and Hrb1, and Tex1 (Strasser et al. 2002). The THO subcomplex facilitates efficient transcription elongation (Rondon et al. 2003). In addition, it prevents hyperrecombination, most likely by preventing the formation of DNA–RNA hybrids, so-called R-loops, during transcription elongation, thus linking transcription to genome stability (Jimeno et al. 2002; Huertas and Aguilera 2003). Furthermore, TREX functions in 3′ end processing and transcription-coupled DNA repair (TCR) (Gaillard et al. 2007; Rougemaille et al. 2008).
THO recruits the other components of the TREX complex to actively transcribed genes (Strasser et al. 2002; Zenklusen et al. 2002). In addition, the 3′ end processing factor Pcf11 has been reported to recruit Yra1 to genes (Johnson et al. 2009). Sub2 and Yra1 are then thought to be transferred to the nascent mRNA and function as intranuclear mRNA export factors that recruit the mRNA export receptor Mex67–Mtr2 to the mRNP (Strasser and Hurt 2000, 2001; Libri et al. 2001; Luo et al. 2001; Zenklusen et al. 2001; Lei and Silver 2002; Abruzzi et al. 2004). TREX leaves the transcribing polymerase downstream from the polyadenylation site (i.e., before RNAPII terminates transcription), most likely together with the mRNP (Hurt et al. 2004; Kim et al. 2004).
In higher eukaryotes, TREX is recruited to the mRNA during splicing (Masuda et al. 2005; Wahl et al. 2009). This might reflect the fact that splicing is more prominent in higher cells. However, since splicing takes place cotranscriptionally, the recruitment of the human TREX complex to spliced mRNAs is indirectly coupled to transcription through splicing. Furthermore, the overall functions of human TREX and Tap-p15, the human homolog of the yeast mRNA export receptor Mex67–Mtr2, are conserved (for review, see Katahira and Yoneda 2009 and references therein). However, despite the well-known and conserved function of the TREX complex, it has been a long-standing question as to how THO/TREX is recruited to transcribed genes.
Here, we show that the Prp19 complex, a non-snRNP protein complex with an essential role in splicing (Will and Lührmann 2006 and references therein), is necessary for TREX occupancy at transcribed genes. Deletion of the C terminus of Syf1, a component of the Prp19 complex, leads to dissociation of the Prp19 complex from RNAPII and the loss of the Prp19 complex and, consequently, TREX from actively transcribed genes. Importantly, Syf1 function is needed for efficient transcription and full processivity of RNAPII. Taken together, the Prp19 complex has a second, independent function in gene expression; namely, transcription elongation by stabilizing the recruitment of TREX to transcribing RNAPII.
Results
Syf1 interacts genetically and biochemically with the THO complex
In S. cerevisiae, the conserved TREX complex couples transcription to mRNP export, is recruited to actively transcribed genes, and travels along the gene together with RNAPII (for review, see Reed and Cheng 2005; Kohler and Hurt 2007; Katahira and Yoneda 2009; Rondon et al. 2010). However, it is not known how TREX is recruited to the transcription machinery or which proteins or protein complexes mediate this interaction. Interestingly, Syf1, a component of the Prp19 splicing complex, is homologous to human XAB2 (XPA-binding protein 2), a protein involved in splicing, but also in transcription and TCR (Nakatsu et al. 2000; Kuraoka et al. 2008). Since XAB2 and TREX have similar functions, we hypothesized that Syf1 functions in transcription in conjunction with TREX.
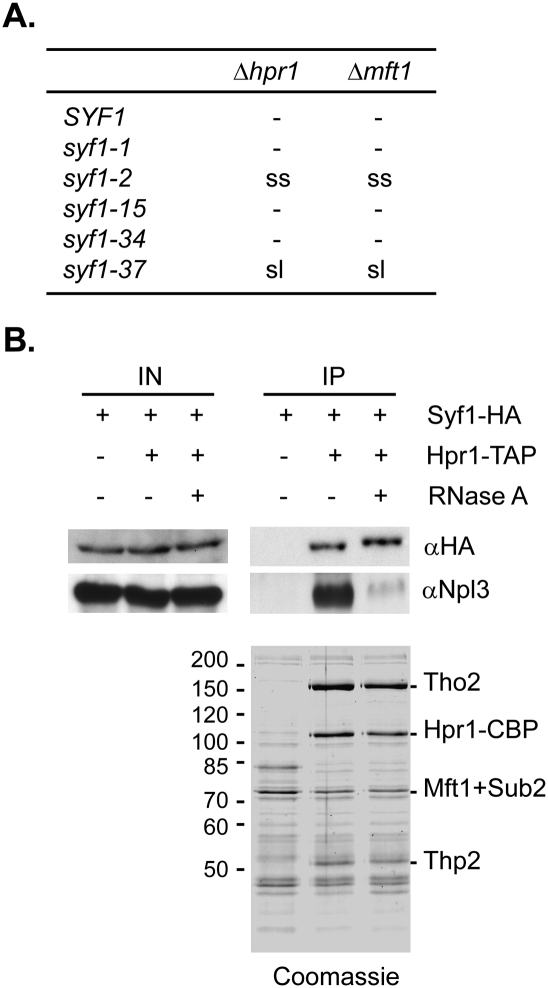
As a first indication for an overlapping function, we assessed whether SYF1 interacts genetically with THO components. To do this, we generated temperature-sensitive (ts) alleles of SYF1 and tested them for synthetic lethality with deletions of HPR1 and MFT1, two of the four components of the THO complex. Five different ts mutants of SYF1 were obtained by random mutagenesis: syf1-1, syf1-2, syf1-15, syf1-34, and syf1-37. All five SYF1 ts mutants grow approximately like wild-type cells at the permissive temperature (30°C), but do not grow at the nonpermissive temperature (37°C) (Supplemental Fig. 1). Interestingly, only one of the five SYF1 ts alleles, syf1-37 (for further information on syf1-37 see the legend for Fig. 1A), is synthetic lethal with Δhpr1 and Δmft1 (Fig. 1A). Thus, SYF1 interacts genetically and in an allele-specific manner with HPR1 and MFT1.
Figure 1.
SYF1 interacts genetically and biochemically with the THO complex. (A) SYF1 interacts genetically with HPR1 and MFT1, two components of the THO complex. SYF1 HPR1 and SYF1 MFT1 shuffle strains were transformed with an empty plasmid or plasmids encoding wild-type HPR1 or MFT1, respectively, and a plasmid encoding wild-type SYF1 or one of the SYF1 ts alleles (syf1-1 [W40G, R319G, K367N, E419G, I544V], syf1-2 [V69A, Y123C, Q596P, E752G], syf1-15 [E27V, V425A, T570P, F615L], syf1-34 [Y729F], and syf1-37 [I5F, R129G, Y453C, N658M, V659G, I660stop; caused by a frameshift mutation]). A synthetic lethal interaction was assessed by restreaking the respective transformants on 5-fluoroorotic acid (5-FOA)-containing plates. (sl) Synthetic lethal; (ss) synthetic sick; (−) no genetic interaction. (B) Syf1 interacts with TREX independently of RNA. The TREX complex was purified using TAP-tagged Hpr1 and copurification of HA-tagged Syf1 assessed by Western blotting with αHA antibodies (αHA). A nontagged strain served as negative control. To assess whether the interaction between TREX and Syf1 is mediated by RNA, extracts were treated with RNaseA prior to purification. Npl3, an SR protein involved in mRNA export that interacts with TREX in an RNA-dependent manner, served as a positive control for the RNase treatment (αNpl3). (IN) Input; (IP) immunoprecipitation; (Coomassie) TEV eluate of the TAP purification.
