Abstract
Alternate polyadenylation affects a large fraction of higher eucaryote mRNAs, producing mature transcripts with 3′ ends of variable length. This variation is poorly represented in the current transcript catalogs derived from whole genome sequences, mostly because such posttranscriptional events are not detectable directly at the DNA level. Alternate polydenylation of an mRNA is better understood by comparision to EST databases. Comparing ESTs to mRNAs, however, is a difficult task subjected to the pitfalls of internal priming, presence of intron sequences, repeated elements, chimerical ESTs or matches with EST from paralogous genes. We present here a computer program that addresses these problems and displays ESTs matches to a query mRNA sequence to predict alternate polyadenylation and to suggest library-specific forms. The output highlights effective polyadenylation signals, possible sources of artifacts such as A-rich stretches in the mRNA sequences, and allows for a direct visualization of EST libraries using color codes. Statistical biases in the distribution of alternative mRNA forms among EST libraries were systematically sought. About 1450 human and 200 mouse mRNAs displayed such biases, suggesting in each case a tissue- or disease-specific regulation of polyadenylation.
Most eukaryotic pre-mRNAs contain long 3′ untranslated regions (UTRs) spanning hundreds of nucleotides, and undergoing cleavage and polyadenylation at one or several polyadenylation sites (PAS). Poly(A) sites are defined by a hexameric polyadenylation signal (AAUAAA or a one-base variant thereof), located ∼15 bases upstream of the cleavage site and, sometimes, a GU (Guanosyl Uridy-R)-rich element located 20–40 bases downstream of the site (for reviews, see Proudfoot 1991; Colgan and Manley 1997). A significant fraction of UTRs has two or more functional, producing mature mRNAs with 3′ regions of variable lengths. As UTRs may contain regulatory elements affecting mRNA stability or translation efficiency, the choice of alternate polyadenylation sites may strongly affect the final expression of the gene. Indeed, differential polyadenylation has been shown repeatedly to occur in a tissue- or disease-specific manner (Edwalds-Gilbert et al. 1997).
Although genome sequencing projects are now polishing complete gene catalogs for several animal species, including human, transcript catalogs covering every polyadenylation or splice variant are still far from completion. Alternate polyadenylation cannot be predicted from the genomic sequence alone, since polyadenylation signals, or GU-rich regions do not carry enough information to constitute useful signatures. The most reliable data on mRNA 3′ ends is experimental, and available in the form of expressed sequence tags (ESTs). The dbEST database (Boguski et al. 1993), currently contains 7.3 million partial cDNAs. These data are highly redundant, the 3 million human ESTs available representing ∼100 times the estimated number of human genes (Lander et al. 2001; Venter et al. 2001). A large fraction of ESTs are sequenced from the 3′ end of mRNAs, and this redundant coverage of the 3′ region often comprises several polyadenylation variants. Computer analyses of EST databases have improved our understanding of polyadenylation signals and alternate polyadenylation (Gautheret et al. 1998; Graber et al. 1999). Studies based on ESTs evaluated that over 29% of human mRNAs had multiple polyadenylation sites (Beaudoing et al. 2000), or >40% if one considers alternative cleavage sites occurring downstream of a single polyadonylation signal. (Pauws et al. 2001).
EST-based annotation requires aligning the mRNA or gene under study to EST sequences. Standard sequence alignment tools such as BLAST (Altschul et al. 1997) can be used for this purpose, provided that certain pitfalls of EST comparisons are dealt with properly. This includes the detection of internally primed ESTs (which can be mistaken for true mRNA 3′ ends), chimeras, and ESTs from paralogous genes. We developed a program (ESTparser) that performs BLAST searches against EST databases and filters the output to produce a general picture of alternatively polyadenylated forms and the in which tissues they occur. We applied this program to a database of human 3′ UTRs (Pesole et al. 1999) and systematically sought instances of tissue-specific 3′ variants. This procedure identified over 3500 events of statistically significant biases. Each bias does not necessarily imply a true differential polyadenylation event because library-specific artifacts may affect the accuracy of ESTs counts. However, outputs of ESTparser show a large number of intriguing cases that combine evidences for alternate poly(A sites and suggestions of tissue- or) disease-specific forms, thus prompting further experimental validations.
