Abstract
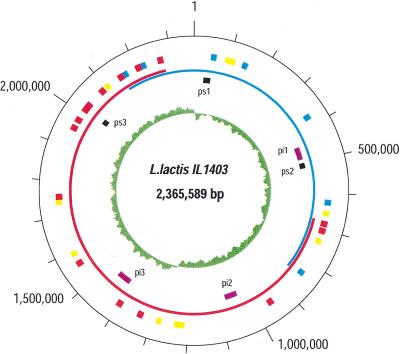
Lactococcus lactis is a nonpathogenic AT-rich gram-positive bacterium closely related to the genus Streptococcus and is the most commonly used cheese starter. It is also the best-characterized lactic acid bacterium. We sequenced the genome of the laboratory strain IL1403, using a novel two-step strategy that comprises diagnostic sequencing of the entire genome and a shotgun polishing step. The genome contains 2,365,589 base pairs and encodes 2310 proteins, including 293 protein-coding genes belonging to six prophages and 43 insertion sequence (IS) elements. Nonrandom distribution of IS elements indicates that the chromosome of the sequenced strain may be a product of recent recombination between two closely related genomes. A complete set of late competence genes is present, indicating the ability of L. lactis to undergo DNA transformation. Genomic sequence revealed new possibilities for fermentation pathways and for aerobic respiration. It also indicated a horizontal transfer of genetic information from Lactococcus to gram-negative enteric bacteria of Salmonella-Escherichia group.
[The sequence data described in this paper has been submitted to the GenBank data library under accession no. AE005176.]
Lactic acid bacteria (LAB) are a heterogeneous group of microorganisms that convert carbohydrates into lactic acid. They comprise both pathogens (such as Streptococcus pneumoniae or Streptococcus pyogenes) and useful bacteria (such as Streptococcus thermophilus and Lactococcus lactis, which were used for millennia in milk fermentation). Determination and analysis of the genome sequence of a representative LAB is therefore of great interest, as it would provide information allowing us to combat the former and use the latter more efficiently. Until now, no complete and annotated genome sequence of either LAB class has been reported.
In nature, L. lactis occupies a niche related to plant or animal surfaces and the animal gastrointestinal tract. It is believed to be dormant on the plant surfaces and to multiply in the gastrointestinal tract after being swallowed by a ruminant. In contrast, “domesticated” species of L. lactis, used by dairy industry as starters in cheese fermentation, live in a different niche, which is defined by technological considerations, such as fast growth and rapid production of lactic acid in milk. The importance of L. lactis for humankind can be appreciated from the estimate that close to 107 tons of cheese are made annually (Fox 1989), leading to human consumption of close to 1018 lactococci.
There are two subspecies of L. lactis, designated initially as Streptococcus lactis and Streptococcus cremoris and reclassified more recently as L. lactis ssp. lactis and L. lactis ssp. cremoris, respectively (Schleifer et al. 1985). The former is preferred for making of soft cheeses and the latter for the hard ones. The two subspecies have been intensely studied, mainly because of their industrial interest, and have became excellent models for research on metabolism, physiology, genetics, and molecular biology of LAB.
The questions addressed in research on useful bacteria are often antithetical to those involving pathogens, because one of the basic objectives is to improve rather than to limit bacterial growth. Efficient use of lactococci by dairy industry requires understanding of many aspects of bacterial physiology, such as use of sugars and proteins from milk for growth, conversion of sugars to lactate, and synthesis of substances involved in cheese flavor, and thus of the relationship between different types of fermentation. The potential for new applications of LAB, such as oral vaccines (Steidler et al. 2000) or production of foreign proteins and metabolites, leads to questions concerning the protein secretion system, biosynthesis of cofactors, and regulation of central metabolism. In addition to questions related to the industrial use of lactococci, fundamental biological questions, such as retrohoming of introns (Cousineau et al. 1998), are also being addressed in L. lactis.
A genetic map of a “laboratory workhorse” L. lactis ssp. lactis strain IL1403, based on a low-fidelity diagnostic genome sequencing, has been reported (Bolotin et al. 1999). Here we present the analysis of the accurate sequence of the IL1403 genome, which is the first such report for any lactic acid bacterium. We focus mainly on features related to the importance of L. lactis for humankind, which is its use in dairy fermentation. Also, several unexpected findings are reported, such as a putative chimerical structure of the genome, the possibility that L. lactis can respire, the existence of genes required for DNA transformation, and a discovery of a transfer of genetic information from lactococci to gram-negative enteric bacteria.
RESULTS AND DISCUSSION
Two-Step Sequencing Strategy
The first step of our strategy, designated diagnostic genome sequencing, was described before (Bolotin et al. 1999). Briefly, it implies cloning of relatively short (1–20 kb) genome fragments in Escherichia coli plasmid and phage vectors, and sequencing of a limited number of randomly chosen clones, to a redundancy of about one. A novel procedure, designated multiplex long accurate PCR (MLA PCR), developed and tested in the course of the Bacillus subtilis genome sequencing project (Sorokin et al. 1996; Kunst et al. 1997), is then applied for connecting the resulting contigs and synthesizing the missing genome regions, sequenced subsequently by standard methods. This approach allowed us to establish the entire L. lactis genome sequence and assemble it in a unique contig, with a sequencing redundancy of less than two (Bolotin et al. 1999). Three- to fourfold fewer sequencing reactions were required to reach this goal than if the fully random approach were used. For comparison, only 10,235 reactions were needed to assemble L. lactis genome sequence, whereas 40,020 were required for the genome of Neisseria meningitidis (Tettelin et al. 2000), which is of a similar size. Diagnostic sequence allowed us to identify all L. lactis genes that encode proteins sufficiently similar to those present in the databases. However, the elevated error rate, estimated to be ∼1%, did not allow us to predict the genes unique for L. lactis or the borders of coding region. To obtain a more complete and reliable description of the L. lactis genome, we carried out a second step of our strategy. It involved random sequencing of additional clones until the overall redundancy of ∼6.4 was reached and then primer walking on PCR-generated templates to ensure that each base was sequenced at least four times and at least once on each strand. We designated this step “shotgun polishing” and concluded that the strategy presented here can be a good alternative to the fully random strategy used in most cases (Fraser and Fleischmann 1997). Its advantages should increase even more when a greater number of completely sequenced and thoroughly annotated bacterial genomes becomes available. Carrying out the diagnostic step and polishing only a very little will then be sufficient to determine a reliable genome sequence of bacteria relatively close to the ones that were already sequenced and annotated.
Gene Content
The circular chromosome of L. lactis IL1403 has 2,365,589 bp and an average G+C content of 35.4%. We detected 2310 open reading frames (ORFs) in the sequence, with an average length of 879 bp. Protein-coding genes represent 86% of the genome, stable RNA 1.4%, and noncoding regions 12.6%. These values are similar to those observed for genomes of other bacteria. We have assigned a biochemical or biological role to 64.2% (1482 ORFs) of the genes and classified them into functional categories (Table 1). There are 20.1% of genes (465 ORFs) that match hypothetical coding sequences of unknown function, and the remaining 15.7% (363 ORFs) represent genes with no similarity to known proteins, which can be considered specific for lactococci.
Table 1.