Second, we assessed whether Syf1 interacts biochemically with the TREX complex. TREX was purified using a strain expressing Hpr1-TAP, and copurification of HA-tagged Syf1 was assessed by Western blotting. Syf1 copurified with the TREX complex in the absence and presence of RNaseA, indicating that Syf1 interacts with TREX in an RNA-independent manner (Fig. 1B, αHA). Nevertheless, the interaction between TREX and Syf1 might be mediated by other proteins. Taken together, Syf1 interacts with TREX genetically and biochemically.
Deletion of the C-terminal domain of Syf1 causes sensitivity to 6-azauracil (6-AU) and synthetic lethality with Δhpr1 and Δmft1
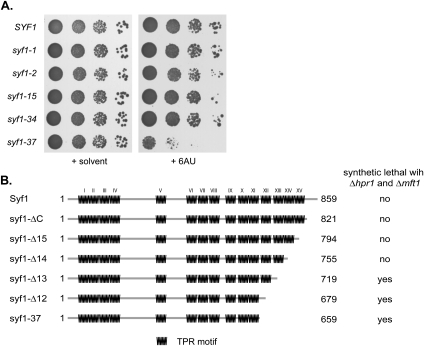
Syf1 and the Prp19 complex are known to function in splicing (Tarn et al. 1994; Ben-Yehuda et al. 2000; Will and Lührmann 2006). However, the function of XAB2, the human homolog of Syf1, in transcription and TCR (Nakatsu et al. 2000; Kuraoka et al. 2008) as well as the genetic interaction between SYF1 and the THO complex components shown here suggest that Syf1 may also function in transcription and/or DNA repair. To obtain further evidence for a function of Syf1 in transcription, we tested syf1-ts cells for sensitivity to 6-AU, a drug that impairs transcription elongation by decreasing the intracellular pools of the nucleotides GTP and UTP. Interestingly, of the five ts alleles, only syf1-37, the allele that is synthetic lethal with Δhpr1 and Δmft1 (Fig. 1A), confers 6-AU sensitivity (Fig. 2A). This further indicates that Syf1 might be involved in transcription elongation, and that the genetic interaction between SYF1 and THO is caused by overlapping functions in transcription elongation.
Figure 2.
Truncation of the Syf1 C terminus leads to 6-AU sensitivity and synthetic lethality with Δhpr1 and Δmft1. (A) The ts mutant syf1-37 is sensitive to 6-AU. Tenfold serial dilutions of yeast cells expressing wild-type SYF1 or one of five different ts alleles of SYF1 and containing pRS316 were spotted onto SDC(−ura) plates containing solvent or 50 μg/mL 6-AU and incubated for 2 d at 30°C. (B) The C terminus of Syf1 is important for the genetic interaction of SYF1 with HPR1 and MFT1. Schematic diagram showing the domain structure of Syf1, with each TPR motif indicated by a zigzag. Deletion of the three most C-terminal TPR motifs (syf1-Δ13) leads to synthetic lethality with Δhpr1 and Δmft1.
Strikingly, of the five syf1 ts alleles, only the syf1-37 allele carries a C-terminal truncation (Fig. 2B, syf1-37, caused by a frameshift mutation, see legend for Fig. 1A). Thus, we assessed whether this C-terminal truncation causes the genetic interaction with HPR1 and MFT1. Syf1 consists mainly of 15 tetratricopeptide repeat (TPR) motifs (Fig. 2B, Syf1). TPR motifs consist of 34 amino acids with the highly degenerate consensus of W-LG-Y-A-F-A-P (Lamb et al. 1995). Despite their sequence diversity, TPR motifs have a conserved three-dimensional structure of a helix–turn–helix motif with adjacent TPR motifs packed into anti-parallel α helices (Das et al. 1998). Proteins containing TPR motifs typically act as scaffolds for the assembly of multiprotein complexes (Blatch and Lassle 1999). In order to test whether and how large a deletion of the C terminus of Syf1 causes the genetic interaction with THO, we progressively truncated Syf1 from the C terminus and tested for synthetic lethality of the resulting syf1 alleles with Δhpr1 and Δmft1. Deletion of the very C-terminal part of Syf1 (syf1-ΔC) and the last (15th) and last two (14th and 15th) TPR motifs (syf1-Δ15 and syf1-Δ14, respectively) did not lead to synthetic lethality with Δhpr1 and Δmft1 (Fig. 2B). However, additional deletion of the 13th TPR motif (syf1-Δ13) caused synthetic lethality in combination with Δhpr1 or Δmft1 (Fig. 2B). Thus, the deletion of the C-terminal part of Syf1 is responsible for the genetic interaction with THO components.
Syf1 functions in transcription
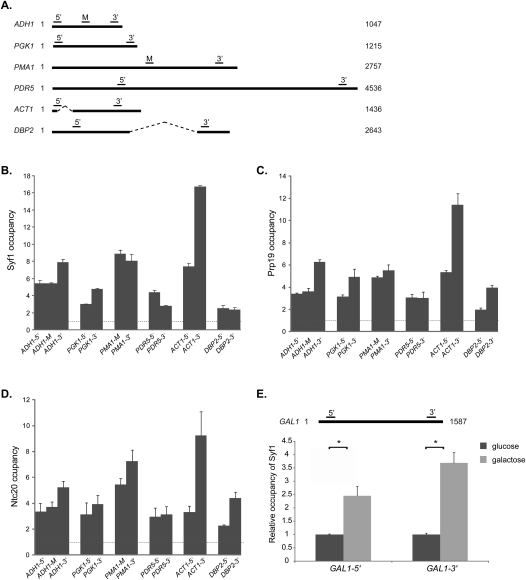
The function of XAB2 in transcription, the genetic interaction of SYF1 with THO, and the 6-AU sensitivity of the syf1-37 mutant indicate a function of Syf1 in transcription. If this is the case, Syf1 should be present at actively transcribed genes. Thus, we assessed recruitment of Syf1 to four intronless genes (ADH1, PGK1, PMA1, and PDR5) and two intron-containing genes (ACT1 and DBP2) by chromatin immunoprecipitation (ChIP) experiments (Fig. 3A). Occupancy of these six genes by Syf1 was calculated as the enrichment of Syf1 at the respective gene relative to a nontranscribed region (NTR), which served as a negative control (NTR1, 174131–174200 on chr. V) (Rother et al. 2010). Syf1 is specifically recruited to all six of these genes (Fig. 3B), but not to two other NTRs (NTR2, NTR3) (Supplemental Fig. 2). Thus, even though Syf1 is a splicing factor, it is recruited to intronless genes, indicating that it might have a general function in transcription. In order to test whether Syf1 alone or the whole Prp19 complex is recruited to genes during transcription, we assessed the occupancy of two further Prp19 complex components, Prp19 and Ntc20, at these six genes. Prp19 and Ntc20 were also present at these genes (Fig. 3C,D), indicating that the whole Prp19 complex is most likely recruited to transcribed genes.
Figure 3.