RESULTS AND DISCUSSION
We analyzed ∼13,000 human and 6000 mouse UTRs using the October 2000 release of dbEST. The number of UTRs displaying two or more putative polyadenylation sites was 5127 for human and 1296 for mouse sequences. From the library information in dbEST (4960 human and 468 mouse libraries), we classified ESTs into 117 tissue-types, subdivided into 14 categories or organ systems (Table 1). Among UTRs with multiple poly(A) sites, we then sought biases in tissue-distribution. Fisher's Exact tests (Agresti 1992) were performed systematically for each pair of poly(A) sites in the same UTR as described in Methods. We observed 3619 biases in polyadenylation site usage in 1438 different human UTRs (Table 2) and 310 biases in 189 different mouse UTRs (Table 3). A single UTR may display several biases as each poly(A) site and library is tested independently. The number of observed biases for each tissue type is roughly proportional to the number of ESTs and/or libraries available for this tissue, which could be expected because biases are sought on a library-by-library basis.
Table 1.
Keyword-Based Classification of EST Libraries into Organ Systems
| Organ system | Main tissue names or keywords used | # Libraries | Total ESTs | ||||
|---|---|---|---|---|---|---|---|
| Homo | Mus | Total | Homo | Mus | Total | ||
| Cell lines | Cell-line, HeLa,… | 198 | 55 | 254 | 131590 | 32389 | 163979 |
| Central nervous system | brain, ear, eye, olfactive, retina | 267 | 75 | 342 | 282448 | 227876 | 510324 |
| Connective tissues and smooth muscle | adipose, connective, fibroblast, smooth-muscle | 72 | 10 | 82 | 28119 | 6301 | 34420 |
| Digestive system | buccal, colon, esophagus, gallbladder, intestine, liver, omentum, pancreas, parotid, stomach | 864 | 36 | 900 | 291371 | 128301 | 419672 |
| Endocrine glands | adrenal, parathyroid, pineal, thyroid | 39 | 6 | 45 | 44240 | 14170 | 58410 |
| Exocrine glands | breast, ductal, mammary | 980 | 13 | 993 | 116127 | 152046 | 268173 |
| Immune system and blood elements | Bcell, blood, bone-marrow, hemopoietic, leukocyte, lymphatic, macrophage, monocyte, spleen, T-cell, thymus, tonsil | 214 | 45 | 269 | 151749 | 194848 | 346597 |
| Mixed and unknown | chromosomes, whole embryo, mediastinum, metastase, mixed, unknown | 132 | 108 | 240 | 353749 | 425154 | 778903 |
| Peripheral nervous system | chord, nervous, oblongata, spinal | 302 | 12 | 314 | 43811 | 65047 | 108858 |
| Respiratory system | bronchi, larynx, lung, pharynx, trachea | 138 | 11 | 149 | 135831 | 28324 | 164155 |
| Bone and skeletic muscle | bone, cartilage, muscle, synovial | 30 | 13 | 43 | 52499 | 37039 | 89538 |
| Skin | cornea, derm | 744 | 8 | 752 | 126789 | 48074 | 174863 |
| Urogenital system | bladder, endometrium, epididym, germinal, gonad, kidney, ovary, oviduct, placenta, prostate, testis, urogenital, uterus, vagina | 937 | 65 | 1002 | 604173 | 246046 | 850219 |
| Vascular system | aorta, endothelium, heart, vein | 43 | 12 | 55 | 90396 | 51952 | 142348 |
| 4960 | 468 | 4886 | 2,452,892 | 1,657,567 | 4,110,459 | ||
Table 2.