Functional Classification of the Lactococcus lactis Protein-Coding Genes#
| AMINO-ACID BIOSYNTHESIS | ||
| Aromatic amino-acid family | ||
| aroA | 1802 | 3-phosphoshikimate 1-carboxyvinyltransferase |
| aroB | 1814 | 3-dehydroquinate synthase |
| aroC | 1811 | chorismate synthase |
| aroD | 1690 | 3-dehydroquinate dehydratase |
| aroE | 1815 | shikimate 5-dehydrogenase |
| aroF | 120 | Tyr-sensitive phospho-2-dehydro-deoxyheptonate aldolase |
| aroH | 1281 | Trp-sensitive phospho-2-dehydro-deoxyheptonate aldolase |
| aroK | 1801 | shikimate kinase |
| pheA | 1801 | prephenate dehydratase |
| trpA | 1494 | tryptophan synthase alpha chain |
| trpB | 1495 | tryptophan synthase beta chain |
| trpC | 1498 | indole-3-glycerol phosphate synthase |
| trpD | 1499 | anthranilate phosphoribosyltransferase |
| trpE | 1501 | anthranilate synthase component I |
| trpF | 1497 | phosphorybosyl-anthranilate isomerase |
| trpG | 1500 | anthranilate synthase component II |
| tyrA | 1803 | prephenate dehydrogenase |
| Aspartate family | ||
| asnB | 357 | asparagine synthetase B |
| asnH | 2312 | asparagine synthetase |
| aspB | 1897 | aspartate aminotransferase |
| aspC | 163 | aspartate aminotransferase |
| ceo | 1265 | N5-carboxyethyl-ornithine synthase |
| dapA | 1665 | dihydrodipicolinate synthase |
| dapB | 1605 | dihydrodipicolinate reductase |
| hom | 1172 | homoserine dehydrogenase |
| lysA | 1314 | diaminopimelate decarboxylase |
| metA | 1997 | homoserine O-succinyltransferase |
| metB1 | 1996 | cystathionine gamma-synthase |
| metB2 | 791 | cystathionine gamma-synthase |
| metE | 1284 | 5-methionine synthase |
| metF | 1282 | 5,10-methylenetetrahydrofolate reductase |
| thrA | 748 | aspartokinase |
| thrB | 1173 | homoserine kinase |
| thrC | 2173 | threonine synthase |
| Branched chain family | ||
| ilvA | 1251 | threonine deaminase |
| ilvB | 1248 | acetolactate synthase large subunit |
| ilvC | 1250 | ketol-acid reductoisomerase |
| ilvD | 1247 | dihydroxy-acid dehydratase |
| ilvN | 1249 | acetolactate synthase small subunit |
| leuA | 1240 | 2-isopropylmalate synthase |
| leuB | 1242 | 3-isopropylmalate dehydrogenase |
| leuC | 1244 | 3-isopropylmalate dehydratase large subunit |
| leuD | 1245 | 3-isopropylmalate dehydratase small subunit |
| Glutamate family | ||
| argB | 808 | acetylglutamate kinase |
| argC | 805 | N-acetyl-gamma-glutamyl-phosphate reductase |
| argD | 807 | acetylornithine aminotransferase |
| argE | 560 | acetylornithine deacetylase |
| argG | 127 | argininosuccinate synthase |
| argH | 129 | argininosuccinate lyase |
| argJ | 806 | ornithine acetyltransferase |
| glnA | 2283 | glutamine synthetase |
| gltA | 668 | citrate synthase |
| gltB | 1319 | glutamate synthase large subunit |
| gltD | 1316 | glutamate synthase small subunit |
| proA | 1651 | gamma-glutamyl phosphate reductase |
| proB | 1652 | glutamate 5-kinase |
| proC | 1953 | pyrroline-5-carboxylate reductase |
| Histidine family | ||
| hisA | 1236 | phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase |
| hisB | 1234 | imidazoleglycerol-phosphate dehydratase |
| hisC | 1229 | histidinol-phosphate aminotransferase |
| hisD | 1232 | histidinol dehydrogenase |
| hisF | 1237 | cyclase HisF |
| hisG | 1231 | ATP phosphoribosyltransferase |
| hisH | 1235 | amidotransferase |
| hisI | 1237 | phosphoribosyl-AMP cyclohydrolase |
| hisK | 1238 | histidinol phosphatase |
| hisZ | 1230 | ATP phosphoribosyltransferase regulatory subunit |
| Serine family | ||
| cysD | 77 | O-acetylhomoserine sulfhydrylase |
| cysE | 1921 | serine acetyltransferase |
| cysK | 792 | cysteine synthase |
| cysM | 527 | cysteine synthase |
| glyA | 592 | serine hydroxymethyltransferase |
| serA | 595 | D-3-phosphoglycerate dehydrogenase |
| serB | 596 | phosphoserine phosphatase |
| serC | 594 | phosphoserine aminotransferase |
| BIOSYNTHESIS OF COFACTORS, PROSTHETIC GROUPS, AND CARRIERS | ||
| Folic acid | ||
| dfrA | 1163 | dihydropholate reductase |
| fhs | 961 | formyltetrahydrofolate synthetase |
| folB | 1166 | dihydroneopterin aldolase |
| folC | 1169 | folylpolyglutamate synthase |
| folD | 877 | tetrahydrofolate dehydrogenase/cyclohydrolase |
| folE | 1167 | GTP cyclohydrolase I |
| folP | 1168 | dihydropteroate synthase |
| pabA | 1349 | para-aminobenzoate synthase component II |
| pabB | 1348 | para-aminobenzoate synthase component I |
| Heme and porphyrin | ||
| hemH | 1609 | ferrohelatase |
| hemK | 589 | protoporphyrinogen oxidase |
| hemN | 1154 | oxygen-independent coproporphyrogen III oxidase |
| Menaquinone and ubiquinone | ||
| ispA | 881 | farnesyl diphosphate synthase |
| ispB | 1380 | heptaprenyl diphosphate synthase component II |
| menB | 735 | dihydroxynaphthonic acid synthase |
| menD | 737 | 2-oxoglutarate decarboxylase |
| menE | 734 | O-succinylbenzoic acid-CoA ligase |
| menF | 739 | menaquinone-specific isochorismate synthase |
| menX | 736 | protein in menaquinone biosynthesis pathway |
| preA | 187 | prenyl transferase |
| ubiE | 1718 | menaquinone biosynthesis methylase |
| yhdB | 732 | racemase |
| Pantothenate | ||
| coaA | 1467 | pantothenate kinase |
| dfpA | 567 | pantothenate metabolism flavoprotein |
| dfpB | 568 | flavoprotein |
| panE | 1358 | ketopantoate reductase |
| Riboflavin and cobalamin | ||
| cobC | 1889 | alpha-ribazole-5'-phosphate phosphatase |
| cobQ | 1115 | cobyric acid synthase |
| ribA | 1024 | GTP cyclohydrolase II / 3,4-dihydroxy-2-butanone 4-phosphate synthase |
| ribB | 1023 | ribiflavin synthase alpha chain |
| ribC | 1142 | riboflavin kinase |
| ribG | 1023 | riboflavin-specific deaminase |
| ribH | 1025 | ribiflavin synthase beta chain |
| Thioredoxin, glutaredoxin, and glutathione | ||
| gpo | 1402 | glutathione peroxidase |
| gshR | 864 | glutathione reductase |
| trxA | 1692 | thioredoxin |
| trxB1 | 966 | thioredoxin reductase |
| trxB2 | 1695 | thioredoxin reductase |
| trxH | 396 | thioredoxin H-type |
| Thiamin | ||
| apbE | 1125 | thiamine biosynthesis lipoprotein |
| thiD1 | 1295 | phosphomethylpyrimidine kinase |