The Prp19 complex is recruited to transcribed genes. (A) Schematic diagram of the ADH1, PGK1, PMA1, PDR5, ACT1, and DBP2 genes. The ORFs are represented by a solid line and the introns are indicated by a hatched line. Bars above the genes show the positions of the primer pairs used for the ChIP analysis. (B) Syf1 is recruited to transcribed genes. The occupancy of Syf1 at the intronless ADH1, PGK1, PDR5, and PMA1 genes and the intron-containing ACT1 and DBP2 genes was calculated as the enrichment of Syf1 at the respective gene relative to its presence at an NTR that served as a negative control (NTR1, 174131–174200 on chr. V). Correspondingly, the occupancy at an NTR is “1” as indicated by a dashed line. (C,D) Experiment as in B for Prp19 and Ntc20, two other components of the Prp19 complex. (E) Recruitment of Syf1 to a gene is transcription-dependent. Occupancy of Syf1 at the GAL1 gene under repressive condition (cells grown in glucose-containing medium; dark-gray bars), which was set to 1, and induced conditions (cells grown in galactose-containing medium; light-gray bars). Primer pairs amplify the 5′ and 3′ region of GAL1, as indicated in the top panel.
To assess whether recruitment of Syf1 to genes depends on active transcription, we assessed the occupancy of Syf1 at the regulatable GAL1 gene (Fig. 3E). The GAL1 gene is repressed when cells are grown in glucose-containing medium and induced when cells are grown in galactose-containing medium. Syf1 is not recruited to the GAL1 gene under repressive conditions (Fig. 3E, glucose, dark-gray bars) relative to a nontranscribed control region (NTR1, 174131–174200 on chr. V). In contrast, Syf1 is present at the GAL1 gene when transcription of GAL1 is induced (Fig. 3E, galactose, light-gray bars), showing that recruitment of Syf1 to an intronless gene is transcription-dependent.
To obtain more direct evidence that the Prp19 complex indeed functions in transcription, we tested whether mutation of SYF1 affects transcription in vivo. Among the SYF1 truncation mutants, syf1-Δ13 is the longest one that is synthetic lethal with Δhpr1 and Δmft1 (Fig. 2B). However, since syf1-Δ13 grows more slowly than the syf1-37 ts mutant (Supplemental Fig. 3A), we chose the syf1-37 allele for further experiments in order to avoid unspecific effects due to slow growth. To ensure that the activity of the syf1-37 allele does not reflect residual activity of wild-type Syf1 produced by frameshifting during translation of the syf1-37 allele, we tested the levels of truncated and full-length Syf1 in syf1-37 and SYF1 cells. Syf1-37 and Syf1 are produced at approximately the same levels, and no expression of full-length Syf1 in the syf1-37 cells could be detected (Supplemental Fig. 3B). Thus, the phenotypes observed for the syf1-37 allele (see below) are most likely due to truncation of the Syf1 C terminus.
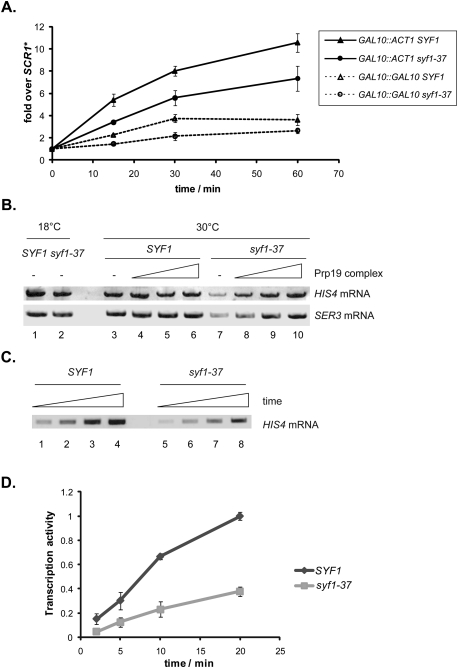
Two reporter constructs were used to assess mRNA synthesis in vivo—ACT1 as an intron-containing gene and GAL10 as an intronless gene—both of which were driven by the galactose-inducible GAL10 promoter. SYF1 and syf1-37 cells were grown in raffinose-containing medium at 30°C (permissive temperature) and expression of either reporter gene induced with galactose. RNA was isolated 0, 15, 30, and 60 min after induction, and the amount of synthesized ACT1 and GAL10 mRNA was quantified. The levels of ACT1 and GAL10 mRNA were decreased in the syf1-37 mutant (Fig. 4A). Since this defect is observed at the permissive temperature, it is not due to a complete functional loss of the syf1-37 ts allele. Thus, Syf1 function is needed to produce normal levels of intron-containing as well as intronless mRNAs in vivo.
Figure 4.
Syf1 function is needed for efficient transcription. (A) Mutation of SYF1 leads to lower mRNA levels in vivo. GAL10::ACT1 and GAL10::GAL10 were used as reporters for an intron-containing and an intronless gene, respectively. The level of ACT1 (solid line) and GAL10 (dotted line) mRNA of cells grown at the permissive temperature (30°C) was assessed by primer extension 0, 15, 30, and 60 min after induction with galactose and normalized to the amount of SCR1 RNA, an RNAPIII transcript. (B) Mutation of SYF1 leads to impaired transcription in vitro. The transcription activity of nuclear extracts from SYF1 and syf1-37 cells was assessed at 18°C (permissive temperature; lanes 1,2) and 30°C (nonpermissive temperature; lanes 3,7) by an in vitro transcription assay using one of two templates containing a HIS4 or SER3 promoter, respectively. (Lanes 7–10) Add-back of 50, 125, or 250 ng of Prp19 complex purified from S. cerevisiae rescues the impaired transcription activity of the syf1-37 extract at the nonpermissive temperature. (C) The transcription activity of a syf1-37 nuclear extract is lower after 2, 5, 10, and 20 min (lanes 5–8) compared with the activity of a SYF1 extract (lanes 1–4) at the nonpermissive temperature. (D) Quantification of the experiment shown in C.
Since Syf1 functions in splicing (Tarn et al. 1994; Ben-Yehuda et al. 2000; Will and Lührmann 2006), we tested whether splicing is affected in the syf1-37 strain under the same conditions as used for the in vivo transcription assay. Expression of the GAL10::ACT1 reporter was induced for 1 h, and the amount of spliced and unspliced ACT1 mRNA was assessed. As expected for a ts mutant of a protein involved in splicing, nonspliced transcripts accumulated in the syf1-37 strain at the nonpermissive temperature (Supplemental Fig. 3C). Importantly, though, the syf1-37 mutant did not display a defect in splicing of the ACT1 mRNA at the permissive temperature (Supplemental Fig. 3C). Thus, the syf1-37 mutation does not cause a severe splicing defect at the permissive temperature, making it unlikely that the decrease in ACT1 mRNA levels in the syf1-37 strain (Fig. 4A) is a secondary effect of defective splicing.