Polyadenylation Site Biases Found in Each Category of Tissue (Human)
| Tissue type | Total EST in tissue | Total libraries in tissue | mRNAs with bias in tissue | Total biases in tissue |
|---|---|---|---|---|
| Mixed | 260442 | 8 | 261 | 374 |
| Brain | 156275 | 18 | 127 | 226 |
| Cell-line | 131590 | 12 | 25 | 32 |
| Lung | 128237 | 18 | 83 | 150 |
| Uterus | 126394 | 26 | 115 | 189 |
| Kidney | 85310 | 12 | 65 | 101 |
| Derm | 69317 | 101 | 170 | 457 |
| Placenta | 66530 | 9 | 33 | 57 |
| Colon | 65066 | 56 | 120 | 271 |
| Germinal | 54513 | 3 | 105 | 152 |
| Ovary | 53488 | 16 | 50 | 101 |
| Prostate | 53269 | 37 | 62 | 111 |
| Tonsil | 50932 | 1 | 64 | 91 |
| Testis | 46846 | 5 | 73 | 92 |
| Liver | 43877 | 3 | 51 | 128 |
| Heart | 43011 | 3 | 26 | 34 |
| Breast | 42361 | 70 | 101 | 245 |
| Pancreas | 37381 | 4 | 47 | 66 |
| Stomach | 35909 | 43 | 87 | 175 |
| Muscle | 35232 | 4 | 32 | 40 |
| Fetus | 27045 | 1 | 17 | 19 |
| Bcell | 26995 | 2 | 17 | 40 |
| Nervous | 22838 | 40 | 83 | 234 |
| Parathyroid | 22412 | 1 | 16 | 20 |
| Retina | 21782 | 3 | 7 | 17 |
| Fibroblast | 13151 | 1 | 5 | 5 |
| Lymphatic | 10857 | 1 | 3 | 3 |
| Ear | 8494 | 1 | 7 | 11 |
| Aorta | 7155 | 2 | 17 | 33 |
| Bladder | 6899 | 1 | 1 | 2 |
| Pineal | 6508 | 1 | 1 | 1 |
| Bone | 5619 | 1 | 2 | 2 |
| Urogenital | 5545 | 1 | 1 | 1 |
| Bone-marrow | 5263 | 3 | 7 | 10 |
| Unknown | 5077 | 4 | 8 | 13 |
| Blood | 4910 | 2 | 12 | 33 |
| Esophag | 3695 | 1 | 1 | 1 |
| Smooth-muscle | 3629 | 14 | 24 | 62 |
| Thymus | 3443 | 1 | 3 | 3 |
| Thyroid | 2459 | 1 | 1 | 1 |
| Endothelium | 1908 | 1 | 1 | 5 |
| Embryo | 1392 | 1 | 1 | 1 |
| Larynx | 1096 | 1 | 1 | 1 |
| Wcell | 1011 | 1 | 3 | 4 |
| Adrenal | 411 | 1 | 1 | 1 |
| Spleen | 311 | 1 | 1 | 2 |
| Vein | 48 | 1 | 1 | 2 |
| Total | 1805933 | 538 | 1939 (1438 distinct) | 3619 |
Table 3.
Polyadenylation Site Biases Found in Each Category of Tissues (Mouse)
| Tissue type | Total EST in tissue | Total libraries in tissue | mRNAs with bias in tissue | Total biases in tissue |
|---|---|---|---|---|
| Embryo | 131214 | 10 | 56 | 74 |
| Breast | 106676 | 5 | 35 | 48 |
| Unknown | 102196 | 9 | 26 | 31 |
| Thymus | 74815 | 3 | 9 | 9 |
| Testis | 54729 | 3 | 18 | 24 |
| Brain | 54255 | 3 | 4 | 4 |
| Derm | 43907 | 3 | 6 | 7 |
| Kidney | 40994 | 4 | 28 | 39 |
| Fetus | 37562 | 2 | 7 | 7 |
| Muscle | 34710 | 2 | 2 | 2 |
| Nervous | 29666 | 2 | 4 | 4 |
| Liver | 29132 | 3 | 9 | 12 |
| Spleen | 24136 | 1 | 5 | 6 |
| Heart | 22345 | 3 | 4 | 6 |
| Tonsil | 20252 | 2 | 2 | 2 |
| Mixed | 16058 | 3 | 3 | 3 |
| Lymphatic | 14753 | 1 | 2 | 2 |
| Vessel | 14572 | 1 | 1 | 1 |
| Cell line | 14510 | 1 | 1 | 1 |
| Germinal | 13979 | 2 | 4 | 4 |
| Buccal | 11489 | 1 | 2 | 4 |
| Colon | 10495 | 1 | 2 | 3 |
| Intestine | 9781 | 1 | 4 | 4 |
| Tcell | 9459 | 1 | 1 | 1 |
| Vagina | 8930 | 1 | 1 | 1 |
| Bone | 8262 | 1 | 1 | 1 |
| Ovary | 7577 | 1 | 2 | 2 |
| Stomach | 7427 | 1 | 2 | 2 |
| Uterus | 6170 | 1 | 2 | 2 |
| Placenta | 5707 | 1 | 1 | 1 |
| Lung | 4406 | 1 | 1 | 1 |
| Adrenal | 3765 | 1 | 1 | 2 |
| Total | 973929 | 75 | 246 (189 distinct) | 310 |
We did not observe a strong positional preference for the differentially polyadenylated forms, except that the shortest UTR form was preferred in two-thirds of the biased libraries. We inspected the UTR sequences between alternate polyadenylation sites for the presence of ARE destabilization elements (AU-rich elements of the type AUUUA or UUAUUUA[U/A][U/A]). The density of ARE in these segments did not differ significantly from that in other UTR regions (data not shown).
