Abstract
A total of 57.8 Mb of publicly available rice (Oryza sativa L.) DNA sequence was searched to determine the frequency and distribution of different simple sequence repeats (SSRs) in the genome. SSR loci were categorized into two groups based on the length of the repeat motif. Class I, or hypervariable markers, consisted of SSRs ≥20 bp, and Class II, or potentially variable markers, consisted of SSRs ≥12 bp <20 bp. The occurrence of Class I SSRs in end-sequences of EcoRI- and HindIII-digested BAC clones was one SSR per 40 Kb, whereas in continuous genomic sequence (represented by 27 fully sequenced BAC and PAC clones), the frequency was one SSR every 16 kb. Class II SSRs were estimated to occur every 3.7 kb in BAC ends and every 1.9 kb in fully sequenced BAC and PAC clones. GC-rich trinucleotide repeats (TNRs) were most abundant in protein-coding portions of ESTs and in fully sequenced BACs and PACs, whereas AT-rich TNRs showed no such preference, and di- and tetranucleotide repeats were most frequently found in noncoding, intergenic regions of the rice genome. Microsatellites with poly(AT)n repeats represented the most abundant and polymorphic class of SSRs but were frequently associated with the Micropon family of miniature inverted-repeat transposable elements (MITEs) and were difficult to amplify. A set of 200 Class I SSR markers was developed and integrated into the existing microsatellite map of rice, providing immediate links between the genetic, physical, and sequence-based maps. This contribution brings the number of microsatellite markers that have been rigorously evaluated for amplification, map position, and allelic diversity in Oryza spp. to a total of 500.
[Clone sequences for 199 markers (RM1–RM88, RM200–RM345) developed in this lab are available as GenBank accessions AF343840–AF343869 and AF344003–AF344169.]
Microsatellites are tandemly arranged repeats of short DNA motifs (1–6 bp in length) that frequently exhibit variation in the number of repeats at a locus. Because of their abundance and inherent potential for variation, these simple sequence repeats (SSRs) have become a valuable source of genetic markers. Previous studies in rice have contributed to the development of several hundred microsatellite markers and a genetic map consisting of 320 SSRs (Wu and Tanksley 1993; Akagi et al. 1996; Panaud et al. 1996; Chen et al. 1997; Temnykh et al. 2000). These markers have been used to analyze diversity (Yang et al. 1994; Olufowote et al. 1997; Cho et al. 2000; Harrington 2000) and to locate genes and QTLs on rice chromosomes using both intra- and interspecific crosses (Xiao et al. 1998; Bao et al. 2000; Zou et al. 2000; Bres-Patry et al. 2001; Moncada et al. 2001). SSRs are increasingly useful for integrating the genetic, physical, and sequence-based maps of rice, and they simultaneously provide breeders and geneticists with an efficient tool to link phenotypic and genotypic variation.
In the past, the advantages of microsatellite markers were partially offset by the difficulty inherent in marker development, as laborious iterations of genomic DNA library screening with SSR probes were required to isolate microsatellite-containing sequences (Panaud et al. 1996; Chen et al. 1997). As random rice EST sequences became available, they provided a new source of SSR markers (Akagi et al. 1996; Temnykh et al. 2000) but the chromosomal positions of these markers had to be determined by genetic mapping. More recently, the growing pool of DNA sequence information being generated by the International Rice Genome Sequencing Project (IRGSP) and by other organizations (e.g., http://www.rice-research.org) allows high-throughput in silico identification of SSR loci in sequenced regions, often with known map position, providing an excellent starting point for marker development. Conversely, mapped SSR markers that have been associated with phenotypes of interest provide a direct link to sequenced regions that can be carefully annotated to identify candidate genes underlying the target trait.
In genomes of eukaryotic organisms, microsatellites are often found in proximity to dispersed repetitive elements such as Alu sequences in primates (Arcot et al. 1995; Jurka and Pethiyagoda 1995) and long terminal repeats of retrotransposons in barley (Ramsay et al. 1999). These associations have immediate practical implications for the success of SSR marker development. The avoidance of flanking sequences corresponding to known repetitive DNA has become a routine procedure during the development of microsatellite markers for mammalian genomes (Fondon et al. 1998; Steen et al. 1999) because positioning PCR primers in repetitive regions generates spurious or nonspecific products. In rice, little is known about the relationships, if any, between microsatellites and different classes of middle repetitive sequences, although several families of transposable elements as well as centromere-associated sequences have been identified in the rice genome (Bureau and Wessler 1994; Aragon-Alcaide et al. 1996; Jiang et al. 1996; Hirochika 1997; Wang et al. 1999; Mao et al. 2000).
We describe the development of microsatellite markers in rice based on publicly available genomic sequence information. We analyzed 57.8 Mb of DNA sequence (roughly equivalent to 13% of the genome) to evaluate different classes of di-, tri- and tetranucleotide microsatellites for their abundance, length distribution, and potential as informative genetic markers. To better understand the genomic domain preferences of SSRs in rice we compared the frequency and distribution of different SSR motifs in a large collection of BAC-end sequences to those identified in completely sequenced BAC and PAC clones or in EST sequence. Associations between microsatellites and repetitive elements were investigated and the implications of these associations for SSR marker design were evaluated. Two hundred new microsatellite markers were surveyed for allelic diversity and added to the existing rice SSR map, bringing the total number of genetically mapped SSR loci to >500.
RESULTS
Frequency, Length Variation, and Domain Specificity of Rice SSRs
To facilitate the identification of SSRs and the development of new microsatellite markers for rice using publicly available DNA sequence information, we used a set of simple Perl scripts for semiautomated identification of nonredundant SSR loci and primer design as described in Methods. Two sources of rice DNA sequences generated by the IRGSP were employed for this purpose: a large set of short BAC-end sequences (500 bp on average) and 27 fully sequenced large-insert clones (150 kb on average).
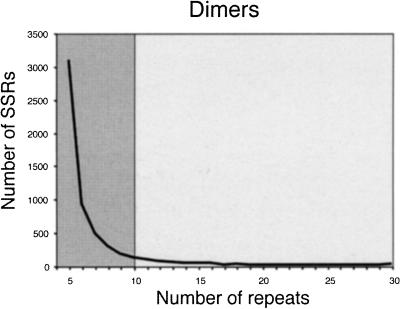
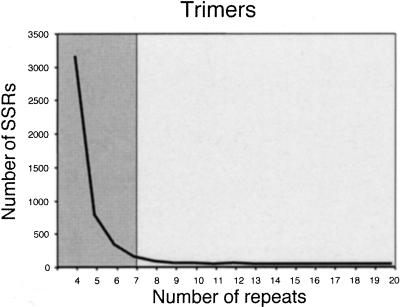
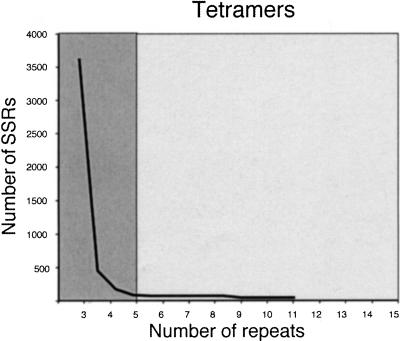
Figure 1 shows the frequency and length distribution of SSRs with different di-, tri-, and tetranucleotide motifs extracted from a set of 74,127 rice BAC-end sequences available in GenBank as of February 2000. 68.8% of the BAC ends originated from a HindIII-digested BAC library, whereas 31.2% came from an EcoRI-digested library (Budiman 1999, http://www.genome.clemson.edu/where/budiman/). A total of 13,989 SSRs with lengths of 12 nucleotides or longer were identified in the 47.43 Mb of BAC-end sequence from cultivar Nipponbare.