In order to assess whether the lower ACT1 and GAL10 mRNA levels in syf1-37 cells (Fig. 4A) are caused by impaired transcription in this mutant, we performed in vitro transcription assays. We used a plasmid-based in vitro transcription assay with a HIS4 or SER3 promoter (Koschubs et al. 2009) and prepared nuclear extracts from SYF1 and syf1-37 cells. At the permissive temperature (18°C), the syf1-37 nuclear extract had a slightly lower transcriptional activity than the wild-type extract, which could be caused by the SYF1 mutation or simply by variations in the extract preparation procedure (Fig. 4B, lanes 1,2). Importantly, though, when the temperature during the transcription reaction was raised (nonpermissive temperature, 30°C) the activity of the nuclear extract prepared from the syf1-37 ts cells decreased significantly relative to the wild-type extract (Fig. 4B, lanes 3,7). This decreased transcriptional activity of the syf1-37 extract at the nonpermissive temperature was observed after different incubation times (Fig. 4 C,D). In order to assure that this decrease in transcriptional activity of the syf1-37 extract is due to impaired Syf1 function, we performed add-back experiments with Prp19 complex purified from S. cerevisiae. Addition of the Prp19 complex rescued transcriptional activity of the syf1-37 extract (Fig. 4B, lanes 7–10) showing that the decreased transcriptional activity of the syf1-37 extract is caused by impaired Syf1 function. Importantly, RNAPII was not associated with the Prp19 complex purified under the conditions used for this experiment (Supplemental Fig. 4), and thus did not cause the stimulatory effect. In addition, since addition of the Prp19 complex did not increase the activity of the wild-type extract (Fig. 4B, lanes 3–6), rescue of the syf1-37 extract is most likely also not caused by addition of other transcription factors copurifying with the Prp19 complex. Thus, Syf1 is required for activated transcription in vitro. Taken together, Syf1 functions in transcription independently of its function in splicing.
Syf1 is necessary for RNAPII processivity
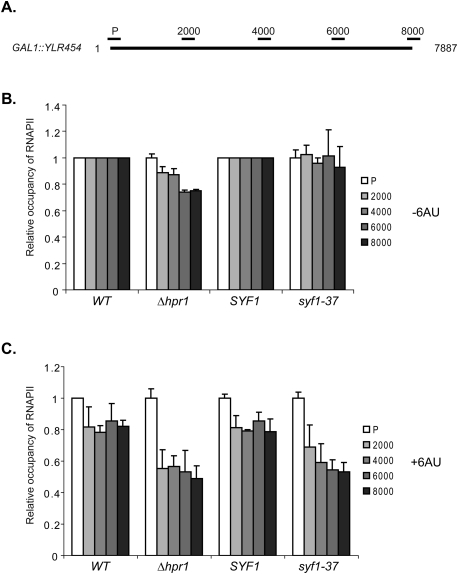
In principle, a decrease in transcription elongation can be caused by a decrease in either the rate of transcription (i.e., the speed of transcribing RNAPII) or the processivity of RNAPII (i.e., the ability of elongating RNAPII to travel the entire length of the gene). Mason and Struhl (2005) developed an assay to measure the rate and processivity of RNAPII in vivo. None of an array of transcription elongation mutants affected the rate of RNAPII in vivo (Mason and Struhl 2005). For RNAPII processivity, the well-known transcription elongation factors fall into three categories: factors the deletion of which (1) causes a decrease in processivity of RNAPII, (2) causes a decrease in processivity only in the presence of 6-AU, and (3) does not affect processivity at all. Interestingly, deletion of THO complex components reduces RNAPII processivity (Mason and Struhl 2005). Since Syf1 interacts genetically with THO, we tested whether mutation of SYF1 also affects RNAPII processivity. Processivity was assessed by measuring the level of RNAPII at five positions along the 8-kb-long YLR454 gene under control of the GAL1 promoter in cells grown in galactose-containing medium at the permissive temperature (30°C) by ChIP (Fig. 5A; Mason and Struhl 2005), and the values for the wild-type strain were normalized to 1.0 at each position.
Figure 5.
RNAPII processivity is decreased in Δhpr1 and syf1-37 cells. (A) Schematic diagram of the GAL1::YLR454 reporter gene used to analyze the processivity of RNAPII in wild-type, Δhpr1, SYF1, and syf1-37 cells grown at 30°C (permissive temperature). Positions of primer pairs used for the ChIP analysis are indicated above the gene. (B) Relative RNAPII occupancy within the YLR454 gene in wild-type, Δhpr1, SYF1, and syf1-37 cells grown in galactose-containing medium. The occupancy in wild-type cells at each position was set to 1. (C) Relative RNAPII occupancy as in B in the presence of 50 μg/mL 6-AU.
As shown previously, deletion of HPR1 caused a decrease in RNAPII processivity (Fig. 5B, Δhpr1; Mason and Struhl 2005), and addition of 6-AU exacerbated the processivity defect of Δhpr1 cells (Fig. 5C, Δhpr1). RNAPII processivity was not affected by the syf1-37 mutation in the absence of 6-AU (Fig. 5B, syf1-37). Importantly, though, in the presence of 6-AU, processivity of RNAPII was reduced in syf1-37 cells at the permissive temperature (30°C) (Fig. 5C, syf1-37). Thus, as observed for other transcription elongation factors (Mason and Struhl 2005), the syf1-37 allele affects RNAPII processivity in the presence of 6-AU (see also the Discussion).
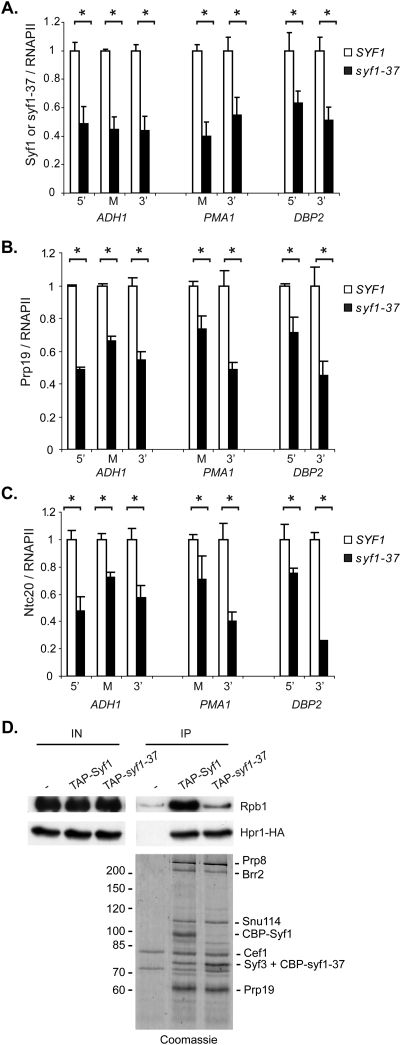
The C terminus of Syf1 is required for recruitment of the Prp19 complex to transcribed genes and the interaction of the Prp19 complex with RNAPII
The syf1-37 allele lacks the four most C-terminal TPR motifs of Syf1, and TPR motifs are known to function as scaffold domains for the interaction with other proteins (Blatch and Lassle 1999). Thus, an explanation for the reduced transcription in syf1-37 cells could be that Syf1 and the Prp19 complex are not recruited to the transcription machinery in this mutant. In order to test this, we assessed recruitment of syf1-37 to genes (Fig. 3A, ADH1, PMA1, and DPB2). Whereas the occupancy of RNAPII did not change in the syf1-37 strain compared with wild type (Supplemental Fig. 5), the occupancy of Syf1 (relative to RNAPII) decreased in the syf1-37 strain to ∼50% compared with wild-type cells (Fig. 6A). Importantly, this effect is observed at the permissive temperature (30°C); i.e., under conditions where function of the syf1-37 ts mutant is not impaired due to elevated temperature. Thus, the C terminus of Syf1 is required to recruit Syf1 efficiently to the transcribed gene. Since Syf1 is a component of the Prp19 complex, we wondered whether recruitment of not only Syf1 but the whole complex is impaired in the syf1-37 strain. Thus, we assessed the occupancy of two further Prp19 complex components (Prp19 and Ntc20) in SYF1 and syf1-37 cells. Indeed, the occupancy of Prp19 and Ntc20 also decreased to ∼50% in syf1-37 compared with wild-type cells (Fig. 6B,C). This indicates that Syf1 functions in transcription elongation most likely as part of the Prp19 complex.