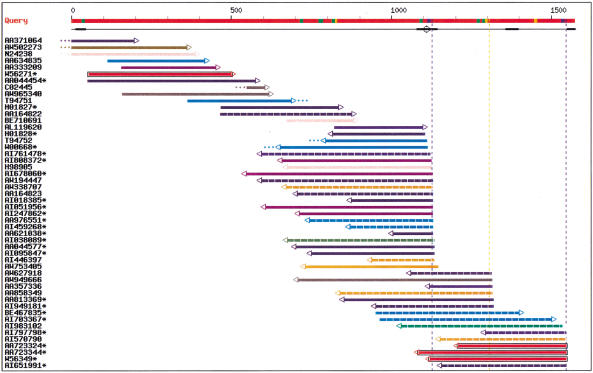
A representative output is shown in Figure 1. In this example, the 3′ UTR sequence of a zinc-finger DNA-binding protein mRNA (Muraosa et al. 1996) was analyzed. The red line on top represents the UTR sequence, numbered from zero at Stop codon. Fifty ESTs (color lines) were found to match this UTR within the required length and identity criteria. Color coding is described in the figure legend. ESTs shown with dashed lines are from cancer libraries. There is evidence for three polyadenylation signals, at positions 1111, 1292, and 1532. The signals at 1111 and 1532 are AATAAA (blue box) and the signal at 1292 is ATTAAA (orange box). The thickened black underlines indicate regions of query masking, which means the program would not consider hits contained entirely in this region as significant because of the presence of a low complexity region, vector sequence, or human repeat such as Alu. The open circle near position 1100 indicates a poly(A) stretch in the query sequence, that is, a possible source of internal priming. Four ESTs (AL119620, H01828, T94752, and WW00668) appear to have been produced by internal priming at this site. Dots at the extremities of ESTs indicate that a fragment larger than 20 nt or 15 nt, respectively at the 3′ or 5′ end of the EST, does not match the query sequence. Dots appearing past the 5′ end of the query indicate ESTs extending into the coding region (e.g., the first three ESTs). Dots present within the limits of the query sequence indicate discrepancies between the EST and query (e.g., EST T94751). The most common explanation for these is the poor sequence quality of EST extremities, but other phenomena, such as chimeras, presence of intronic sequences, or alternative exons may also produce such mismatches. Therefore, these dubious ESTs should not be considered in alternative form counts.
Figure 1.
EST-parser output for the 3′ untranslated region of a zinc-finger DNA-binding protein mRNA (EMBL accession no. D45132, Muraosa et al. 1996). The red line on top represents the query sequence. Potential poly(A) signals are shown with colored boxes: blue, AAUAAA signals; orange, AUUAAA signals; green, other alternate signals. The next line indicates regions masked for their unspecific content (low complexity, vectors, mammalian repeats) using a thickened line, and potential internal priming sites (adenine stretches) are indicated by open circles. Vertical broken lines indicate putative polyA sites. When a signal is present, the vertical line has the same color as the signal box, otherwise, the line is grey. Each EST is then represented by a horizontal line incorporating information by means of a color code. EST coloring is made according to the organ system of the EST library (see Table 1). Color coding is as follows: olive, cell line; lime, central nervous system; fuschia, connective tissues; orange, digestive system; green, endocrine glands; dark slate blue, exocrine glands; blue, immune system; purple, mixed tissues; yellow, peripheral nervous system; aqua, respiratory system; maroon, squelettic; pink, skin; grey, unknown; navy, uro-genital; red, vascular system. The EST line also shows dangling ends of 20 nt or more (dots at extremities); 5′ to 3′ direction of EST sequence (arrow at extremity); and possible evidence of library-specific 3′ end (black box around EST line). Asterisks indicate ESTs from normalized or subtracted libraries. In the Web interface, additional library information is available by sliding the mouse over any EST in the chart. Organ name and Library Id. Will appear in a pop-up box (using Microsoft Internet Explorer) or at the bottom of the window (using Netscape), along with various information on the EST match, such as: Genbank ID of EST, dbEST library Id, tissue name, disease/normal state, EST length, percent identity with query sequence, coordinates for query and EST, signal type, signal position on query, and presence or absence of A/T tail on EST.