Figure 1.
Observed number of microsatellites with di-, tri-, and tetranucleotide motifs in 74,127 BAC-end sequences. Lighter fields correspond to Class I simple sequence repeats (SSRs) (>20 bp), darker areas correspond to Class II SSRs (12 nucleotides ≤ n < 20 nucleotides).
Microsatellites were categorized into two groups based on length of SSR tracts and their potential as informative genetic markers: Class I microsatellites contain perfect SSRs ≥20 nucleotides in length and Class II contain perfect SSRs >12 nucleotides and <20 nucleotides in length (indicated by differential shading in Fig. 1). The rationale for these two categories is that longer perfect repeats (Class I) are highly polymorphic, as evidenced by the experimental data originally reported for human (Weber 1990) and then confirmed by studies in many other organisms, including rice (Cho et al. 2000; Temnykh et al. 2000). Microsatellites in Class II tended to be less variable, representing sites where SSR expansion may occasionally occur but its probability is limited due to a smaller chance of slipped-strand mispairing over the shorter SSR template. Microsatellites shorter than 12 bp have a mutation potential that is no different than that of most unique sequences, and therefore demonstrate stochastic variation as has been shown in yeast (Pupko and Graur 1999).
Of the total number of SSRs identified in BAC-end sequences, 1178 (8.4%) were defined as Class I and 12,811 as Class II (91.6%) microsatellites. On average, the estimated frequency of Class I microsatellites in the BAC ends was one SSR per 40.3 kb, whereas the frequency of Class II was one SSR every 3.7 kb. When 27 completely sequenced PAC and BAC clones comprising 4.04 Mb were screened for SSRs, a total of 2368 microsatellites were identified, with 266 (11.2%) belonging to Class I and 2102 (88.8%) to Class II. Although the relative frequencies of Class I and II SSRs were similar, the absolute frequencies differed markedly in these two sources of sequence data. The fully sequenced clones contained an average of one Class I SSR every 15.8 kb and one Class II SSR every 1.9 kb. Interestingly, the estimated frequency of Class I SSRs in the rice EST data — on average, one Class I SSR per 19 Kb in a random set of 12,532 ESTs (6.3 Mb) analyzed previously by Temnykh et al. (2000) — was very similar to that in the fully sequenced large-insert clones.
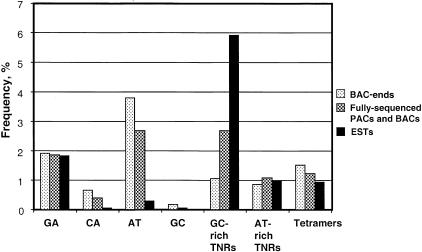
When the relative frequencies of different Class I di-, tri- and tetranucleotide motifs extracted from the three independent sources of sequence data were compared, i.e., BAC-end sequences, completely sequenced BAC and PAC clones and rice ESTs, very obvious differences and patterns were observed (Fig. 2). The frequency of GC-rich trinucleotide repeats (TNR) (those which contain ≥2 G and/or C in their repeating units) were the most variable, ranging from a low of 10.5% in BAC-end sequence to a high of 59% of all SSRs identified in ESTs, with intermediate frequencies (∼27%) observed in fully sequenced BAC and PAC clones. These differences were mirrored in reverse when (AT)n dinucleotide repeat (DNR) frequencies were compared, with an estimated 38.2% in BAC-end sequence, 27% in fully sequenced clones, and only 2.9% in EST sequences. Tetranucleotide SSRs and (CA)n DNR SSRs demonstrated the same pattern as (AT)n DNR microsatellites, in that they were most abundant in BAC ends, followed by fully sequenced BAC and PAC clones, and were least frequent in ESTs. The pattern of variation demonstrated that SSR motif categories are not randomly distributed in the rice genome.
Figure 2.
Relative frequency of Class I microsatellites with different simple sequence repeat motifs in three sets of DNA sequence data.
Further, it was observed that the frequencies of GC-rich TNRs and (AT)n DNRs were almost identical in data from completely sequenced BAC and PAC clones (∼27%) whereas the frequency of these motifs varied greatly and inversely in sequences from BAC ends and ESTs. This data led to the conclusion that the BAC ends and the ESTs represented different genomic domains, neither of which reflected the SSR composition in the genome as a whole. The fully sequenced BACs and PACs harbored a more balanced combination of microsatellites of all types.
To characterize the spatial relationship between Class I microsatellites and genes, we evaluated the distribution of SSRs in four fully annotated PAC and BAC sequences relative to the occurrence of: (a) coding DNA sequences (CDSs), (b) untranslated regions (UTRs), (c) introns, and (d) intergenic regions. From this analysis, we observed that ∼80% of GC-rich TNRs occurred in predicted exons, whereas AT-rich TNRs were distributed roughly evenly in all four genomic components in this data set. DNRs and tetranucleotide SSRs were predominantly situated in noncoding, mainly intergenic, regions (Table 1).
Table 1.
Gene context of 67 Rice Mirosatellites from Four Fully-Sequenced PAC Clones Comprising 654,596 nt of Genomic DNA Sequence
| SSR class | Total no.SSRs in this class | SSRs of this class in CDSs | SSRs of this class in UTRs | SSRs of this class in introns | SSRs of this class in intergenic spaces | ||||
|---|---|---|---|---|---|---|---|---|---|
| no. | % | no. | % | no. | % | no. | % | ||
| (AT)n | 14 | 0 | 0.0 | 0 | 0.0 | 3 | 21.4 | 11 | 78.6 |
| (GA)n | 13 | 1 | 7.7 | 0 | 0.0 | 2 | 15.4 | 10 | 76.9 |
| (CA)n | 4 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 4 | 100.0 |
| AT-rich TNRs | 11 | 2 | 18.2 | 4 | 36.4 | 2 | 18.2 | 3 | 27.3 |
| GC-rich TNRs | 21 | 17 | 80.1 | 1 | 4.8 | 1 | 4.8 | 2 | 9.6 |
| Tetranucleotides | 4 | 0 | 0.0 | 1 | 25.0 | 0 | 0.0 | 3 | 75.0 |
When this data is considered together, it can be concluded that: (a) the 27 fully sequenced clones in this study represent regions of relatively high gene density, in keeping with the prevalence of GC-rich trinucleotide repeats (known to be frequently associated with genes and ESTs), whereas the BAC ends represent regions of lower gene density, as evidenced by the prevalence of (AT)n and (AC)n DNRs and tetranucleotide repeat motifs (which are most abundant in noncoding, intergenic regions), and (b) that regions with relatively high gene density also harbor higher densities of both Class I and Class II SSR attributable mostly to the abundance of GC-rich TNRs in genes. These observations have important implications for the use of SSRs as genetic markers.