Figure 6.
The C terminus of Syf1 is needed for the interaction of the Prp19 complex with the transcription machinery. (A) Deletion of the C terminus of Syf1 leads to loss of Syf1 from the transcribed gene. ChIP of TAP-Syf1 or TAP-Syf1-37 and Rpb3-TAP (RNAPII) in SYF1 and syf1-37 cells grown at permissive temperature (30°C) were quantified by real-time PCR using the primer pairs shown in Figure 3A. The ratio of Syf1 over RNAPII occupancy over an NTR (NTR1) was calculated, and the values for SYF1 were set to 1. (B,C) The C terminus of Syf1 is required to recruit the Prp19 complex to the transcribed gene. Experiment as in A, but the occupancy of Prp19 (B) and Ntc20 (C) relative to RNAPII on the indicated genes was assessed in SYF1 and syf1-37 cells grown at permissive temperature (30°C). (D) The C terminus of Syf1 mediates the interaction between the Prp19 complex and RNAPII. The interaction of syf1-37 with RNAPII is strongly reduced, while its interaction with the Prp19 and TREX complex is not affected. TAP-Syf1 and TAP-syf1-37 were purified from cells expressing Hpr1-HA and copurification of RNAPII and Hpr1 assessed by Western blotting using the 8WG16 antibody, which recognizes Rpb1, the largest subunit of RNAPII, or an αHA antibody (Hpr1-HA), respectively. Copurifying proteins were identified by mass spectrometry and are indicated to the right: Prp8, Brr2, and Snu114 are components of the U5 snRNP, and Cef1, Syf3, and Prp19 are components of the Prp19 complex. The other components of the Prp19 complex (Isy1, Cwc2, Syf2, Snt309, and Ntc20) have a molecular weight between 16 and 38 kD, and thus are not present on this gel. A nontagged strain served as negative control. (IN) Input; (IP) immunoprecipitation; (Coomassie) EGTA eluates of the TAP purifications.
The syf1-37 mutation results in lower levels of the Prp19 complex at the site of transcription. In order to find out whether this is due to a compromised interaction of syf1-37 with TREX or RNAPII we assessed their respective interaction by purifying TAP-tagged versions of Syf1 and syf1-37. Both Syf1 and syf1-37 copurified equal levels of Prp19 complex components and Hpr1 (Fig. 6D, Coomassie and Hpr1-HA), suggesting that syf1-37 interacts with the Prp19 complex and TREX. However, copurification of Rpb1, the largest subunit of RNAPII, with syf1-37 was greatly reduced (Fig. 6D, Rpb1), indicating that the direct or indirect interaction of the Prp19 complex with RNAPII is impaired by truncation of the C terminus of Syf1. Taken together, the C terminus of Syf1 is necessary for the interaction between the Prp19 complex and RNAPII and thus for recruitment of the Prp19 complex to transcribed genes. In addition, these findings indicate that the Prp19 complex functions in transcription elongation, since its occupancy on genes is reduced in the syf1-37 mutant that displays the transcription defects.
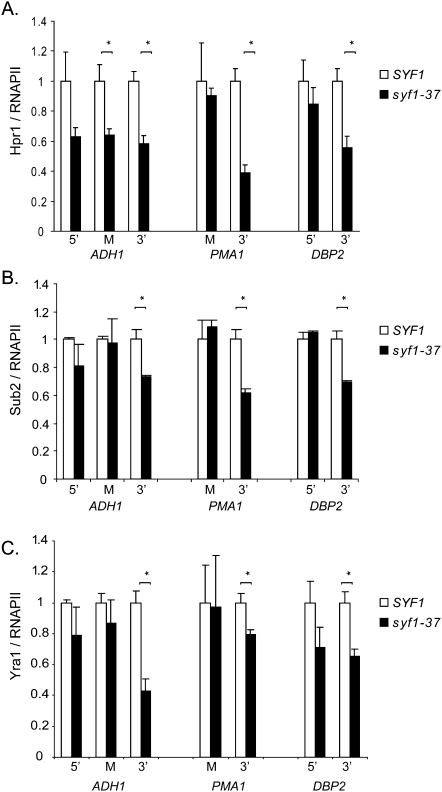
The Prp19 complex is required for TREX occupancy at genes
The function of Syf1 in transcription and its genetic interaction with THO suggest that the Prp19 and TREX complexes might have a related function in transcription. Thus, we tested whether the occupancy of Hpr1, one of the THO components, depends on Syf1 and vice versa. The occupancy of Syf1 relative to RNAPII was not affected by deletion of HPR1 (Supplemental Fig. 6). In contrast, Hpr1 occupancy relative to RNAPII decreased in syf1-37 cells to ∼50% compared with wild-type cells (Fig. 7A), reflecting the reduced recruitment of the Prp19 complex in these cells (Fig. 6A–C). Again, this effect is observed at the permissive temperature (30°C) and thus is not caused by a general loss of function of the syf1-37 ts allele, but by deletion of its C terminus. Loss of THO is especially observed at the 3′ end of genes (also see the Discussion). Importantly, this decrease of THO recruitment in syf1-37 cells is not caused by a general, and thus nonspecific, loss of transcription factors from the transcription machinery, since recruitment of two other transcription elongation factors (Paf1 [a component of the PAF complex] and Spt5) was not affected by mutation of SYF1 (Supplemental Fig. 7). Therefore, the Prp19 complex is required for THO occupancy at transcribed genes.
Figure 7.
Syf1 ensures TREX occupancy at genes. (A) Mutation of SYF1 leads to loss of the THO complex from the transcribed gene. ChIP of Hpr1-TAP (THO) in SYF1 and syf1-37 cells grown at permissive temperature (30°C) were quantified by real-time PCR using the primer pairs shown in Figure 3A. The ratio of Hpr1 relative to RNAPII occupancy over an NTR (NTR1) was calculated, and the values for SYF1 were set to 1. (B,C) Syf1 is required to recruit the TREX complex to the gene. Experiment as in A to determine the occupancy of the TREX components Sub2 (B) and Yra1 (C) relative to RNAPII in SYF1 and syf1-37 cells.
The THO subcomplex is needed to recruit the other TREX components—such as the intranuclear mRNA export factors Sub2 and Yra1—to the gene. In order to test whether recruitment of the entire TREX complex is affected by the syf1-37 mutation and the resulting loss of THO from genes, we assessed recruitment of Sub2 and Yra1 in syf1-37 cells. Indeed, like Hpr1, the occupancy of Sub2 and Yra1 was decreased in syf1-37 cells (Fig. 7B,C). However, the observed decrease in occupancy of Sub2 and Yra1 at genes is lower than the decrease for Hpr1. This is expected, since Sub2 and Yra1 are still partially recruited to genes even in the complete absence of Hpr1 (Zenklusen et al. 2002). In summary, the Prp19 complex functions in transcription by recruiting TREX to transcribing RNAPII.