| thiD2 | 485 | phosphomethylpyrimidine kinase |
| thiE | 1294 | thiamin-phosphate pyrophosphorylase |
| thiM | 1295 | hydroxyethylthiazole kinase |
| Pyridine nucleotides | ||
| nadE | 1110 | NAD-synthetase |
| yvdG | 2139 | pyridine nucleotide-disulfide oxidoreductase |
| CELL ENVELOPE | ||
| Membranes, lipoproteins, and porins | ||
| bmpA | 1462 | basic membrane protein A |
| cdsA | 2200 | phosphatidate cytidylyltransferase |
| clsA | 988 | cardiolipin synthase |
| clsB | 1188 | cardiolipin synthase |
| dgkA | 1095 | diacylglycerol kinase |
| lgt | 606 | prolipoprotein diacylglycerol transferase |
| pgsA | 2047 | CDP-diacylglycerol-phosphate phosphatidyltransferase |
| plpA | 318 | outer membrane lipoprotein precursor |
| plpB | 319 | outer membrane lipoprotein precursor |
| plpC | 320 | outer membrane lipoprotein precursor |
| plpD | 321 | outer membrane lipoprotein precursor |
| yfjC | 596 | acylphosphate phosphohydrolase |
| Murein sacculus and peptidoglycan | ||
| acmA | 269 | N-acetylmuramidase |
| acmB | 1977 | N-acetylmuramidase |
| acmC | 1403 | N-acetylmuramidase |
| acmD | 528 | N-acetylmuramidase |
| asd | 1667 | aspartate-semialdehyde dehydrogenase |
| dacA | 2356 | D-alanyl-D-alanine carboxypeptidase |
| dacB | 976 | D-alanyl-D-alanine carboxypeptidase |
| dal | 862 | alanine racemase |
| ddl | 341 | D-alanine-D-alanine ligase |
| glmU | 1952 | UDP-N-acetylglucosamine pyrophosphorylase |
| mraY | 892 | phospho-N-acetylmuramoyl-pentapeptide transferase |
| mreC | 2316 | cell shape determining protein |
| mreD | 2315 | cell shape determining protein |
| murA1 | 1314 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase |
| murA2 | 535 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase |
| murB | 1175 | UDP-N-acetylenolpyruvoylglucosamine reductase |
| murC | 2119 | UDP-N-acetylmuramate-alanine ligase |
| murD | 1634 | UDP-N-acetylmuramoylalanine D-glutamate ligase |
| murE | 1871 | UDP-MurNac-tripeptide synthetase |
| murF | 342 | D-Ala-D-Ala adding enzyme |
| murG | 1633 | peptidoglycan synthesis protein MurG |
| murI | 1313 | glutamate racemase |
| pbp1B | 393 | penicillin-binding protein 1B |
| pbp2A | 2178 | penicillin-binding protein 2a |
| pbp2B | 339 | penicillin-binding protein 2B |
| pbpX | 890 | penicillin-binding protein |
| ponA | 530 | penicillin-binding protein 1A |
| racD | 2310 | aspartate racemase |
| uppS | 2201 | undecaprenyl pyrophosphate synthetase |
| Surface polysaccharides, lipopolysaccharides and antigens | ||
| dltA | 1293 | D-alanine activating enzyme |
| dltB | 1291 | peptidoglycan biosynthesis protein |
| dltC | 1290 | D-alanyl carrier protein |
| dltD | 1290 | D-alanine transfer protein DltD |
| dltE | 145 | oxidoreductase |
| floL | 746 | flotillin-like protein |
| hasC | 1378 | UTP-glucose-1-phosphate uridylyltransferase |
| icaA | 681 | glycosyl transferase |
| icaB | 683 | intercellular adhesion protein IcaB |
| icaC | 684 | collagen adhesin |
| kdtB | 2239 | lipopolysaccharide core biosynthesis protein |
| mvaA | 1611 | hydroxymethylglutaryl-CoA reductase |
| mycA | 981 | myosin-crossreactive antigen |
| pspA | 2304 | glucosyltransferase-S |
| pspB | 2306 | glucosyltransferase-S |
| rgpA | 202 | rhamnosyltransferase |
| rgpB | 203 | rhamnosyltransferase |
| rgpE | 207 | glycosyltransferase |
| rgpF | 209 | polysaccharide biosynthesis protein |
| tagB | 953 | teichoic acid biosynthesis protein B |
| tagD1 | 220 | glycerol-3-phosphate cytidiltransferase |
| tagD2 | 951 | glycerol-3-phosphate cytidyltransferase |
| tagF | 952 | teichoic acid biosynthesis protein F |
| tagL | 936 | exopolysaccharide biosynthesis protein |
| tagX | 948 | teichoic acid biosynthesis protein |
| tagY | 945 | teichoic acid biosynthesis protein |
| tagZ | 943 | teichoic acid biosynthesis protein |
| ycbB | 212 | glycosyltransferase |
| ycbD | 213 | UDP-glucose 4-epimerase |
| ycbF | 215 | LPS biosynthesis protein |
| ycbG | 216 | LPS biosynthesis protein |
| ycbH | 217 | sugar transferase |
| ycbI | 218 | sugar transferase |
| ycbJ | 219 | LPS biosynthesis protein |
| ycbK | 214 | polysaccharide biosynthesis export protein |
| yijG | 899 | glycosyl transferase |
| yjeF | 949 | lipopolysaccharide biosynthesis protein |
| ymjE | 1297 | glycosyl transferase |
| ymjF | 1299 | UDP-N-acetylglucosamine 2-epimerase |
| yohH | 1478 | lipopolysaccharide biosynthesis protein |
| yohJ | 1479 | lipopolysaccharide biosynthesis protein |
| ysfC | 1853 | polysacharide biosynthesis protein |
| ywaF | 2206 | glycosyltransferase |
| ywaG | 2207 | lipopolysaccharide biosynthesis protein |
| CELLULAR PROCESSES | ||
| Cell division | ||
| ezrA | 2225 | cell division regulator |
| ftsA | 1940 | cell division protein FtsA |
| ftsE | 1000 | cell-division ATP-binding protein FtsE |
| ftsH | 27 | cell division protein FtsH |
| ftsK | 1705 | cell division protein FtsK |
| ftsQ | 1632 | cell division protein FtsQ |
| ftsW1 | 663 | cell division protein FtsW |
| ftsW2 | 908 | cell division protein FtsW |
| ftsX | 1001 | cell division protein |
| ftsY | 825 | cell division protein FtsY |
| ftsZ | 1938 | cell division protein FtsZ |
| gidA | 1915 | glucose inhibited division protein GidA |
| gidB | 1381 | glucose-inhibited division protein GidB |
| gidC | 1257 | glucose inhibited division protein GidC |
| mesJ | 24 | cell cycle protein MesJ |
| parA | 99 | chromosome partitioning protein |
| rodA | 917 | rod-shape determining protein |
| smc | 812 | chromosome segregation SMC protein |
| Cell killing | ||
| hly | 498 | hemolysin like protein |
| Chaperones | ||
| dnaK | 979 | DnaK protein |
| groEL | 400 | 60 KD chaperonin |
| groES | 399 | 10 KD chaperonin |
| sugE | 25 | SugE protein |
| Detoxification | ||
| ahpC | 336 | alkyl hydroperoxide reductase |
| ahpF | 337 | alkyl hydroperoxide reductase |
| sodA | 413 | superoxide dismutase |
| Protein and peptide secretion | ||
| ffh | 1658 | signal recognition particle protein Ffh |
| lspA | 1026 | lipoprotein signal peptidase |
| secA | 118 | preprotein translocase SecA subunit |