Strategies for High-Efficiency SSR Marker Development
We aimed to find the most efficient approaches to the development of new microsatellite markers based on public genomic sequence information, incorporating empirically derived data related to the frequency, size variation potential, and PCR-amplification properties of different types of rice SSRs. First, primers were designed for most of the Class I nonredundant di-, tri- and tetranucleotide SSRs contained in the 74,127 BAC-end sequences. A total of 362 primer pairs were first evaluated for successful PCR amplification on genomic DNA from two rice cultivars. Two hundred sixty six successfully amplified SSRs were then tested for allelic diversity, as described in Methods. The results are summarized in Table 2. The highest rate of successful amplification was achieved for (GA)n, (GAA)n, and (CAT)n microsatellites. The former two classes were also highly polymorphic, which was in accordance with the high mean number of repeat units and wider range of allele size variation observed for these SSRs. Markers with the poly(AT)n motif amplified poorly in general, although primers for compound (TA)n(CA)n blocks, which are a fairly common class of dinucleotide tracts in the rice genome, performed better. The complex (TA)n(CA)n microsatellites have the longest runs of uninterrupted repeats and demonstrate the highest level of allelic diversity in our panel. Three remaining groups — poly(CA)n, polytetranucleotides, and GC-rich TNRs — amplified with moderate success but tended to have fewer alleles and lower polymorphism information content (PIC) values than the other classes.
Table 2.
Characteristics of Rice Microsatellites and Efficiency of SSR Marker Development
| Class of microsatellite markers | No. primer pairs ordereda | Mean no.repeats in SSRb | Mean GC% in flanking regionsc | Rate of successful amplification | Percent of polymorphic SSRs | No. mapped markers |
|---|---|---|---|---|---|---|
| GA | 130 | 15.0 | 44 | 83.8% | 80.7 | 88 |
| CA | 32 | 12.1 | 38 | 71.8% | 73.9 | 17 |
| AT | 63 | 22.3 | 39 | 31.7% | 80.0 | 16 |
| GC-rich TNRs | 45 | 7.8 | 54 | 64.4% | 65.5 | 18 |
| ATT | 23 | 17.6 | 36 | 78.3% | 72.2 | 13 |
| GAA | 23 | 12.7 | 47 | 87.0% | 80.0 | 16 |
| CAT and CAA | 12 | 8.8 | 50 | 83.3% | 70.0 | 7 |
| Tetranucleotides | 42 | 6.1 | 41 | 71.4% | 60.0 | 18 |
| (TA)n(CA)n | 12 | 39.8 | 39 | 58.3% | 100 | 7 |
| TOTAL | 382 | 200 |
The number includes 362 primer pairs ordered for BAC-end derived SSRs and 20 markers selected from the fully-sequenced BACs and PACs.
The mean number of repeats was calculated for the sequences selected for the primer design.
The mean GC content in flanking regions was estimated based on a total number of sequences in this class analyzed in this study.
The poly(AT)n blocks are the most abundant and variable microsatellite sequences in the rice genome, which makes them a major potential source of polymorphic SSR markers. However, they frequently failed to amplify, and we sought an explanation for this behavior. An overwhelming majority of poly(AT)n SSRs reside in noncoding regions and it is possible that the AT richness and the repetitive nature of the DNA sequence flanking the poly(AT)n blocks adversely affect the PCR-based assays. To test this possibility, we determined the GC and AT content (as percentages) in genomic sequences (up to 500 bp in length) immediately flanking each SSR. The results clearly demonstrate that microsatellites with the poly(AT)n and poly(ATT)n motifs are preferentially found in AT-rich genomic regions (Table 2). Nevertheless, the rate of successful amplification was much higher for (ATT)n (78.3%) than for (AT)n SSRs (31.7%), suggesting that the AT richness alone could not explain the poor amplification of the (AT)n-containing markers. Interestingly the flanking regions of (CA)n DNRs, which are successfully PCR-amplified at a higher frequency than the (AT) DNRs, are also relatively AT-rich (38% GC content), whereas (GA)n, (GAA)n, and (CAT)n-containing blocks are typically located in more GC-rich genomic regions (44%–50% GC content). The mean GC content in the flanking regions of GC-rich TNRs is 54%, which is only slightly below the 58% figure obtained by averaging the GC content of 208 rice gene sequences (Carels et al. 1998). This analysis indicates that the occurrence of particular microsatellite motifs is associated strongly with the GC content of the DNA sequences immediately adjoining each repeat in genomic DNA. However, it did not explain the low amplification rate of poly(AT)n containing markers. We therefore hypothesized that the presence of specific sequences in the vicinity of SSRs may influence the PCR success rate for markers derived from these SSRs.
Association of Microsatellites with Other Classes of Repetitive DNA
A possible association between rice SSRs and dispersed repetitive elements in this study was first suggested by BLAST analysis, where sequences flanking the SSR motif were used as a query. The original goal of this analysis was the elimination of redundant SSR-containing sequences from the data set prior to primer design. Numerous sequences flanking (AT)n DNR motifs showed similarity to other BAC ends (these matches were considered significant when one or both sides of the SSR target matched other BAC-end sequences in the database at a cutoff of E < 10−20). Some of the database hits also had poly(AT)n blocks, but with a different number of repeat units. Other database hits matching the same SSR-containing query did not contain any SSRs at all. This observation suggested that (AT)n DNRs in rice are adjoined frequently by other repetitive (non-SSR) sequences that occur at a detectable frequency throughout the genome. Because this definition fits interspersed repetitive elements, we subjected the SSR-containing BAC-end sequences to a BLAST search against a rice repeat database containing known transposable elements and other classes of repetitive DNA (http://www.tigr.org/tdb/rice/blastsearch.html). The search revealed that ∼45% of (AT)n-containing BAC ends from the EcoRI-digested library showed significant homology to Micropon sequences, a new family of miniature inverted-repeat transposable elements (MITEs) discovered by H. Akagi, Y. Yokozeki, A. Inagaki, and T. Fujimura (unpubl.). The Micropon-4 element was most common in the EcoRI BAC-end sequences (Table 3). On the contrary, sequences from the HindIII library rarely showed sequence similarity with the Micropon-4 element and were more frequently associated with other types of repeats, including retrotransposon-derived, centromeric, and ribosomal sequences. The association of (AT)n DNRs with dispersed repetitive elements seems to be a likely reason for poor amplification of these repeats, as primers from their flanking sequences recognize many targets and do not amplify cleanly from a unique site.