ESTs from libraries with a 3′ end bias are shown boxed. Here, three ESTs from Soares fetal heart library NbHH19W have their 3′ end at signal 1532 (red, boxed ESTs), whereas no EST from this library ends at signal 1111 or 1292. When combining all other tissues, the number of ESTs with a 3′ end at 1111 and 1532 is 17 and 3, respectively. Fisher's exact value for the quadruplet (0,3,17,4) is 0.017. Thus there is a statistically significant bias for ESTs from Soares fetal heart library NbHH19W to use the polyadenylation signal at position 1532 rather than the signal at 1111. Comparing sites 1532 and 1292 would not give a significant bias.
Among the most interesting cases of differential polyadenylation are those linked to human pathologies. Distinct causes, such as alterations of the 3′ regions of genes or changes in the expression of UTR-binding proteins, induce variations in polyadenylation site selection and processing or stability of transcripts that have been linked to a number of diseases (for review, see Conne et al. 2000). These different phenomena may all affect the distribution of alternate mRNA forms and should be detectable when transcriptional profiles from affected and unaffected tissues are compared. ESTs from the Cancer Genome Anatomy Project (CGAP; Strausberg et al. 1997) and other EST sequencing efforts (e.g., Simpson 1999; Sese et al. 2001) now offer this opportunity. CGAP has produced, to date, >2.4 million EST sequences from cancer and normal cells, constituting an invaluable source of expression data in pathological tissues. Our analysis identified 1030 biases involving human cancer libraries, distributed in 504 UTRs.
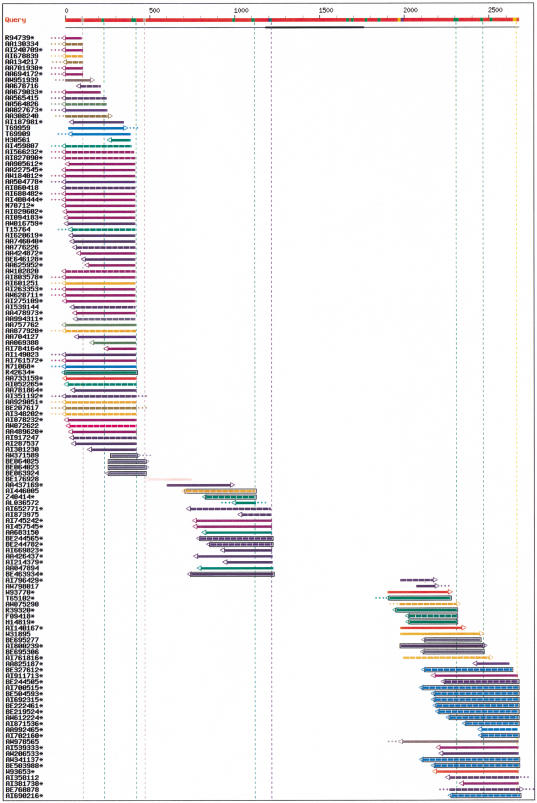
An example of potential cancer-specific polyadenylation is shown in Figure 2 for mRNA KIAA0764, coding for an unknown protein (Nagase et al. 1998). The UTR is 2673 bp long and shows multiple polyadenylation signals. The strongest sites are observed after signals AATATA 404, AATAAA 1199, and ATTAAA 2644. Minor sites are also observed around positions 102 (no signal), 215 (GATAAA), 465 (no signal), 1100 (AATATA), 2290 (GATAAA), and 2450 (AATACA). Interestingly, most of the polyadenylation signals in this UTR differ from the canonical AATAAA and ATTAAA sequences and would have been overlooked in the absence of EST information. The most significant bias involves ESTs from lung carcinoid tissue library NCI_CGAP_Lu24 (Strausberg et al. 1997), represented with dashed light-blue lines. Eleven ESTs from this library and eight ESTs from other libraries use the poly(A) signal at 2644. In comparison, the poly(A) signal at position 404 has no EST from library NCI_CGAP_Lu24 (or from another lung cancer library) and has 47 ESTs from other libraries. This distribution obtains a Fisher's Exact P value <10−6. Approximately one-half of the biases in our analysis involve cancer libraries similar to the ones in this case.