| secE | 2175 | preprotein translocase SecE subunit |
| secG | 967 | protein-export protein SecG |
| secY | 2159 | preprotein translocase SecY subunit |
| sipL | 2351 | signal peptidase I |
| tig | 536 | trigger factor |
| Transformation | ||
| coiA | 1785 | competence protein CoiA |
| comC | 2104 | type 4 prepilin-like protein specific leader peptidase |
| comEA | 1833 | competence protein ComEA |
| comEC | 1832 | competence protein ComEC |
| comFA | 1098 | competence protein ComFA |
| comFC | 1097 | competence protein ComFC |
| comGA | 2189 | competence protein ComGA |
| comGB | 2188 | competence protein ComGB |
| comGC | 2187 | competence protein ComGC |
| comGD | 2187 | competence protein ComGD |
| comX | 2224 | competence regulator ComX |
| dprA | 1254 | DNA processing SMF protein |
| radA | 2150 | DNA repair protein RadA |
| recQ | 1874 | ATP-dependent DNA helicase RecQ |
| CENTRAL INTERMEDIARY METABOLISM | ||
| General | ||
| metK | 1971 | S-adenosylmethionine synthetase |
| pcaC | 2052 | gamma-carboxymuconolactone decarboxylase |
| Amino sugars | ||
| femD | 436 | phosphoglucosamine mutase |
| glmS | 1035 | glucosamine-fructose-6-phosphate aminotransferase |
| nagA | 1374 | N-acetylglucosamine-6-phosphate deacetylase |
| nagB | 1615 | glucosamine-6-P isomerase |
| ylfH | 1157 | N-acetylglucosamine catabolic protein |
| ypcD | 1524 | endo-beta-N-acetylglucosaminidase |
| Degradation of polysaccharides | ||
| agl | 1732 | alpha-glucosidase |
| amyL | 1278 | alpha-amylase |
| amyY | 1734 | alpha-amylase |
| apu | 703 | amylopullulanase |
| chiA | 2027 | chitinase |
| dexA | 1736 | oligo-1,6-glucosidase |
| dexB | 1526 | alpha 1-6-glucosidase |
| dexC | 1738 | neopullulanase |
| lnbA | 1527 | lacto-N-biosidase |
| xynD | 282 | endo-1,4-beta-xylanase D |
| yucG | 2028 | chitin binding protein |
| Phosphorus compounds | ||
| apl | 719 | alkaline phosphatase |
| Polyamine biosynthesis | ||
| yqfF | 1657 | spermidine acetyltransferase |
| Other | ||
| glgA | 699 | glycogen synthase |
| glgB | 147 | 1,4-alpha-glucan branching enzyme |
| glgC | 697 | glucose-1-phosphate adenylyltransferase |
| glgD | 698 | glucose-1-phosphate adenylyltransferase |
| glgP | 701 | glycogen phosphorylase |
| mapA | 1730 | maltosephosphorylase |
| xylH | 568 | 4-oxalocrotonate tautomerase |
| ENERGY METABOLISM | ||
| Aerobic | ||
| cbr | 144 | carbonyl reductase |
| noxA | 841 | NADH dehydrogenase |
| noxB | 842 | NADH dehydrogenase |
| noxC | 795 | NADH oxidase |
| noxD | 2195 | NADH oxidase |
| noxE | 397 | NADH oxidase |
| poxL | 2130 | pyruvate oxidase |
| yahI | 78 | short-chain type dehydrogenase |
| ybdE | 134 | oxidoreductase |
| ybiE | 186 | oxidoreductase |
| ycdG | 234 | oxidoreductase |
| ycgD | 264 | oxidoreductase |
| ycgG | 267 | oxidoreductase |
| yddB | 333 | oxidoreductase |
| ygcA | 620 | oxidoreductase |
| yhgA | 760 | oxidoreductase |
| yiaB | 802 | oxidoreductase |
| ymgK | 1268 | oxidoreductase |
| yneD | 1343 | oxidoreductase |
| ypaI | 1509 | oxidoreductase |
| ypgB | 1562 | oxidoreductase |
| yphA | 1571 | NADH dehydrogenase |
| yphC | 1574 | oxidoreductase |
| ypiA | 1580 | oxidoreductase |
| ypjA | 1591 | dehydrogenase |
| ypjF | 1595 | oxidoreductase |
| ypjH | 1599 | oxidoreductase |
| yrbA | 1711 | oxidoreductase |
| yrfB | 1751 | NADH-dependent oxidoreductase |
| yrjB | 1791 | oxidoreductase |
| yrjC | 1792 | iron-binding oxidase subunit |
| ysjB | 1892 | oxidoreductase |
| yteC | 1944 | oxidoreductase |
| yudI | 2036 | oxidoreductase |
| yugB | 2066 | oxidoreductase |
| yugC | 2068 | dehydrogenase |
| yxdE | 2338 | oxidoreductase |
| Amino acids and amines | ||
| ansB | 743 | L-asparaginase |
| araT | 57 | aromatic amino acid specific aminotransferase |
| arcA | 2115 | arginine deiminase |
| arcB | 2114 | ornithine carbamoyltransferase |
| arcC1 | 2111 | carbamate kinase |
| arcC2 | 2110 | carbamate kinase |
| arcC3 | 1752 | carbamate kinase |
| arcT | 2109 | aminotransferase |
| argF | 809 | ornithine carbamoyltransferase |
| bcaT | 1322 | branched-chain amino acid aminotransferase |
| gadB | 1325 | glutamate decarboxylase |
| hicD | 490 | L-2-hydroxyisocaproate dehydrogenase |
| ipd | 1340 | indole-3-pyruvate decarboxylase |
| otcA | 1757 | ornithine carbamoyltransferase |
| pdc | 2011 | phenolic acid decarboxylase |
| pfs | 1950 | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase |
| sdaA | 845 | alpha-subuni L-serine dehydratase |
| sdaB | 844 | beta-subunit of L-serine dehydratase |
| yciA | 281 | amino acid amidohydrolase |
| yjiB | 983 | amino acid aminohydrolase |
| ytjE | 1995 | aminotransferase |
| ywjF | 2299 | 3-hydroxyisobutyrate dehydrogenase |
| Anaerobic | ||
| dhaK | 245 | dihydroxyacetone kinase |
| dhaL | 246 | dihydroxyacetone kinase |
| dhaM | 247 | dihydroxyacetone kinase |
| glpD | 1271 | glycerol-3-phosphate dehydrogenase |
| glpK | 1273 | glycerol kinase |
| gpdA | 1377 | glycerol-3-phosphate dehydrogenase |
| lctO | 1280 | L-lactate oxidase |
| ylbE | 1120 | oxidoreductase |
| ATP-proton motive force interconversion | ||
| atpA | 1826 | ATP synthase alpha subunit |
| atpB | 1829 | ATP synthase subunit a |
| atpD | 1824 | ATP synthase alpha subunit |
| atpE | 1823 | ATP synthase epsilon subunit |
| atpF | 1828 | ATP synthase subunit b |
| atpG | 1825 | ATP synthase gamma subunit |
| atpH | 1828 | ATP synthase delta subunit |
| Electron transport | ||
| cydA | 708 | cytochrome D ubiquinol oxidase subunit I |
| cydB | 710 | cytochrome D ubiquinol oxidase subunit II |
| fer | 1762 | ferredoxin |
| ndrH | 1006 | glutaredoxin-like protein NrdH |
| ndrI | 1005 | ribonucleotide reductase |
| nifJ | 431 | pyruvate-flavodoxin oxidoreductase |
| nifS | 1928 | pyridoxal-phosphate dependent aminotransferase NifS |
| nifU | 1848 | NifU protein |
| nifZ | 523 | pyridoxal-phosphate dependent aminotransferase |
| qor | 724 | quinone oxidoreductase |
| yfiJ | 588 | NADPH-flavin oxidoreductase |
| yfjE | 598 | flavodoxin |
| yviC | 2181 | FMN-binding protein |
| Entner-Doudoroff | ||
| kdgA | 1672 | 2-dehydro-3-deoxyphosphogluconate aldolase |