Table 3.
Association between Rice Microsatellites and Other Types of Repetitive Elements in BAC-end Sequences
| Number of sequences in this class | ||||||||
|---|---|---|---|---|---|---|---|---|
| Type of homology | BAC ends from the HindIII library | BAC ends from EcoRI library | ||||||
| (AT)n | (CA)n | (AT)n | (CA)n | |||||
| no. | % | no. | % | no. | % | no. | % | |
| >80% with Micropon-4 | 3 | 1.0 | 0 | 47 | 29.7 | 0 | ||
| >80% with other Micropons | 0 | 0 | 4 | 2.5 | 2 | 5.5 | ||
| 54%–80% with any Microponsa | 40 | 13.1 | 10 | 15.4 | 26 | 16.5 | 9 | 25.0 |
| >80% with non-Micropon MITEs | 11 | 3.6 | 2 | 3.1 | 0 | 0 | ||
| 54%–80% with non-Micropon MITEsa | 29 | 9.5 | 11 | 16.9 | 23 | 14.6 | 11 | 30.6 |
| Any retroelement homology | 13 | 4.3 | 5 | 7.7 | 5 | 3.2 | 1 | 2.8 |
| Other (centromeric, ribosomal, etc.) | 18 | 5.9 | 2 | 3.1 | 3 | 1.9 | 0 | |
| Unique, no homology with any repeats in TIGR database on either end of SSR | 188 | 61.6 | 35 | 53.8 | 50 | 31.6 | 13 | 36.1 |
| TOTAL | 304 | 65 | 158 | 36 | ||||
Integration of Genetic and Physical Maps with SSR Markers
Two hundred new microsatellite markers showing polymorphism between parents of the IR64/Azucena doubled haploid (DH) mapping population have been integrated into the existing genetic map (Fig. 3). Among these, 12 primer pairs amplified complex patterns of several independently segregating bands, which in some cases were mapped at multiple loci (RM456, RM464, RM465, RM473, RM476, and RM558). The rest produced PCR fragments that could be mapped as single-locus codominant markers. Information about the new markers (RM400–RM600) is available in the Gramene database (http://www.gramene.org) including the source GenBank ID, clone name, SSR motif description, primer sequences, and polymorphism survey results.
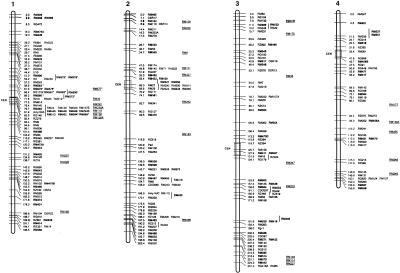
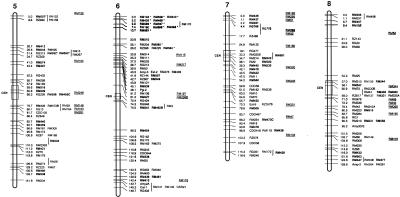
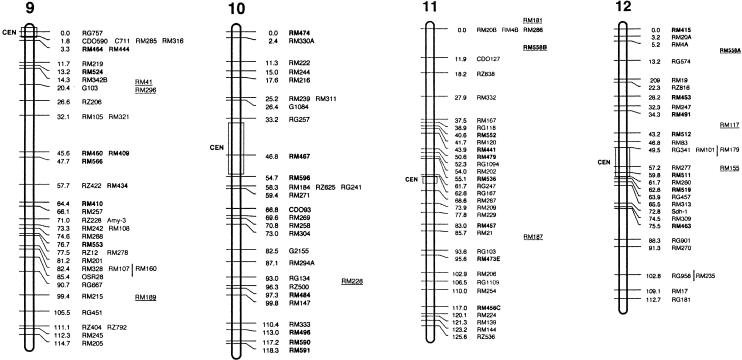
Figure 3.
Molecular linkage map of rice. The framework is based on the IR64/Azucena doubled haploid (DH) population. Short arms of chromosomes are at the top. Approximate positions of centromeres are indicated by CEN with an open box. Framework markers (those ordered at LOD score >2.0) have tick marks on chromosome bars. Cosegregating markers with absolute linkage are in the same row. Vertical lines delimit probable intervals for markers mapped with low LOD score. Markers mapped onto other populations are underlined and placed to the right side of the DH map based on their position in relation to common markers. Abbreviation RM is used for RiceMicrosatellite Markers developed in this lab (Cornell University). OSR loci correspond to microsatellite markers reported previously by Akagi et al. (1996). New RM markers derived from BAC-end sequences are shown in bold; those from fully sequenced large-insert clones are in bold and marked by asterisks. (Figure continues on following pages.)
The current rice microsatellite map shown in Figure 3 has an average density of one SSR marker every 4 cM. To estimate the average density of hypervariable SSR loci that can be reliably targeted for primer design in fully sequenced regions of the rice genome, we identified all the SSR loci in 10 randomly selected, completely sequenced PAC and BAC clones, and then selected the most appropriate targets for experimental analysis. Long (GA)n DNRs and AT-rich TNRs were targeted because they amplified with the highest efficiency and revealed the most polymorphism overall. We avoided Class II SSRs of all types and also avoided Class I poly(AT)n motifs, due to the low polymorphism of the former and the high PCR failure rate of the latter. All primer pairs selected from fully sequenced PAC and BAC clones based on these criteria worked well and detected a high level of allelic diversity among the 13 genotypes in our diversity panel. This experiment demonstrated that polymorphic microsatellite markers could be easily designed to obtain a density of one SSR per 50–75 kb, which translates into a density of one SSR marker every 0.2–0.5 cM on the rice genetic map. For relatively gene-rich regions, similar to those represented by our set of 10 fully sequenced clones, significantly higher densities of microsatellite markers can be achieved if all types of Class I and Class II SSRs are considered and a trial and error approach to marker development is adopted.
In addition to random mapping, a targeted approach was used to fill gaps in the genetic map. Several new markers were designed from completely sequenced PAC clones already assigned to the high-density genetic map (http://rgp.dna.affrc.go.jp/GenomeSeq.html). Markers RM587 and RM510 were targeted to close a gap of 23.6 cM between markers RM190 and RM204 in the vicinity of the waxy locus on the short arm of chromosome 6. Similarly, new microsatellite markers derived from fully sequenced PACs were designed to saturate the region between the RFLP anchor markers RG532 and R210 on the short arm of chromosome 1 and near the telomere on the long arm of chromosome 10 (markers with asterisks in Fig. 3). These examples illustrate the utility of SSRs for establishing connections between genomic sequence, the physical map (http://rgp.dna.affrc.go.jp/GenomeSeq.html), high-density RFLP maps (Causse et al. 1994; Harushima et al. 1998), and the current microsatellite map.