Figure 2.
EST-parser output for the 3′ untranslated region of mRNA for KIAA0764 protein (EMBL entry AB018307). See Figure 1 legend for color codes.
Conclusion
Even though reasonably accurate gene models can now be obtained from complete genome sequences, reconstructing the 3′ UTR and its alternative forms remains a challenging task. To date, this task is best performed using the experimental expression data available in the form of ESTs. The present software should help in identifying actual polyadenylation sites and in providing insight into possible tissue-specific 3′ ends. Running the program in batch mode on complete mRNA datasets from the newly sequenced eucaryotic genomes, we also expect to acquire a better understanding of alternate polyadenylation in general and its functional implications.
METHODS
Polyadenylation Site Identification
Human 3′ UTR sequences were obtained from UTRdb-nr release 13 (Pesole et al. 2000), a nonredundant database of eukaryotic UTRs generated by parsing the Feature table in the EMBL database (ftp://area.ba.cnr.it/pub/embnet/database/utr). We compared the 13,681 human and 6016 mouse UTRs to 2,452,892 human and 1,657,567 mouse ESTs from dbEST (October 2000 release) based on the sequence comparison procedure defined previously (Gautheret et al. 1998; Beaudoing et al. 2000) and summarized hereafter. UTR sequences were masked for common repeats and low complexity sequences using Repbase, Nov. 2000 release (Jurka 2000), and for vector sequences. ESTs were required to match the UTR sequence with at least 95% identity, encompassing the entire length of the EST sequence (at least 40 nucleotides), except for allowed 25 nt and 5 nt mismatches at the EST 5′ and 3′ sides, respectively, as revealed by the boundaries of the BLAST hit. This was intended to dismiss probable chimerical ESTs, ESTs produced from alternatively spliced or unspliced RNAs and ESTs exhibiting lane tracking errors or high error rates in the terminal region. Poly(A) and poly(T) trailers were removed from EST sequences prior to BLAST runs to avoid additional dangling regions. Internal priming (cDNA primers hybridized to internal poly(A) stretches instead of the actual poly(A) tail) was assessed by seeking adenine stretches in the UTR region flanking the 3′ extremity of the EST. Polyadenylation sites flanking eight or more consecutive adenines, or nine adenines in a 10-nucleotide window within +/−15 bases of a poly(A) signal were considered artifactual, except when the poly(A) stretch formed the tail of the query sequence. Further, one of the two following conditions was required to validate a polyadenylation site:(1) two or more ESTs ending within 30 nt downstream of an AAUAAA polyadenylation signal or any single-base variant described by Beaudoing et al. (2000). In this case, the 3′ base of the signal was selected as the transcript end; (2) in the absence of signal, two or more ESTs ending at the exact same 3′ position. In this case, the transcript end was taken as the EST extremity (such signal-less polyadenylation sites are frequent and should be allowed (Beaudoing et al. 2000).
Finally, when two or more predicted poly(A) sites occurred <30 nt from each other, only the one with the largest number of associated ESTs was retained. Since alternative poly(A) sites have been observed <30 nt apart (see Pauws et al. 2001), we left this minimal distance as a user-defined parameter on the Web interface. However, nearby poly(A) sites are less likely to be functionally important and their analysis will be hampered by error-prone 3′ ends in nonpolyadenylated ESTs.
Tissue Biases in 3′ End Usage
Organ and tissue data in dbEST reports are present under the “Library Description” section. These data, however, are inconsistently annotated in fields “Name,” “Organ,” “Development Stage,” “Cell line,” or “Tissue.” We extracted this information using a Perl script identifying a number of representative keywords, and categorized it into 117 tissues and 14 tissue categories or organ systems, as described in Table 1. For each EST, the library name, tissue, and organ system were recorded.