Discussion
The Prp19 complex has a well-established and conserved function in splicing (Will and Lührmann 2006 and references therein). It forms a heteromeric complex that is also known as the NTC (nineteen complex) in S. cerevisiae (Tarn et al. 1994) and the hPrp19/CDC5L complex in humans (Ajuh et al. 2000; Makarova et al. 2004). The Prp19 complex is one of numerous non-snRNP proteins that play an essential role during splicing (Will and Lührmann 2006), and is required for the activation of the spliceosome (Ohi and Gould 2002; Chan et al. 2003).
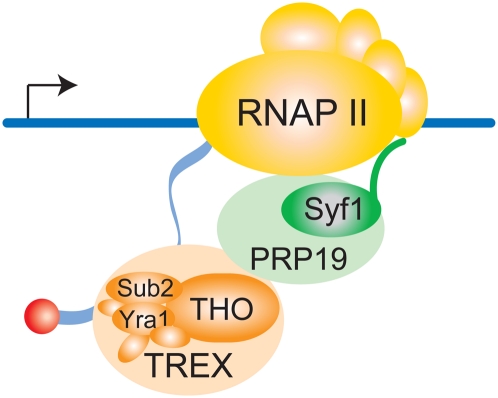
Here, we show that, in yeast, the Prp19 complex has a second function in gene expression; namely, in transcription elongation. The Prp19 complex is recruited to intron-containing as well as intronless genes in a transcription-dependent manner (Fig. 3). In addition, mutation of SYF1, one of the conserved components of the Prp19 complex, causes a reduced presence of the Prp19 complex at transcribed genes and a concomitant decrease in transcriptional activity (Figs. 4A,B, 6A–D). The syf1-37 mutation decreases RNAPII processivity, suggesting that the Prp19 complex most likely functions in transcription elongation (Fig. 5). This novel function of the Prp19 complex is most likely independent of its function in splicing, since splicing is not affected in syf1-37 cells at the permissive temperature (Supplemental Fig. 3C), where all of the transcription defects are evident (Figs. 2A, 4–7). Furthermore, the occupancy of the TREX complex, which functions in transcription elongation and mRNA export, is specifically reduced in syf1-37 cells (Fig. 7). Thus, we identify for the first time a factor required for full occupancy of TREX on transcribed genes. Interestingly, for some genes such as PMA1 and DBP2, the Prp19 complex is not required for TREX occupancy at the 5′ end (Fig. 7). Thus, there might be another factor responsible to recruit TREX to the transcription machinery at the 5′ end of genes. Importantly, though, corresponding to the decrease of the Prp19 complex, the occupancy of the TREX components Hpr1, Sub2, and Yra1 is reduced to ∼50% in the syf1-37 mutant. Thus, Prp19 complex function is essential for the presence of TREX on the transcribed gene. Based on this data, we propose the model that the Prp19 complex functions in transcription elongation by ensuring occupancy of the TREX complex at the transcribed gene (Fig. 8).
Figure 8.
The Prp19 complex functions in transcription. The Prp19 complex functions in transcription elongation by stabilizing the recruitment of TREX to transcribed genes. TREX components then bind—together with other proteins—to the nascent mRNA to form an mRNP. Thus, the Prp19 complex links transcription to mRNP formation. The Prp19 complex is recruited to the transcription machinery by the C terminus of its component, Syf1. Illustration by Susanne Röther.
The transcription defect observed in the syf1 mutant is most likely due to the reduced presence of the THO complex at genes in this mutant. In contrast to THO, the occupancy of two other transcription elongation factors (Paf1 and Spt5) is not affected by mutation of SYF1 (Supplemental Fig. 7). It is interesting to note that the PAF complex leaves the transcribing polymerase downstream from the polyadenylation site, as does TREX (Kim et al. 2004), but obviously depends on factors other than TREX for recruitment. THO is important for efficient transcription elongation by ensuring the stability of the RNAPII elongation machinery on the chromatin template (Mason and Struhl 2005). Consistent with a requirement of the THO complex for RNAPII processivity, syf1-37 cells show impaired processivity of RNAPII in the presence of 6-AU (Fig. 5). Thus, the reduced amount of THO/TREX in the SYF1 mutant seems to be sufficient for full RNAPII processivity under optimal conditions, but causes reduced RNAPII processivity when transcription is impaired in the presence of 6-AU. Likewise, even though deletion mutants of THO components show a mild mRNA export defect, we did not observe an mRNA export defect in the syf1-37 mutant (data not shown). Thus, the 50% of TREX present at genes in syf1-37 cells are sufficient for mRNA export.
Interestingly, Prp19 and hPrp19 have E3 ubiquitin ligase activity in vitro (Hatakeyama et al. 2001; Ohi et al. 2003). Recently, it has been shown that this catalytic activity of Prp19 is important for structural rearrangements during the splicing reaction (Song et al. 2010). Prp19 ubiquitylates Prp3, a component of the U4 snRNP (Nottrott et al. 2002), forming nonproteolytic K63-linked chains. This modification of Prp3 increases the affinity of Prp3 for the U5 component Prp8 necessary to stabilize the U4/U6.U5.snRNP. The interaction of the Prp19 complex with the U5 snRNP does not seem to be affected in the syf1-37 mutant (Fig. 6D); i.e., when transcription is impaired. Thus, the E3 ligase activity of the Prp19 complex is most likely not affected in this SYF1 mutant. Nevertheless, it is tempting to speculate that the E3 ligase activity of the Prp19 complex might be important for its function in transcription, in addition to its structural role to recruit the TREX complex. Notably, Hpr1 is polyubiquitylated and degraded, but ubiquitylated Hpr1 interacts with the UBA domain of the mRNA export receptor Mex67, an interaction that recruits Mex67 to transcribed genes and transiently protects Hpr1 from degradation (Gwizdek et al. 2006). However, when we purified Hpr1 from wild-type and syf1-37 cells, Hpr1 ubiquitylation was not affected (data not shown), consistent with the notion that the Prp19 E3 ligase activity is not affected in syf1-37 cells (see above). Thus, it remains an open question as to whether the E3 ligase activity of Prp19 is important for efficient transcription and what the target protein(s) might be.
Deletion of the last three TPR motifs (XIII–XV) of Syf1 significantly weakens the direct or indirect interaction between the Prp19 complex and RNAPII, and leads to a reduced Prp19 occupancy at transcribed genes in vivo (Fig. 6). Thus, the C-terminal domain of Syf1 is an important interface for the interaction between the Prp19 complex and RNAPII. However, since ∼50% of the Prp19 complex is still recruited to the active transcription machinery in this mutant, another domain of the Prp19 complex for interaction with RNAPII must exist. Within the C terminus of Syf1, the 13th of the 15 TPRs of Syf1 seems to be specifically important for the interaction with RNAPII. Whereas deletion of the two most C-terminal TPRs (XV + XIV, syf1-Δ14) does not cause synthetic lethality with Δhpr1, additional deletion of the 13th TPR (syf1-Δ13) leads to synthetic lethality with Δhpr1 (Fig. 2B). Fusion of six different combinations of two “other” TPR motifs (I + II, III + IV, VI + VII, VII + VIII, IX + X, or XI + XII) of Syf1 to the syf1-Δ13 deletion mutant—with the same spacing between TPR 12 and the TRP motif C-terminal of it—are still synthetic lethal with Δhpr1 (data not shown). Thus, the mere addition of TPR motifs to the 3′ end of the truncated Syf1 does not rescue functionality of Syf1 in transcription, indicating that different TPR motifs might have specific functions. Similarly, deletion of the N-terminal five TPRs of Syf1 (I—V, syf1-aa408–859) does not lead to a synthetic lethal relationship with Δhpr1 or Δmft1 (data not shown), indicating that these TPRs are not important for the interaction of the Prp19 complex with the transcription machinery and the function of the Prp19 complex in transcription. Taken together, the 13th TPR of Syf1 might be important for the interaction of the Prp19 complex with RNAPII.