DISCUSSION
Integration of Physical and Genetic Maps with New SSR Markers
To best utilize rice as a model system in plant genomics, it is necessary to integrate classical genetic resources, functional analysis, and sequence information. Saturating the existing rice genetic map with highly informative, technically efficient, sequence-based markers will accelerate the integration. Hypervariable microsatellites provide a useful source of polymorphic DNA markers for connecting genetic maps with genomic sequences and ultimately with phenotypic variation. We report the semiautomated detection and design of SSR markers using genomic sequences of rice. Using this approach, 266 new Class I microsatellite markers were developed, of which 200 have been integrated into the existing microsatellite map, bringing the total number of mapped SSR loci to >500. A subset of 160 of the microsatellite markers was used to develop chromosome-specific multiplex panels of SSR markers for efficient fluorescent-based detection for use in genetics and breeding programs (J.R. Coburn, S.V. Temnykh, and S.R. McCouch, in prep.).
Diversity studies have shown that a high proportion of primers (>80%) designed for Oryza sativa also successfully amplify in cultivated rice from Africa, Oryza glaberrima (Panaud et al. 1996; Lorieux et al. 2000; T. Cadalen and M. Semon, unpubl.), as well as in five other wild AA-genome species (Wu and Tanksley 1993; Panaud et al. 1996; Harrington 2000). The high rate of successful amplification of SSR markers across related species, together with the single-locus nature of these markers, assures their validity as sequence-based connectors for future consolidation of genetic and physical maps and provides the foundation for association of these maps with phenotypes of interest. In particular, it provides an opportunity to use SSR markers for investigating the wide range of genetic diversity that exists in wild relatives outside of the gene pool of O. sativa.
PCR Efficiency, Marker Utility, and Genomic Context of Selected SSR Classes
GC-Rich Trinucleotide SSRs
Comparison of the three data sets — BAC-end sequences, completely sequenced PACs and BACs, and partially sequenced cDNAs (ESTs) — showed that Class I and Class II microsatellites are more frequent in gene-rich regions of the rice genome (represented by ESTs and fully sequenced PACs and BACs) than in randomly sequenced BAC ends, and that the difference is attributable largely to the greater abundance of GC-rich TNRs in exons. This observation was in good agreement with findings in maize and human, where GC-rich polytrinucleotides represented the largest proportion of SSRs detected in cDNAs (Jurka and Petiyagoda 1995; Chin et al. 1996). On the contrary, in the genomes of Arabidopsis and yeast Saccharomyces cerevisiae, the majority of TNRs found in exons are AT-rich (Cardle et al. 2000; Young et al. 2000). It is intriguing that that these TNRs were found preferentially in yeast genes involved in the regulation of transcription, signal transduction, and cell growth and division, but only rarely in genes controlling common metabolic functions such as glycolysis and respiration. It follows that these associations between different TNRs and certain types of genes may be genome-specific. This knowledge, when used in the context of known frequencies of particular TNRs in specific functional categories of genes, might be useful in characterizing novel TNR-containing genes during genome-scale annotation.
(GA)n Dinucleotide SSR
Microsatellite sequences with DNRs usually reside outside of coding regions of genes and are known to be the best source of highly polymorphic SSR markers. In rice, (GA)n polydinucleotides usually occur in regions with a balanced GC content (close to 50%), which favors robust PCR amplification. As demonstrated in earlier studies by Temnykh et al. (2000) and Cho et al. (2000), they are frequently situated in 5′- or 3′-flanking regions of genes and do not appear to be commonly associated with transposable elements (this study). Only a small proportion (10%) of (GA)n DNRs isolated previously from a physically sheared small-insert library was found in close proximity to Gypsy/Ty3 retrotransposon LTRs (S.V. Temnykh, D.M. Larkin, and S.R. McCouch, unpubl.).
Other Types of SSRs
We attempted to develop more SSR markers based on the (AT)n motif, which is the most abundant DNR in rice as well as in other plant species (for review, see Powell et al. 1996; Cardle et al. 2000). Although some new AT-based markers performed well, the PCR success rate was low. This was explained partly by the AT-richness of their flanking regions and partly by the frequent associations of AT-rich SSRs with interspersed repetitive elements, particularly Micropons. Interestingly, the occurrence of (CA)n DNRs is similar to that of the (AT)n DNRs across data sets. They are most frequent in random BAC ends, rare in gene-rich regions, and almost completely absent within or very near genes. We also confirmed our earlier observation that (CA)n repeats are frequently associated with (AT)n blocks and sometimes with tetranucleotide repeats (Temnykh et al. 2000). The observed higher frequency of (CA)n and (AT)n DNRs and tetranucleotide repeats in BAC ends, relative to the much lower proportion of these SSRs in gene-rich BACs and PACs, is likely attributable to a bias for AT-richness in the BAC-end sequences. This bias can be explained by the fact that EcoRI and HindIII, the two restriction enzymes used to construct the libraries, recognize AT-rich sites (GAATTC and AAGCTT, respectively). It remains to be seen whether this association of specific SSR motifs with the AT-rich portion of the rice genome has any relationship to high-order chromatin structure similar to that documented for other plant species (Pedersen et. al. 1996; Schmidt and Heslop-Harrison 1996).
Microsatellite Evolution: From Dispersed Repeats to Evolutionary Constraints
The possible association of rice microsatellites with AT-rich dispersed repeats should be considered in the context of lineage-specific associations between SSRs and middle repetitive elements, which have been only recently elucidated (Arcot et al. 1995; Ramsay et al. 1999). In primates, the close association of SINE repeats and microsatellites has been hypothesized to be due to a preferential retroinsertion of SINEs at staggered nicks within AT-rich genomic regions, followed by mutation-driven conversion of the terminal poly(A) tracts and middle A-rich regions of Alu repeats into microsatellites (Arcot et al. 1995). In avian genomes, notable for their lack of SINEs, no association between SSRs and dispersed repeats is observed, and the overall frequency of SSRs is significantly lower than in the human genome, likely caused in part by the absence of the poly(A)-tailed SINEs whose middle and terminal sequences provide a source for SSR evolution in the human (Primmer et al. 1997). Although the rice genome contains some putative SINEs (such as the pSINE-r family) (Mochizuki et al. 1992) and numerous retrotransposons (Hirochika 1997; Wang et al. 1999) these two classes of repetitive DNA were only rarely associated with SSRs in this study.