After putative poly(A) sites were identified in a given UTR, biased site usage with respect to EST libraries were sought as follows: Let Si, Sj a pair of polyadenylation sites and Ni, Nj their respective number of ESTs (that is, the ESTs that permitted to identify the sites). Let any EST library L, represented by ni ESTs at site Si and nj ESTs at site Sj. A preference for polyadenylation site Si in library L is computed using Fisher's Exact test (2-tail) on the quadruplet {ni, Ni-ni, Nj, Nj-nj} This actually compares the occurrence of library L to that of all other libraries combined. This turned out to be more practicable than comparing all libraries pairwise, which increased considerably the number of tests and produced too many uninteresting hits. Also, we treated poly(A) sites independently instead of comparing one site against the others. This last option would probably have brought to light a few more interesting cases, but it would have masked others: for instance when one library is overrepresented at more than one site. Fisher's exact test calculations were performed using the C code provided by T. Kadosawa (http://infofarm.cc.affrc.go.jp/∼kadosawa/fishertest.htm). Any value <0.05 was considered significant and was highlighted in the graphical user interface. Detailed output for all significant biases was observed in human and mouse 3′ UTR are available at http://tagc.univ-mrs.fr/bioinfo/ESTparser.
Graphical User Interface
A graphical user interface (GUI) has been specifically designed to highlight polyadenylation signals/sites and tissue biases. Any cDNA or mRNA sequence (intronless) can be used as input. An example output is shown in Figure 1. Graphical and color symbols are explained in Figure 1 legend. A Web server (http://tagc.univ-mrs.fr/bioinfo/ESTparser) allows a user to perform the whole analysis on any user-defined mRNA sequence. The sequence analysis program and GUI were both developed in Perl on Linux workstations.
Acknowledgments
E.B. was supported by a Ph.D. studentship from Association pour la Recherche sur le Cancer. The authors thank Rémi Houlgatte for critical reading of the manuscript
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.190501.
REFERENCES
- Agresti A. A survey of exact inference for contingency tables. Stat Sci. 1992;7:131–153. [Google Scholar]
- Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoing E, Freier S, Wyatt J, Claverie JM, Gautheret D. Patterns of variant polyadenylation signals in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, Lowe TM, Tolstoshev CM. dbEST—database for expressed sequence tags. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes & Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: A molecular ‘hotspot’ for pathology? Nat Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: mean to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautheret D, Poirot O, Lopez F, Audic S, Claverie JM. Expressed sequence tag (EST) clustering reveals the extent of alternate polyadenylation in human mRNAs. Genome Res. 1998;8:524–530. doi: 10.1101/gr.8.5.524. [DOI] [PubMed] [Google Scholar]
- Graber JH, Cantor CR, Mohr SC, Smith TF. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc Natl Acad Sci. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Repbase Update, a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- Lander E, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHuge W, et al. Initial sequencing and analysis of the human genome 2001. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Muraosa Y, Takahashi K, Yoshizawa M, Shibahara S. cDNA cloning of a novel protein containing two zinc-finger domains that may function as a transcription factor for the human heme-oxygenase-1 gene. Eur J Biochem. 1996;235:471–479. doi: 10.1111/j.1432-1033.1996.00471.x. [DOI] [PubMed] [Google Scholar]
- Nagase T, Ishikawa K, Suyama M, Kikuno R, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. XI. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1998;5:277–286. doi: 10.1093/dnares/5.5.277. [DOI] [PubMed] [Google Scholar]
- Pauws E, van Kampen AH, van De Graaf SA, de Vijlder JJ, Ris-Stalpers C. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: Implications for SAGE analysis. Nucleic Acids Res. 2001;29:1690–4. doi: 10.1093/nar/29.8.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole G, Liuni S, Grillo G, Licciulli F, Larizza A, Makałowski W, Saccone C. UTRdb and UTRsite: Specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2000;28:193–196. doi: 10.1093/nar/28.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Sese J, Nikaidou H, Kawamoto S, Minesaki Y, Morishita S, Okubo K. BodyMap incorporated PCR-based expression profiling data and a gene ranking system. Nucl Acids Res. 2001;29:156–158. doi: 10.1093/nar/29.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg RL, Dahl CA, Klausner RD. New opportunities for uncovering the molecular basis of cancer. Nat Genet. 1997;15:415–416. doi: 10.1038/ng0497supp-415. [DOI] [PubMed] [Google Scholar]
- Simpson AGJ. The FAPESP/LICR Human Cancer Genome Project. 1999. http://www.ludwig.org.br/ORESTES http://www.ludwig.org.br/ORESTES. [Google Scholar]
- Venter C, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]