| kdgK | 1673 | 2-dehydro-3-deoxygluconokinase |
| Fermentation | ||
| ackA1 | 2091 | acetate kinase |
| ackA2 | 2089 | acetate kinase |
| adhA | 1873 | alcohol dehydrogenase |
| adhE | 2231 | alcohol-acetaldehyde dehydrogenase |
| aldB | 1253 | alpha-acetolactate decarboxylase |
| aldC | 1117 | alpha-acetolactate decarboxylase |
| als | 1201 | alpha-acetolactate synthase |
| butA | 919 | acetoin reductase |
| butB | 918 | 2,3-butanediol dehydrogenase |
| frdC | 1139 | fumarate reductase flavoprotein subunit |
| mae | 1204 | malate oxidoreductase |
| mleS | 923 | malolactic enzyme |
| pfl | 659 | pyruvate-formate lyase |
| pflA | 1881 | pyruvate-formate lyase activating enzyme |
| pta | 1709 | phosphate acetyltransferase |
| yseE | 1846 | 2-nitropropane deoxygenase |
| Gluconeogenesis | ||
| fbp | 255 | fructose-1,6-bisphosphatase |
| Glycolysis | ||
| enoA | 634 | enolase |
| enoB | 275 | 2-phosphoglycerate dehydratase |
| fbaA | 1980 | fructose-bisphosphate aldolase |
| gapA | 554 | glyceraldehyde 3-phosphate dehydrogenase |
| gapB | 2333 | glyceraldehyde 3-phosphate dehydrogenase |
| ldh | 1370 | L-lactate dehydrogenase |
| ldhB | 380 | L-lactate dehydrogenase |
| ldhX | 1143 | L-lactate dehydrogenase |
| pfk | 1372 | 6-phosphofructokinase |
| pgiA | 2245 | glucose-6-phosphate isomerase A |
| pgk | 243 | phosphoglycerate kinase |
| pgmB | 442 | beta-phosphoglucomutase |
| pmg | 335 | phosphoglycerate mutase |
| pycA | 665 | pyruvate carboxylase |
| pyk | 1371 | pyruvate kinase |
| tpiA | 1149 | triosephosphate isomerase |
| yjhF | 975 | phosphoglycerate mutase |
| yrjI | 1800 | phosphoglycerate mutase |
| Pentose phosphate pathway | ||
| dxsA | 1510 | 1-deoxyxylulose-5-phosphate synthase |
| dxsB | 1725 | 1-deoxyxylulose-5-phosphate synthase |
| gnd | 609 | decarboxylating 6-phosphogluconate dehydrogenase |
| ptk | 1540 | phosphoketolase |
| rpe | 2004 | ribulose-phosphate 3-epimerase |
| rpiA | 2317 | ribose 5-phosphate isomerase A |
| tkt | 1670 | transketolase |
| zwf | 2302 | glucose-6-phosphate 1-dehydrogenase |
| Pyruvate dehydrogenase | ||
| pdhA | 64 | PDH E1 component alpha subunit |
| pdhB | 63 | PDH E1 component beta subunit |
| pdhC | 61 | dihydrolipoamide acetyltransferase component of PDH complex |
| pdhD | 60 | lipoamide dehydrogenase component of PDH complex |
| Sugars | ||
| bglA | 423 | phospho-beta-glucosidase |
| bglH | 1490 | beta-glucosidase |
| bglS | 180 | beta-glucosidase A |
| galE | 2055 | UDP-glucose 4-epimerase |
| galK | 2061 | galactokinase |
| galM | 2062 | aldose 1-epimerase |
| galT | 2060 | galactose-1-phosphate uridylyltransferase |
| glk | 2101 | glucose kinase |
| gntK | 2269 | gluconate kinase |
| gntZ | 2271 | 6-phosphogluconate dehydrogenase |
| lacC | 985 | tagatose-6-phosphate kinase |
| lacZ | 2057 | beta-galactosidase |
| maa | 1735 | maltose O-acetyltransferase |
| malQ | 695 | 4-alpha-glucanotransferase |
| mtlD | 34 | mannitol 1-phosphate 5-dehydrogenase |
| pmi | 780 | mannose-6-phosphate isomerase |
| rbsK | 1687 | ribokinase |
| scrK | 1518 | fructokinase |
| thgA | 2058 | thiogalactoside acetyltransferase |
| uxaC | 1674 | glucuronate isomerase |
| uxuA | 1678 | D-mannonate dehydratase |
| uxuB | 1679 | fructuronate reductase |
| xylA | 1550 | xylose isomerase |
| xylB | 1548 | xylulose kinase |
| xylM | 1547 | aldose 1-epimerase |
| xylX | 1543 | acetyltransferase hypothetical protein |
| xynB | 1544 | beta-1,4-xylosidase |
| yeeB | 443 | sugar hydrolase |
| ygjD | 694 | 4-alpha-glucanotransferase |
| yidC | 834 | beta-glucosidase |
| yncA | 1321 | acetyltransferase |
| ypbG | 1519 | sugar kinase |
| ypcA | 1521 | beta-glucosidase |
| ypdB | 1532 | sugar hydrolase |
| ypdD | 1537 | sugar hydrolase |
| yrcA | 1722 | phospho-beta-glucosidase |
| TCA cycle | ||
| citB | 670 | aconitate hydratase |
| citC | 1207 | acetate-SH-citrate lyase ligase |
| citD | 1208 | citrate lyase acyl-carrier protein |
| citE | 1209 | citrate lyase beta chain |
| citF | 1210 | citrate lyase alpha chain |
| citG | 1211 | CitG protein |
| icd | 672 | isocitrate dehydrogenase |
| FATTY ACID AND PHOSPHOLIPID METABOLISM | ||
| General | ||
| accA | 790 | acetyl-CoA carboxylase carboxyl transferase subunit alpha |
| accB | 786 | biotin carboxyl carrier protein of acetyl-CoA carboxylase |
| accC | 788 | biotin carboxylase |
| accD | 789 | acetyl-CoA carboxylase carboxyl transferase subunit betta |
| acpA | 782 | acyl carrier protein |
| acpD | 116 | acyl carrier protein phosphodiesterase |
| acpS | 862 | acyl carrier protein synthase |
| cfa | 1972 | cyclopropane fatty acid synthase |
| fabD | 783 | malonyl CoA-acyl carrier protein transacylase |
| fabF | 785 | 3-oxoacyl-acyl carrier protein synthase II |
| fabG1 | 784 | 3-oxoacyl-acyl carrier protein reductase |
| fabG2 | 1845 | 3-oxoacyl-acyl carrier protein reductase |
| fabH | 782 | 3-oxoacyl-acyl-carrier-protein synthase III |
| fabI | 562 | NADH-dependent enoyl-ACP reductase |
| fabZ1 | 561 | hydroxymyristoyl-acyl carrier protein dehydratase |
| fabZ2 | 787 | 3R-hydroxymyristoyl-acyl carrier protein dehydratase |
| fadA | 1843 | acetyl coenzyme A acetyltransferase |
| fadD | 655 | long-chain acyl-CoA synthetase |
| hmcM | 1614 | hydroxymethylglutaryl-CoA synthase |
| lplL | 65 | lipoate-protein ligase |
| plsX | 72 | fatty acid/phospholipid synthesis protein |
| thiL | 1613 | acetyl coenzyme A acetyltransferase |
| ydiD | 386 | acyl carrier protein phosphodiesterase |
| yeaG | 408 | mevalonate kinase |
| yeaH | 410 | diphosphomevalonate decarboxylase |
| yebA | 411 | mevalonate kinase |
| yscE | 1830 | lipase |
| PURINES, PYRIMIDINES, NUCLEOSIDES AND NUCLEOTIDES | ||
| 2'-deoxyribonucleotide metabolism | ||
| dcdA | 1156 | dCMP deaminase |
| nrdE | 1004 | ribonucleoside-diphosphate reductase alpha chain |
| nrdF | 1002 | ribonucleoside-diphosphate reductase beta chain |
| Nucleotide and nucleoside interconversions | ||
| cmk | 1761 | cytidine monophosphate kinase |
| dukA | 494 | deoxynucleoside kinase |
| dukB | 1171 | deoxynucleoside kinase |