Although our results confirm the association of (AT)n DNR microsatellites with the Micropon-family of MITEs reported by H. Akagi, Y. Yokozeki, A. Inagaki, and T. Fujimura (unpubl.), our analysis of the sequences adjoining the poly(AT)n tracts indicates that the genomic expansion of Micropon elements in rice is ancient in evolutionary terms. This conclusion is based on the fact that these elements are frequently so diverged from the consensus sequence that they show <60% similarity. It was also noted that the majority of Micropon-like elements associated with (AT)n repeats display stronger homology to Micropon-4 than to other Micropon family members. We hypothesize that the differential genomic expansion of MITE families in rice, which more recently favored active Micropon-4 copies, is a major cause of the current abundance of (AT)n DNRs in this genome. This association may support SSR evolutionary mechanisms similar to those observed for DNRs in the vicinity of mammalian SINEs.
MITE elements are very widespread in rice and other plant species (Bureau and Wessler 1994; Bureau et al. 1996; Mao et al. 2000) but are only a minor component of the dispersed repeat content of other genomes, including that of the human. Therefore, their peculiar association with (AT)n DNRs in rice is expected to be a somewhat unique observation. As few comprehensive studies of transposable elements (TE) in any plant genome are available at this time, a conclusion on this matter awaits further characterization of TE family identity, frequency, and distribution in rice and other plant genera.
The properties of other SSR motifs in rice suggest that their origin and evolution are unlikely to involve dispersed repeats. For instance, the widespread occurrence of GC-rich TNRs in expressed sequences of rice and maize may simply reflect the overall high GC content of coding sequences in species from the Gramineae family (Carels et al. 1998) and the stochastic nature of these SSRs. In fully sequenced large-insert clones of the rice genome, GC-rich TNRs usually occur in larger regions of quasirepetitive GC-rich sequences, which frequently code for homopolymeric runs of amino acids in the corresponding predicted proteins. This observation suggests other mechanisms that operate to maintain these DNA structures via selection for homopolymeric tracts at the protein level. It has been suggested that TNRs could have an important role in speciation, as their instability may lead to rapid evolution of new domains in regulatory proteins (Young et al. 2000). Obviously, an extreme expansion of TNRs in genes can have drastic phenotypic effects as has been documented for several human disorders (McMurray 1995; Cummings and Zoghbi 2000).
The position of numerous rice TNRs in a GC-rich, low-complexity sequence context suggests that short TNRs with a few perfect repeats could have originated purely as a result of stochastically occurring nucleotide substitutions. Statistical characterization of microsatellites in the fully sequenced genome of the yeast S. cerevisiae provides evidence that short tracts of SSRs occur simply by chance due to random nucleotide substitutions. Only after reaching a certain minimum length do they acquire the ability to expand, most likely by the slipped-strand mispairing mechanism. For yeast microsatellites, this length is close to 10 nucleotides, which translates into five repeat units for DNRs, at least four repeat units for TNRs, and at least three repeats for tetranucleotides (Pupko and Graur 1999). Our data demonstrate that in rice, as in yeast, potential sites for microsatellite expansion are generally longer than 10 nucleotides. The long, hypervariable SSR sequences of the rice genome, the type needed for map construction and linkage mapping, represent only a small portion of the total set of potential slippage-driven expansion sites. Therefore, the set of 500 long, polymorphic SSRs that is currently available for rice (Akagi et al. 1996; Panaud et al. 1996; Chen et al. 1997; Temnykh et al. 2000; this study) is of great value, as it is enriched for useful DNA landmarks. In addition, the availability of a script that enables rapid and reliable extraction of additional Class I microsatellites from the emerging genomic sequence (http://www.gramene.org) will greatly expand the repertoire of known SSRs in rice. Although rare in the overall SSR fraction of the rice genome, the more polymorphic microsatellites can be effectively exploited for mapping and annotation of the rice genome, leading to a better utilization of rice, both as a model for genome studies and as a staple food crop.
METHODS
Accessing Rice Sequence Information and Detection of SSRs
A total of 74,127 rice BAC-end sequences (Clemson University BAC End Sequencing Project, http://www.genome.clemson.edu/projects/rice/), with an average length of 500 bp each, were acquired in bulk from GenBank using Batch Entrez (http://www.ncbi.nlm.nih.gov/Entrez/batch.html). In addition, five complete BAC sequences generated by the US Rice Genome Sequencing Program (USRGSP) (http://www.tigr.org/tdb/rice/) and 22 complete PAC sequences developed by the Rice Genome Research Program in Japan (JRGP) (http://rgp.dna.affrc.go.jp/GenomeSeq.html) were used. A program was written in the Perl scripting language for SSR identification and characterization. The script uses regular expressions to locate SSR patterns in FASTA-formatted sequence files and reports the GenBank ID, SSR motif, number of repeats, sequence coordinates for each SSR and GC% in DNA sequences (up to 500 bp in length) immediately adjoining SSR. This searching routine can be used to identify SSRs in different types of genomic DNA sequences, varying in size from several hundred nucleotides (BAC-end reads) up to 1 Mb of long contigs assembled from fully sequenced BACs and PACs. The script is available at URL: http://www.gramene.org
Sequence Redundancy Check and Primer Design
To eliminate redundancy (and therefore, the possibility that multiple sets of primers would be designed for the same locus), all SSR-containing sequences were subjected to a BLAST search (Altschol et al.) against a local BLAST database that contained all the downloaded BAC-end and full-length BAC and PAC sequences, in addition to sequences of short-insert genomic library clones previously used for microsatellite marker development in this laboratory. Default BLAST parameters included in the stand-alone BLAST software were used for all redundancy searches. After redundant sequences were removed from the sequence pool, unique SSR-containing sequences were passed to PRIMER 0.5 for primer design (Daly et al. 1991).
Annotation Tools and Search for Associations between SSRs and Other Classes of Repetitive DNA
The comprehensive annotation package Seqhelp (Lee et al. 1998) was used for gene prediction and for the identification of potential dispersed repeats in fully sequenced large-insert clones. Locations of SSRs were indicated in relation to open reading frames (ORFs) to estimate the frequency of occurrence of SSR tracts in genes and in intergenic spaces. Four fully sequenced clones (GenBank AC007789 and AP000969 on chr. 1; AP000616 and AP001129 on chr. 6) were annotated in this manner. Homology searches against the rice repeat database at TIGR (http://www.tigr.org/tdb/rice/blastsearch.html) were performed to reveal candidate repetitive elements in the vicinity of SSRs.