Interestingly, in higher cells, TREX is recruited to the mRNA during the splicing reaction (Masuda et al. 2005; Wahl et al. 2009), and, in yeast (where TREX is recruited in a transcription-dependent manner), a splicing complex recruits TREX to the gene. The spliceosome is a highly dynamic molecular machine that is assembled in a stepwise manner onto the mRNA to be spliced, leading to the formation of assembly intermediates called complex E, A, B, B*, and C (Wahl et al. 2009 and references therein). Briefly, binding of the U1 snRNP to the 5′ splice site defines the E complex. Recognition of the pre-mRNA's branch-point sequence by the U2 snRNP leads to the A complex, and subsequent binding of U4, U5, and U6 as a preassembled tri-snRNP results in the B complex. The recruitment of additional factors as well as extensive structural rearrangements that destabilize the association of U1 and U4 snRNPs with the catalytic core lead to the catalytically activated B* complex. The first transesterification reaction generates the C complex, which catalyzes the second step of splicing. Interestingly, the hPrp19 and hPrp19-related complexes containing XAB2/hSyf1 are present within the B complex, whereas TREX components are only present later, when the C complex has been formed (Wahl et al. 2009 and references therein). Since the Prp19 complex is part of the spliceosome before TREX components join, it might recruit TREX to the mRNA during the splicing reaction. Thus, the role of the Prp19 complex in recruiting TREX to the mRNA may be conserved in higher eukaryotes.
In summary, we identified the Prp19 splicing complex as a novel transcription elongation factor. In addition, we establish the Prp19 complex as the first factor essential for TREX complex occupancy at transcribed genes. In this respect, the Prp19 complex links transcription to mRNP formation.
Materials and methods
Yeast strains and plasmids
Yeast strains and plasmids are listed in Supplemental Tables S1 and S2, respectively.
ChIP experiments
ChIP experiments were performed essentially as described previously (Rother et al. 2010) with some modifications. Briefly, cross-linked cells were lysed with an equal volume of glass beads by vortexing six times for 3 min with 3-min breaks, on ice. Chromatin lysate was used for immunoprecipitation with 15 μL of IgG-coupled or anti-Yra1-coupled (Yra1 ChIP) Dynabeads for 3 h at 20°C. After washing and elution, the immunoprecipitation eluates as well as input samples were treated with proteinase K overnight at 65°C. DNA was purified by phenol/chloroform extraction. Quantitative PCR with input and immunoprecipitation samples was done on an Applied Biosystems StepOnePlus cycler, using Applied Biosystems' Power SYBR Green PCR Master Mix. As negative control, primers for three NTRs amplifying 174131–174200 on chr. V (NTR1) (Rother et al. 2010), 116689–116758 on chr. X (NTR2), and 131523–131582 on chr. V (NTR3) were used. PCR efficiencies (E) were determined with standard curves. The occupancy of each factor at the respective gene was calculated as its enrichment at the respective gene relative to NTR1 according to [E^(CTIP-CTInput)]NTR/[E^(CTIP-CTInput)]gene.
Protein purification
Affinity purification of TAP-tagged proteins was performed as described previously (Puig et al. 2001; Strasser et al. 2002). Briefly, cells were lysed with glass beads in 2 vol of TAP buffer (50 mM Tris-Hcl at pH 7.5, 100 mM NaCl, 1.5 mM MgCl2, 0.15% NP40, 1 mM DTT, 1.3 μg/mL pepstatin A, 0.28 μg/mL leupeptin, 170 μg/mL PMSF, 330 μg/mL benzamidine). Crude extracts were centrifuged and supernatants were incubated with IgG-coupled sepharose beads. Subsequently, the IgG beads were washed with the same buffer and eluted with TEV protease. The TEV eluates were then incubated with calmodulin beads prewashed with TAP buffer containing 2 mM CaCl2, and calmodulin beads washed and eluted with calmodulin elution buffer (10 mM Tris-HCl at pH 8.0, 5 mM EGTA). Copurifying proteins were analyzed by SDS-PAGE and Coomassie staining, and were identified by mass spectrometry or Western blotting using antibodies directed against HA (Roche Applied Science), Npl3 (Siebel and Guthrie 1996), the N terminus (yN-18, Santa Cruz Biotechnology), and the CTD of Rpb1 (8WG16, Covance).
RNA analysis
Total cellular RNA was isolated by acidic phenol extraction from cells grown in raffinose or shifted to 2% galactose for the indicated time points. The primer extension analysis was performed according to a standard method. Briefly, 1 pmol of 32P-labeled primer (4 × 106 counts per minute [cpm]/pmol) specific for ACT1 mRNA, GAL10 mRNA, or SCR1 RNA (an RNAPIII transcript that served as a loading control) was mixed with 5 μg of RNA, hybridized for 5 min at 37°C, extended by reverse transcriptase for 1 h at 42°C, and analyzed on a 6% urea/acrylamide sequencing gel. Quantification was performed with a STORM 860 and ImageQuant software (GE Healthcare). Analysis of splicing efficiency was performed by RT–PCR. Products were separated on 2% agarose gels, images were recorded digitally, and ratios between spliced and unspliced products were quantified using ImageJ (NIH).
In vitro transcription assays
Yeast nuclear extracts were prepared from 3 L of culture as described previously. Plasmid transcription was performed essentially as described (Ranish et al. 1999). Transcription reactions were carried out in a 25-μL volume. The reaction mixture contained 200 μg of nuclear extract, 150 ng of plasmid template containing the yeast HIS4 or SER3 promoter (sequence available on request) (Koschubs et al. 2009), 1× transcription buffer (10 mM HEPES at pH 7.6, 50 mM potassium acetate, 0.5 mM EDTA, 2.5 mM magnesium acetate), 2.5 mM DTT, 192 μg of phosphocreatine, 0.2 μg of creatine phosphokinase, 10 U of Ribolock RNase inhibitor (Fermentas), 100 μM nucleoside triphosphates (NTPs), and 200 ng of recombinant Gcn4. The reaction was incubated for 20 min at 18°C or 30°C or for 2, 5, 10, and 20 min at 30°C for the time-course experiment, and total RNA was isolated using the RNeasy MinElute Cleanup kit (Qiagen). Transcripts were analyzed by primer extension essentially as described (Ranish et al. 1999). Instead of the 32P-labeled lacI oligo, 0.125 pmol of a fluorescently labeled 5′ Cy5-oligo was used. Quantification was performed with a Typhoon 9400 and ImageQuant software (GE Healthcare). For the add-back experiment, the Prp19 complex was purified to the TEV eluate using a SYF1-TAP strain by TAP as described above, except that the TAP buffer contained 500 mM NaCl and the HIS-tagged TEV protease was depleted using Ni-NTA agarose.
Statistical analyses
Data are means ± SEM from n = 3. Asterisks indicate statistical significance (Student's t-test; [*] P < 0.05).