| nucA | 1101 | nucleotidase |
| pyrH | 2088 | UMP-kinase |
| ycjM | 301 | phosphatase |
| Purine ribonucleotide biosynthesis | ||
| guaA | 1517 | GMP synthase |
| guaB | 222 | IMP dehydrogenase |
| guaC | 1159 | GMP reductase |
| hprT | 1561 | hypoxanthine-guanine phosphoribosyltransferase |
| purA | 2029 | adenylosuccinate synthase |
| purB | 1689 | adenylosuccinate lyase |
| purC | 1578 | phosphoribosylaminoimidazole-succinocarboxamide synthetase |
| purD | 1554 | phosphoribosylamine-glycine ligase |
| purE | 1553 | phosphoribosylaminoimidazole carboxylase |
| purF | 1572 | phosphoribosylpyrophosphate amidotransferase |
| purH | 1560 | bifunctional purine biosynthesis protein PurH |
| purK | 1552 | phosphoribosylaminoimidazole carboxylase |
| purL | 1575 | phosphoribosyl formylglycinamidine synthase II |
| purM | 1566 | phosphoribosyl-aminoimidazole synthetase |
| purN | 1565 | phosphoribosylglycinamide formyltransferase |
| purQ | 1577 | phosphoribosyl formylglycinamidine synttase I |
| Pyrimidine ribonucleotide biosynthesis | ||
| carA | 1645 | glutaminase of carbamoyl-phosphate synthase |
| carB | 1400 | carbamoylphosphate synthetase |
| dut | 181 | deoxyuridine 5'-triphosphate nucleotidhydrolase |
| pydA | 1593 | dihydroorotate dehydrogenase A |
| pydB | 1383 | dihydroorotate dehydrogenase B |
| pyrB | 1646 | aspartate carbamoyltransferase |
| pyrC | 1082 | dihydroorotase |
| pyrE | 1081 | orotate phosphoribosyltransferase |
| pyrF | 1382 | orotidine-phosphate decarboxylase |
| pyrZ | 1384 | dihydroorotate dehydrogenase electron transfer subunit |
| thyA | 1583 | thymidylate synthase |
| yeaB | 404 | thymidylate kinase |
| Salvage of nucleosides and nucleotides | ||
| add | 288 | adenosine deaminase |
| adk | 2158 | adenylate kinase |
| apt | 623 | adenine phosphoribosyltransferase |
| cdd | 1463 | cytidine deaminase |
| deoB | 956 | phosphopentomutase |
| deoC | 1464 | deoxyribose-phosphate aldolase |
| deoD | 957 | purine nucleoside phosphorylase |
| gmk | 1967 | guanylate kinase |
| hpt | 25 | hypoxantine-guanine phosphorybosyltransferase |
| nrdD | 272 | anaerobic ribonucleoside-triphosphate reductase |
| nrdG | 273 | anaerobic ribonucleoside-triphosphate reductase activating protein |
| pdp | 1465 | pyrimidine-nucleoside phosphorylase |
| prsA | 826 | ribose-phosphate pyrophosphokinase |
| prsB | 1926 | ribose-phosphate pyrophosphokinase |
| udk | 1710 | uridine kinase |
| udp | 855 | uridine phosphorylase |
| upp | 1992 | uracil phosphoribosyltransferase |
| xpt | 1160 | xanthine phosphoribosyltransferase |
| yfiG | 585 | thymidine kinase |
| Sugar-nucleotide biosynthesis and interconversions | ||
| cpsM | 199 | dTDP-4-keto-6-deoxyglucose-3,5-epimerase |
| rmlA | 197 | glucose-1-phosphate thymidylyltransferase |
| rmlB | 200 | dTDP-glucose 4,6-dehydratase |
| rmlC | 201 | dTDP-L-rhamnose synthase |
| REGULATORY FUNCTIONS | ||
| General | ||
| ahrC | 883 | transcriptional regulator |
| aldR | 1253 | regulatory protein AldR |
| argR | 2118 | arginine catabolic regulator |
| birA1 | 1840 | bifunctional protein BirA |
| birA2 | 1973 | bifunctional protein BirA |
| codY | 164 | transcriptional regulator |
| codZ | 1865 | transcriptional regulator |
| copR | 845 | transcriptional regulator |
| fur | 1506 | ferric uptake regulator |
| gadR | 1327 | positive regulator |
| glnB | 1636 | nitrogen regulatory protein P-II |
| glnR | 2284 | glutamine synthetase repressor |
| gntR | 2272 | transcription regulator |
| nadR | 2067 | transcriptional regulator |
| phoU | 1771 | phosphate transport system regulator |
| purR | 2351 | regulator of purine biosynthetic genes |
| pyrR | 1648 | pyrimidine operon regulator |
| rarA | 1649 | transcriptional regulator |
| rcfA | 2083 | transcriptional regulator |
| rcfB | 2318 | transcriptional regulator |
| relA | 108 | ppGpp synthetase I |
| rmeA | 1947 | transcriptional regulator |
| rmeB | 1508 | transcriptional regulator |
| rmeC | 237 | transcriptional regulator |
| rmeD | 2053 | transcriptional regulator |
| tagR | 936 | transcriptional regulator |
| tenA | 1839 | transcriptional regulator TenA |
| yabA | 11 | transcriptional regulator |
| yabB | 13 | transcriptional regulator |
| ybdA | 131 | transcription regulator |
| ybdG | 135 | transcriptional regulator |
| ybeD | 146 | transcriptional regulator |
| ycdF | 235 | transcriptional regulator |
| ycfA | 250 | transcriptional regulator |
| ydbF | 316 | transcriptional regulator |
| ydcG | 327 | transcriptional regulator |
| yebF | 418 | transcriptional regulator |
| yecA | 420 | transcriptional regulator |
| yecE | 427 | transcriptional regulator |
| yeeG | 446 | transcriptional regulator |
| yfbM | 518 | transcriptional regulator |
| yfeA | 548 | transcription regulator |
| yfjG | 600 | transcriptional regulator |
| ygfC | 654 | transcriptional regulator |
| yhgC | 763 | transcriptional regulator |
| yidA | 831 | transcription regulator |
| yjaD | 904 | transcriptional regulator |
| yjaJ | 910 | transcriptional regulator |
| yjfE | 955 | transcription regulator |
| yjjB | 992 | transcriptional regulator |
| ykhI | 1078 | transcriptional regulator |
| yleF | 1148 | transcription regulator |
| yliA | 1180 | positive transcriptional regulator |
| yljC | 1193 | transcriptional regulator |
| ymcE | 1223 | transcriptional regulator |
| ymiA | 1285 | metalloregulator |
| ynaB | 1303 | transcriptional regulator |
| yogL | 1469 | transcriptional regulator |
| yohC | 1472 | transcriptional regulator |
| ypfD | 1555 | transcriptional regulator |
| ypgC | 1563 | transcription regulator |
| yqbH | 1618 | transcriptional regulator |
| yrbI | 1717 | transcriptional regulator |
| yrfA | 1750 | transcription regulator |
| yrfE | 1760 | transcription regulator |
| ysfD | 1854 | regulatory protein |
| ysfG | 1857 | transcriptional regulator |
| ysgA | 1866 | transcriptional regulator |
| yugA | 2065 | transcription regulator |
| ywdE | 2237 | transcription regulator |
| ywiI | 228 | transcriptional regulator |
| ywjD | 2297 | transcription regulator |
| yxcB | 2326 | transcriptional regulator |