Amplification Test and Evaluation of Allelic Diversity
Primers were synthesized by Research Genetics and tested for amplification using DNA from the parents of the IR64 × Azucena DH mapping population. Thirteen diverse O. sativa genotypes widely used as mapping parents in rice research programs in the US, China, Japan, Korea, and the Philippines comprised the panel used for the evaluation of microsatellite allelic diversity as described in Cho et al. (2000). This panel included seven varieties of indica type (IR36, IR64, N22, Zhai-Ye-Qing 8, Teqing, Kasalath, BS125), five varieties of japonica type (Azucena, Jing-Xi 17, Gihobyeo, Lemont, Nipponbare) and one variety derived form a indica/japonica cross (Milyang 23). DNA was extracted from fresh leaves by a method based on the protocol described by Causse et al. (1994). PCR was performed in a PTC100 96V thermocycler (MJ Research) as described by Temnykh et al. (2000). The basic profile was: 5 min at 94°C, 35 cycles of 1 min at 94°C, 1 min at 55°C, 2 min at 72°C, and 5 min at 72°C for final extension. PCR products were separated on 4% polyacrylamide denaturing gels and marker bands were revealed using the silver staining protocol as described by Panaud et al. (1996). Allele scoring and estimation of expected heterozygosity (PIC value) was done as described by Cho et al. (2000).
Mapping of SSRs
A population of DH lines derived from the intersubspecific cross between IR64 (O. sativa ssp. indica) and Azucena (tropical japonica) via another culture (Guiderdoni et al. 1992; Huang et al. 1994) was used as the basis for placing microsatellite markers onto rice chromosomes. The last version of the SSR map reported by Temnykh et al. (2000) included 237 microsatellite markers in addition to 145 RFLP anchor markers mapped previously on this population by Huang et al. (1994). Two hundred new markers developed in this study were incorporated into the existing DH map using a subset of 96 individuals from the original mapping population. MAPMAKER 2.0 (Lander et al. 1987) was run on a Macintosh computer using the Kosambi mapping function. The “ripple” test was used to confirm marker order as determined by multipoint analysis. Markers with a ripple of LOD > 2.0 were integrated into the framework maps, and those mapping with LOD < 2.0 were assigned to the most likely intervals.
Acknowledgments
We gratefully acknowledge the Rockefeller Foundation for financial support (RF98001#663 and RF99001#726), and Angela Baldo and Doreen Ware for presentation of our data in the RiceGenes/Gramene database. We also thank Richard Durrett, Mark Sorrells, Ramesh Kantety, and Angela Baldo for critical reviews of the manuscript; and Lois Swales for help with formatting.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Corresponding author.
E-MAIL SRM4@cornell.edu; FAX (607) 255-6683.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.184001.
REFERENCES
- Akagi H, Yokozeki Y, Inagaki A, Fujimura T. Microsatellite DNA markers for rice chromosomes. Theor Appl Genet. 1996;94:61–67. doi: 10.1007/BF00230127. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aragon-Alcaide L, Miller T, Schwarzacher T, Reader S, Moore G. A cereal centromeric sequence. Chromosoma. 1996;105:261–268. doi: 10.1007/BF02524643. [DOI] [PubMed] [Google Scholar]
- Arcot SS, Wan Z, Weber JL, Deininger PL, Batzer MA. Alu repeats: A source for the genesis of primate microsatellites. Genomics. 1995;29:136–144. doi: 10.1006/geno.1995.1224. [DOI] [PubMed] [Google Scholar]
- Bao JS, Zheng XW, Xia YW, He P, Shu QY, Lu X, Chen Y, Zhu LH. QTL mapping for the paste viscosity characteristics in rice (Oryza sativa L.) Theor Appl Genet. 2000;100:280–284. [Google Scholar]
- Bres-Patry C, Loreux M, Clement G, Bangratz M, Ghesquiere A. Heredity and genetic mapping of domestication-related traits in a temperate japonica weedy rice. Theor Appl Genet. 2001;102:118–126. [Google Scholar]
- Budiman MA. “Construction and characterization of deep coverage BAC libraries for two model crops: Tomato and rice, and initiation of a chromosome walk to jointless-2 in tomato”. PhD. thesis. College Station, TX: Texas A & M University; 1999. [Google Scholar]
- Bureau TE, Wessler SR. Stowaway: A new family of inverted repeat elements associated with the genes of both monocotylenous and dicotyledonous plants. Plant Cell. 1994;6:907–916. doi: 10.1105/tpc.6.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau TE, Ronald PC, Wessler SR. A computer-based systemic survey reveals the predominance of small inverted-repeat elements in wild-type rice genes. Proc Natl Acad Sci. 1996;93:8524–8529. doi: 10.1073/pnas.93.16.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings CJ, Zoghbi HY. Fourteen and counting: Unraveling trinucleotide repeat diseases. Hum Mol Genet. 2000;9:909–916. doi: 10.1093/hmg/9.6.909. [DOI] [PubMed] [Google Scholar]
- Carels N, Hatey P, Jabbari K, Bernardi G. Compositional properties of homologous coding sequences from plants. J Mol Evol. 1998;46:45–53. doi: 10.1007/pl00006282. [DOI] [PubMed] [Google Scholar]
- Cardle L, Ramsay L, Milbourne D, Macaulay M, Marshall D, Waugh R. Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics. 2000;156:847–854. doi: 10.1093/genetics/156.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causse MA, Fulton TM, Cho YG, Ahn SN, Chunwongse J, Wu K, Yu Z, Ronald PC, Harrington SE, Second G, et al. Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics. 1994;138:1251–1274. doi: 10.1093/genetics/138.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Temnykh S, Xu Y, Cho YG, McCouch SR. Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.) Theor Appl Genet. 1997;95:553–567. [Google Scholar]
- Chin ECL, Senior ML, Shu H, Smith JSC. Maize simple repetitive DNA sequences: abundance and allele variation. Genome. 1996;39:866–873. doi: 10.1139/g96-109. [DOI] [PubMed] [Google Scholar]
- Cho YG, Ishii T, Temnykh S, Chen X, Lipovich L, Park WD, Ayres N, Cartinhour S, McCouch SR. Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.) Theor Appl Genet. 2000;100:713–722. [Google Scholar]
- Daly MJ, Lincoln SE, Lander ES. PRIMER unpublished software, Whitehead Institute/MIT Center for Genome Research. 1991. http://www-genome.wi.mit.edu/ftp/pub/software/primer.0.5ftp-genome.wi.mit.edu Available at http://www-genome.wi.mit.edu/ftp/pub/software/primer.0.5, and via anonymous ftp to ftp-genome.wi.mit.edu, directory/pub/software/primer.0.5. , and via anonymous ftp to , directory/pub/software/primer.0.5. , directory/pub/software/primer.0.5. [Google Scholar]