Acknowledgments
We are thankful to Yuh Min Chook and Dietmar Martin for critical reading of the manuscript, Susanne Röther for artwork, and the Zentrallabor für Proteinanalytik (ZfP) for mass spectrometry. This research was supported by the Boehringer Ingelheim Fonds and the Elitenetzwerk Bayern (M.S.), and by grants of the Sonderfoschungsbereich SFB646 and the EMBO Young Investigator Program (K.S.).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.623411.
Supplemental material is available for this article.
References
- Abruzzi KC, Lacadie S, Rosbash M 2004. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J 23: 2620–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SH, Kim M, Buratowski S 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 [DOI] [PubMed] [Google Scholar]
- Ajuh P, Kuster B, Panov K, Zomerdijk JC, Mann M, Lamond AI 2000. Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J 19: 6569–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Dix I, Russell CS, McGarvey M, Beggs JD, Kupiec M 2000. Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics 156: 1503–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch GL, Lassle M 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21: 932–939 [DOI] [PubMed] [Google Scholar]
- Carmody SR, Wente SR 2009. mRNA nuclear export at a glance. J Cell Sci 122: 1933–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC 2003. The Prp19p-associated complex in spliceosome activation. Science 302: 279–282 [DOI] [PubMed] [Google Scholar]
- Das AK, Cohen PW, Barford D 1998. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J 17: 1192–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard H, Wellinger RE, Aguilera A 2007. A new connection of mRNP biogenesis and export with transcription-coupled repair. Nucleic Acids Res 35: 3893–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwizdek C, Iglesias N, Rodriguez MS, Ossareh-Nazari B, Hobeika M, Divita G, Stutz F, Dargemont C 2006. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc Natl Acad Sci 103: 16376–16381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI 2001. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem 276: 33111–33120 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL 2000. RNA polymerase II and the integration of nuclear events. Genes Dev 14: 1415–1429 [PubMed] [Google Scholar]
- Huertas P, Aguilera A 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721 [DOI] [PubMed] [Google Scholar]
- Hurt E, Luo MJ, Rother S, Reed R, Strasser K 2004. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci 101: 1858–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N, Stutz F 2008. Regulation of mRNP dynamics along the export pathway. FEBS Lett 582: 1987–1996 [DOI] [PubMed] [Google Scholar]
- Jimeno S, Rondon AG, Luna R, Aguilera A 2002. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 21: 3526–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Cubberley G, Bentley DL 2009. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol Cell 33: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Yoneda Y 2009. Roles of the TREX complex in nuclear export of mRNA. RNA Biol 6: 149–152 [DOI] [PubMed] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J 23: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Hurt E 2007. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8: 761–773 [DOI] [PubMed] [Google Scholar]
- Komili S, Silver PA 2008. Coupling and coordination in gene expression processes: a systems biology view. Nat Rev Genet 9: 38–48 [DOI] [PubMed] [Google Scholar]
- Koschubs T, Seizl M, Lariviere L, Kurth F, Baumli S, Martin DE, Cramer P 2009. Identification, structure, and functional requirement of the Mediator submodule Med7N/31. EMBO J 28: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraoka I, Ito S, Wada T, Hayashida M, Lee L, Saijo M, Nakatsu Y, Matsumoto M, Matsunaga T, Handa H, et al. 2008. Isolation of XAB2 complex involved in pre-mRNA splicing, transcription, and transcription-coupled repair. J Biol Chem 283: 940–950 [DOI] [PubMed] [Google Scholar]
- Lamb JR, Tugendreich S, Hieter P 1995. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci 20: 257–259 [DOI] [PubMed] [Google Scholar]
- Lei EP, Silver PA 2002. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes Dev 16: 2761–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Graziani N, Saguez C, Boulay J 2001. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev 15: 36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413: 644–647 [DOI] [PubMed] [Google Scholar]
- Makarova OV, Makarov EM, Urlaub H, Will CL, Gentzel M, Wilm M, Luhrmann R 2004. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J 23: 2381–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506 [DOI] [PubMed] [Google Scholar]
- Mason PB, Struhl K 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell 17: 831–840 [DOI] [PubMed] [Google Scholar]
- Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev 19: 1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu Y, Asahina H, Citterio E, Rademakers S, Vermeulen W, Kamiuchi S, Yeo JP, Khaw MC, Saijo M, Kodo N, et al. 2000. XAB2, a novel tetratricopeptide repeat protein involved in transcription-coupled DNA repair and transcription. J Biol Chem 275: 34931–34937 [DOI] [PubMed] [Google Scholar]
- Nottrott S, Urlaub H, Luhrmann R 2002. Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. EMBO J 21: 5527–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Gould KL 2002. Characterization of interactions among the Cef1p–Prp19p-associated splicing complex. RNA 8: 798–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL 2003. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol 10: 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D 2000. RNA polymerase II elongation through chromatin. Nature 407: 471–475 [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D 2002. A unified theory of gene expression. Cell 108: 439–451 [DOI] [PubMed] [Google Scholar]
- Perales R, Bentley D 2009. ‘Cotranscriptionality’: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell 36: 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Hannett NM, Young RA 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell 9: 799–809 [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Ranish JA, Yudkovsky N, Hahn S 1999. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev 13: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R 2003. Coupling transcription, splicing and mRNA export. Curr Opin Cell Biol 15: 326–331 [DOI] [PubMed] [Google Scholar]
- Reed R, Cheng H 2005. TREX, SR proteins and export of mRNA. Curr Opin Cell Biol 17: 269–273 [DOI] [PubMed] [Google Scholar]
- Rondon AG, Jimeno S, Garcia-Rubio M, Aguilera A 2003. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J Biol Chem 278: 39037–39043 [DOI] [PubMed] [Google Scholar]
- Rondon AG, Jimeno S, Aguilera A 2010. The interface between transcription and mRNP export: from THO to THSC/TREX-2. Biochim Biophys Acta 1799: 533–538 [DOI] [PubMed] [Google Scholar]
- Rother S, Burkert C, Brunger KM, Mayer A, Kieser A, Strasser K 2010. Nucleocytoplasmic shuttling of the La motif-containing protein Sro9 might link its nuclear and cytoplasmic functions. RNA 16: 1393–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, Boulay J, Jensen TH, Stutz F, Devaux F, et al. 2008. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell 135: 308–321 [DOI] [PubMed] [Google Scholar]
- Siebel CW, Guthrie C 1996. The essential yeast RNA binding protein Np13p is methylated. Proc Natl Acad Sci 93: 13641–13646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song EJ, Werner SL, Neubauer J, Stegmeier F, Aspden J, Rio D, Harper JW, Elledge SJ, Kirschner MW, Rape M 2010. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev 24: 1434–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Hurt E 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J 19: 410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Hurt E 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413: 648–652 [DOI] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- Stutz F, Izaurralde E 2003. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol 13: 319–327 [DOI] [PubMed] [Google Scholar]
- Tarn WY, Hsu CH, Huang KT, Chen HR, Kao HY, Lee KR, Cheng SC 1994. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J 13: 2421–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R 2006. Spliceosome structure and function. In The RNA world (ed. Gesteland RF et al. ), 3rd ed, pp. 369–400 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Zenklusen D, Vinciguerra P, Strahm Y, Stutz F 2001. The yeast hnRNP-Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol Cell Biol 21: 4219–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Wyss JC, Stutz F 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol 22: 8241–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]