| yxdD | 2337 | transcriptional regulator |
| Two-component systems | ||
| kinA | 1638 | sensor protein kinase |
| kinB | 1460 | sensor protein kinase |
| kinC | 402 | sensor protein kinase |
| kinD | 912 | sensor protein kinase |
| kinE | 1032 | sensor protein kinase |
| kinF | 1726 | sensor protein kinase |
| kinG | 1804 | sensor protein kinase |
| llrA | 1639 | two-component system regulator |
| llrB | 1458 | two-component system regulator |
| llrC | 403 | two-component system regulator |
| llrD | 913 | two-component system regulator |
| llrE | 1031 | two-component system regulator |
| llrF | 1727 | two-component system regulator |
| llrG | 1805 | two-component system regulator |
| llrH | 1758 | two-component system regulator |
| LacI-family regulators | ||
| ccpA | 1696 | catabolite control protein A |
| rbsR | 1688 | ribose operon repressor |
| rliA | 1728 | transcriptional regulator |
| rliB | 1536 | transcriptional regulator |
| rliC | 731 | transcriptional regulator |
| rliDA | 2215 | transcriptional regulator |
| rliDB | 2218 | transcriptional regulator |
| LysR-family regulators | ||
| fhuR | 331 | fhu operon transcriptional regulator |
| mleR | 896 | malolactic fermentation system transcriptional activator |
| mtlR | 32 | transcriptional regulator |
| rlrA | 1264 | transcriptional regulator |
| rlrB | 1946 | transcriptional regulator |
| rlrC | 1341 | transcriptional regulator |
| rlrD | 381 | transcriptional regulator |
| rlrE | 1598 | transcriptional regulator |
| rlrG | 378 | transcriptional regulator |
| AraC-family regulators | ||
| adaA | 519 | methylphosphotriester-DNA alkyltransferase |
| xylR | 1551 | xylose operon regulator |
| yneE | 1344 | transcriptional regulator |
| GntR-family regulators | ||
| busR | 1476 | transcriptional regulator |
| kdgR | 1680 | transcriptional regulator |
| rgrA | 437 | transcriptional regulator |
| rgrB | 1461 | transcriptional regulator |
| DeoR-family regulators | ||
| citR | 1206 | citrate lyase regulator |
| lacR | 984 | lactose transport regulator |
| rdrA | 797 | transcriptional regulator |
| rdrB | 1332 | transcriptional regulator |
| MarR-family regulators | ||
| rmaA | 750 | transcriptional regulator |
| rmaB | 715 | transcriptional regulator |
| rmaC | 1503 | transcriptional regulator |
| rmaD | 115 | transcriptional regulator |
| rmaE | 1511 | transcriptional regulator |
| rmaF | 1341 | transcriptional regulator |
| rmaG | 781 | transcriptional regulator |
| rmaH | 932 | transcriptional regulator |
| rmaI | 1583 | transcriptional regulator |
| rmaJ | 584 | transcriptional regulator |
| zitR | 2185 | zinc transport transcriptional regulator |
| BglG-family regulators | ||
| bglR | 1493 | beta-glucoside operon antiterminator |
| GTP-binding proteins | ||
| eraL | 355 | GTP-binding protein Era |
| hflX | 225 | GTP-binding protein HflX |
| obgL | 1630 | GTP-binding protein Obg |
| thdF | 2328 | GTP-binding protein ThdF |
| typA | 2094 | GTP-binding protein TypA/BipA |
| ylqL | 1330 | GTP-binding protein |
| yphL | 762 | GTP-binding protein |
| yqeL | 224 | GTP-binding protein |
| ysxL | 1165 | GTP-binding protein |
| yyaL | 12 | GTP-binding protein |
| REPLICATION | ||
| Degradation of DNA | ||
| exoA | 799 | exodeoxyribonuclease A |
| nth | 1084 | endonuclease III |
| recJ | 622 | single-stranded DNA specific exonuclease |
| rexA | 8 | subunit A of ATP-dependent exonuclease |
| rexB | 5 | subunit B of ATP-dependent exonuclease |
| sbcC | 1354 | ATP-dependent dsDNA exonuclease |
| sbcD | 1357 | exonuclease SbcD |
| uvrA | 1887 | excinuclease ABC subunit A |
| uvrB | 557 | excinuclease ABC subunit B |
| uvrC | 857 | excinuclease ABC subunit C |
| xseA | 878 | exonuclease VII large subunit |
| xseB | 879 | exonuclease VII small subunit |
| DNA replication, restriction, modification, recombination, and repair | ||
| cshA | 100 | chromosome segregation helicase |
| dinG | 1900 | ATP-dependent helicase DinG |
| dnaA | 1 | replication initiation protein DnaA |
| dnaB | 758 | replication protein DnaB |
| dnaC | 754 | replicative DNA helicase |
| dnaD | 1083 | DNA replication protein DnaD |
| dnaE | 496 | DNA polymerase III, alpha chain 2 |
| dnaG | 545 | DNA primase |
| dnaH | 2279 | DNA polymerase III, subunits beta and tau |
| dnaI | 759 | primosomal protein DnaI |
| dnaJ | 2308 | DnaJ protein |
| dnaN | 2 | DNA polymerase III, beta chain |
| dnaQ | 1010 | DNA polymerase III, epsilon chain |
| gyrA | 1123 | DNA gyrase subunit A |
| gyrB | 929 | DNA gyrase subunit B |
| hexA | 2294 | mismatch repair protein MutS |
| hexB | 2291 | DNA mismatch repair protein MutL |
| holB | 405 | DNA polymerase III, delta' subunit |
| hsdM | 645 | type I restriction enzyme M protein |
| hsdR | 642 | type I restriction enzyme R protein |
| hsdS | 646 | type I restriction enzyme specificity protein |
| hslA | 502 | HU like DNA-binding protein |
| hslB | 903 | HU-like DNA-binding protein |
| ligA | 425 | DNA ligase |
| mutM | 358 | formamidopyrimidine-DNA glycosylase |
| mutS | 1693 | DNA mismatch repair protein MutS |
| mutX | 1136 | mutator protein MutT |
| mutY | 859 | A/G-specific adenine glycosylase |
| ogt | 519 | 6-O-methylguanine-DNA methyltransferase |
| parC | 1012 | topoisomerase IV subunit B |
| parE | 1008 | topoisomerase IV subunit B |
| pcrA | 1135 | ATP-dependent helicase PcrA |
| polA | 2212 | DNA polymerase I |
| polC | 2192 | DNA polymerase III, alpha chain |
| priA | 1965 | primosomal protein N' |
| radC | 1036 | DNA repair protein RadC |
| recA | 359 | RecA protein |
| recD | 1798 | exodeoxyribonuclease V alpha chain |
| recF | 2052 | RecF protein |
| recG | 2331 | ATP-dependent DNA helicase RecG |
| recM | 340 | RecM protein |
| recN | 884 | DNA repair protein RecN |
| ruvA | 2290 | DNA helicase RuvA |
| ruvB | 2289 | DNA helicase RuvB |
| snf | 2122 | SWI/SNF family helicase |
| ssbA | 398 | single-strand binding protein |
| ssbB | 2274 | single-strand binding protein |
| tag | 1137 | DNA-3-methyladenine glycosidase I |
| topA | 1256 | DNA topoisomerase I |