- Fondon JW, III, Mele GM, Brezinschek RI, Cummings D, Pande A, Wren J, O'Brien KM, Kupfer KC, Wei M-H, Lerman M, et al. Computerized polymorphic marker identification: Experimental validation and a predicted human polymorphism catalog. Proc Natl Acad Sci. 1998;95:7514–7519. doi: 10.1073/pnas.95.13.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiderdoni E, Galinato E, Luistro J, Vergara G. Anther culture of tropical japonica x indica hybrids of rice (Oryza sativa L) Euphytica. 1992;62:219–224. [Google Scholar]
- Harrington S. “A survey of genetic diversity of eight AA genome species of Oryza using microsatellite markers.” MS thesis. Ithaca, NY: Cornell University; 2000. [Google Scholar]
- Harushima Y, Jano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin SY, Antonio BA, Parco A, et al. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics. 1998;148:479–494. doi: 10.1093/genetics/148.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H. Retrotransposons of rice: Their regulation and use for genome analysis. Plant Mol Biol. 1997;35:231–240. [PubMed] [Google Scholar]
- Huang N, McCouch SR, Mew MT, Parco A, Guiderdoni E. Development of a RFLP map from a double haploid population in rice. Rice Geneti Newsl. 1994;11:134–137. [Google Scholar]
- Jiang J, Nasuda S, Dong F, Scherrer CW, Woo S-S, Wing RA, Gill BS, Ward DC. A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc Natl Acad Sci. 1996;93:14210–14213. doi: 10.1073/pnas.93.24.14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Pethiyagoda C. Simple repetitive DNA sequences from primates: Compilation and analysis. J Mol Evol. 1995;40:120–126. doi: 10.1007/BF00167107. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow MJ, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lee MK, Lynch ED, King MC. SeqHelp: A program to analyze molecular sequences using common computational resources. Genome Res. 1998;8:306–312. doi: 10.1101/gr.8.3.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieux M, Ndjiondjop M-N, Ghesquiere A. A first interspecific Oryza sativa x Oryza glaberrima microsatellite-based genetic map. Theor Appl Genet. 2000;100:591–601. [Google Scholar]
- Mao L, Wood TC, Yu Y, Budiman MA, Tomkins J, Woo S-S, Sasinowski M, Presting G, Frisch D, Goff S, et al. Rice Transposable Elements: A Survey of 73,000 Sequence-Tagged-Connectors. Genome Res. 2000;10:982–990. doi: 10.1101/gr.10.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray CT. Mechanisms of DNA expansion. Chromosoma. 1995;104:2–13. doi: 10.1007/BF00352220. [DOI] [PubMed] [Google Scholar]
- Moncada P, Martinez CP, Borrero J, Chatel M, Gauch H, Jr, Guimaraes E, Tohme J, McCouch SR. Quantitative trait loci for yield and yield components in an Oryza sativa x Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor Appl Genet. 2001;102:41–52. [Google Scholar]
- Mochizuki K, Umeda M, Ohtsubo H, Ohtsubo E. Characterization of plant SINE, p-SINE1, in rice genomes. Jpn J Genet. 1992;67:155–166. doi: 10.1266/jjg.67.155. [DOI] [PubMed] [Google Scholar]
- Olufowote JO, Xu Y, Chen X, Park WD, Beachell HM, Dilday RH, Goto M, McCouch SR. Comparative evaluation of within-cultivar variation of rice (Oryza sativa L.) using microsatellite and RFLP markers. Genome. 1997;38:1170–1176. doi: 10.1139/g97-050. [DOI] [PubMed] [Google Scholar]
- Panaud O, Chen X, McCouch SR. Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.) Mol Gen Genet. 1996;252:597–607. doi: 10.1007/BF02172406. [DOI] [PubMed] [Google Scholar]
- Pedersen C, Rasmussen SK, Linde-Laursen I. Genome and chromosome identification in cultivated barley and related species of the Triticeae (Poaceae) by in situ hybridization with the GAA-satellite sequence. Genome. 1996;39:93–104. doi: 10.1139/g96-013. [DOI] [PubMed] [Google Scholar]
- Powell W, Machray GC, Provan J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996;1:215–222. [Google Scholar]
- Primmer CR, Raudsepp T, Chowdhary BP, Moller AP, Ellegren H. Low frequency of microsatellites in the avian genome. Genome Res. 1997;7:471–482. doi: 10.1101/gr.7.5.471. [DOI] [PubMed] [Google Scholar]
- Pupko T, Graur D. Evolution of microsatellites in the yeast Saccharomyces cerevisiae: Role of length and number of repeated units. J Mol Evol. 1999;48:313–316. doi: 10.1007/pl00006474. [DOI] [PubMed] [Google Scholar]
- Ramsay L, Macaulay M, Cardle L, Mongante M, Ivanissevich S, Maestri E, Powell W, Waugh R. Intimate association of microsatellite repeats with retrotransposons and other dispersed repetitive elements in barley. Plant J. 1999;17:415–425. doi: 10.1046/j.1365-313x.1999.00392.x. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Heslop-Harrison JS. The physical and genomic organization of microsatellites in sugar beet. Proc Natl Acad Sci. 1996;93:8761–8765. doi: 10.1073/pnas.93.16.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen RG, Kwitek-Black AE, Glenn C, Gullings-Hindley J, Van Etten W, Atkinson S, Appel D, Twiggler S, Muir M, Mull T, et al. A high-density integrated genetic linkage and radiation hybrid map of the laboratory rat. Genome Res. 1999;9:1–9. [PubMed] [Google Scholar]
- Temnykh S, Park WD, Ayers N, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishii T, McCouch SR. Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.) Theor Appl Genet. 2000;100:697–712. [Google Scholar]
- Wang S, Liu N, Peng K, Zhang Q. The distribution and copy number of copia-like retrotransposons in rice (Oryza sativa L.) and their implications in the organization and evolution of the rice genome. Proc Natl Acad Sci. 1999;96:6824–6828. doi: 10.1073/pnas.96.12.6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JL. Informativeness of human (dC-dA)n (dG-dT)n polymorphisms. Genomics. 1990;7:524–530. doi: 10.1016/0888-7543(90)90195-z. [DOI] [PubMed] [Google Scholar]
- Wu KS, Tanksley SD. Abundance, polymorphism and genetic mapping of microsatellites in rice. Mol Gen Genet. 1993;241:225–235. doi: 10.1007/BF00280220. [DOI] [PubMed] [Google Scholar]
- Xiao J, Li J, Grandillo S, Ahn SN, Yuan L, Tanksley SD, McCouch SR. Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Orysa rufipogon. Genetics. 1998;150:899–909. doi: 10.1093/genetics/150.2.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang GP, Saghai Maroof MA, Xu CG, Zhang Q, Biyashev RM. Comparative analysis of microsatellite DNA polymorphism in landraces and cultivars of rice. Mol Gen Genet. 1994;245:187–194. doi: 10.1007/BF00283266. [DOI] [PubMed] [Google Scholar]
- Young ET, Sloan JS, Van Riper K. Trinucleotide repeats are clustered in regulatory genes in Saccharomyces cerevisiae. Genetics. 2000;154:1053–1068. doi: 10.1093/genetics/154.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou JH, Pan XB, Chen ZX, Xu JY, Lu JF, Zhai WX, Zhu LH. Mapping quantitative trait loci controlling sheath blight resistance into rice cultivars (Oryza sativa L.) Theor Appl Genet. 2000;101:569–573. [Google Scholar]