| umuC | 581 | SOS responce UmuC protein |
| ung | 233 | uracil-DNA glycosilase |
| xerD | 635 | integrase-recombinase |
| ybaH | 101 | acetyl transferase |
| yffD | 556 | diadenosine 5',5”'-P1,P4-tetraphosphate hydrolase |
| ymgA | 1260 | integrase-recombinase |
| TRANSCRIPTION | ||
| Degradation of RNA | ||
| pnpA | 1923 | polyribonucleotide nucleotidyltransferase |
| rnc | 810 | ribonuclease III |
| rnhA | 2350 | ribonuclease HII |
| rnhB | 1329 | ribonuclease HII |
| vacB1 | 968 | ribonuclease |
| vacB2 | 1227 | ribonuclease |
| RNA synthesis, modification, and DNA transcription | ||
| greA | 626 | transcription elongation factor GreA |
| mfd | 19 | transcription-repair coupling factor |
| nusA | 774 | transcription termination protein NusA |
| nusB | 693 | transcription termination protein NusB |
| nusG | 2174 | transcription antitermination protein |
| papL | 1603 | poly(A) polymerase |
| queA | 1617 | S-adenosylmethionine tRNA ribosyltransferase |
| rluA | 2182 | pseudouridine synthase |
| rluB | 1308 | pseudouridine synthase |
| rluC | 1390 | pseudouridine synthase |
| rluD | 1027 | pseudouridine synthase |
| rluE | 368 | pseudouridine synthase |
| rpoA | 2153 | DNA-directed RNA polymerase alpha chain |
| rpoB | 1863 | DNA-directed RNA polymerase beta chain |
| rpoC | 1859 | DNA-directed RNA polymerase beta' chain |
| rpoD | 547 | major RNA polymerase sigma factor |
| rpoE | 624 | DNA-directed RNA polymerase delta chain |
| rrmA | 1365 | rRNA methyltransferase |
| rsuA | 2327 | ribosomal small subunit pseudouridine synthase A |
| sigX | 2243 | RNA polymerase ECF sigma factor |
| smpB | 1777 | tmRNA-binding protein SmpB |
| sunL | 1958 | rRNA methylase |
| trmD | 1607 | tRNA methyltransferase |
| yfjD | 597 | tRNA/rRNA methyltransferase |
| RNA processing | ||
| rheA | 354 | ATP-dependent RNA helicase |
| rheB | 416 | ATP-dependent RNA helicase |
| rimM | 1607 | 16S rRNA processing protein |
| TRANSLATION | ||
| Amino acyl tRNA synthetases | ||
| alaS | 1780 | alanyl-tRNA synthetase |
| argS | 2117 | arginyl-tRNA synthetase |
| asnS | 1896 | asparaginyl-tRNA synthetase |
| aspS | 2041 | aspartyl-tRNA synthetase |
| cysS | 1919 | cysteinyl-tRNA synthetase |
| gltX | 2141 | glutamyl-tRNA synthetase |
| glyS | 1102 | glycyl-tRNA synthetase alpha chain |
| glyT | 1104 | glycyl-tRNA synthetase beta chain |
| hisS | 2043 | histidyl-tRNA synthetase |
| ileS | 1933 | isoleucyl-tRNA synthetase |
| leuS | 829 | leucyl-tRNA synthetase |
| lysS | 377 | lysyl-tRNA synthetase |
| metS | 800 | methyonyl-tRNA synthetase |
| pheS | 2010 | phenylalanil-tRNA synthetase alpha chain |
| pheT | 2008 | phenylalanyl-tRNA synthetase beta chain |
| proS | 2197 | prolyl-tRNA synthetase |
| serS | 1768 | seryl-tRNA synthetase |
| thrS | 1988 | theronyl-tRNA synthetase |
| trpS | 68 | tryptophanyl-tRNA synthetase |
| tyrS | 391 | tyrosyl-tRNA synthetase 1 |
| valS | 2250 | valyl-tRNA synthetase |
| Degradation of proteins, peptides, and glycopeptides | ||
| gcp | 294 | O-sialoglycoprotein endopeptidase |
| htrA | 2205 | exported serine protease |
| pepA | 394 | glutamyl aminopeptidase |
| pepC | 1948 | aminopeptidase C |
| pepDA | 249 | dipeptidase |
| pepDB | 1601 | dipeptidase |
| pepF | 1784 | oligoendopeptidase F |
| pepM | 601 | mathionine aminopeptidase |
| pepN | 304 | aminopeptidase N |
| pepO | 1867 | neutral endopeptidase |
| pepP | 691 | aminopeptidase P |
| pepQ | 1698 | proline dipeptidase |
| pepT | 1878 | tripeptidase |
| pepV | 861 | dipeptidase |
| pepXP | 2136 | X-prolyl dipeptidyl aminopeptidase |
| yueE | 2049 | protease |
| yueF | 2050 | protease |
| yugD | 2069 | protease |
| yuhB | 2071 | protease |
| Protein modification | ||
| def | 555 | polypeptide deformylase |
| pknB | 1956 | serine/threonine protein kinase |
| pmpA | 1782 | protein maturation protein |
| pmsR | 2085 | peptide methionine sulfoxide reductase |
| pmsX | 1594 | peptide methionine sulfoxide reductase |
| ppiA | 369 | peptidyl-prolyl cis-trans isomerase |
| ppiB | 914 | peptidyl-prolyl cis-trans isomerase |
| pppL | 1957 | protein serine/threonine phosphatase |
| ptpL | 2284 | protein-tyrosine phosphatase |
| ytaD | 1905 | protein-tyrosine phosphatase |
| Ribosomal proteins: synthesis and modification | ||
| prmA | 105 | methyltransferase |
| rplA | 2079 | 50S ribosomal protein L1 |
| rplB | 2168 | 50S ribosomal protein L2 |
| rplC | 2170 | 50S ribosomal protein L3 |
| rplD | 2169 | 50S ribosomal protein L4 |
| rplE | 2164 | 50S ribosomal protein L5 |
| rplF | 2162 | 50S ribosomal protein L6 |
| rplI | 753 | 50S ribosomal protein L9 |
| rplJ | 1302 | 50S ribosomal protein L10 |
| rplK | 2080 | 50S ribosomal protein L11 |
| rplL | 1301 | 50S ribosomal protein L7/L12 |
| rplM | 2347 | 50S ribosomal protein L13 |
| rplN | 2165 | 50S ribosomal protein L14 |
| rplO | 2160 | 50S ribosomal protein L15 |
| rplP | 2166 | 50S ribosomal protein L16 |
| rplQ | 2152 | 50S ribosomal protein L17 |
| rplR | 2161 | 50S ribosomal protein L18 |
| rplS | 898 | 50S ribosomal protein L19 |
| rplT | 1911 | 50S ribosomal protein L20 |
| rplU | 1091 | 50S ribosomal protein L21 |
| rplV | 2167 | 50S ribosomal protein L22 |
| rplW | 2169 | 50S ribosomal protein L23 |
| rplX | 2165 | 50S ribosomal protein L24 |
| rpmA | 1091 | 50S ribosomal protein L27 |
| rpmB | 196 | 50S ribosomal protein L28 |
| rpmC | 2166 | 50S ribosomal protein L29 |
| rpmD | 2160 | 50S ribosomal protein L30 |
| rpmE | 1640 | 50S ribosomal protein L31 |
| rpmF | 96 | 50S ribosomal protein L32 |
| rpmGA | 662 | 50S ribosomal protein L33 |
| rpmGB | 96 | 50S ribosomal protein L33 |
| rpmGC | 2175 | 50S ribosomal protein L33 |
| rpmH | 134 | 50S ribosomal protein L34 |
| rpmI | 1912 | 50S ribosomal protein L35 |
| rpmJ | 2154 | 50S ribosomal protein L36 |
| rpsA | 854 | 30S ribosomal protein S1 |
| rpsB | 2228 | 30S ribosomal protein S2 |
| rpsC | 2166 | 30S ribosomal protein S3 |
| rpsD | 284 | 30S ribosomal protein S4 |
| rpsE | 2161 | 30S ribosomal protein S5 |
| rpsF | 2275 | 30S ribosomal protein S6 |
| rpsG | 2355 | 30S ribosomal protein S7 |
| rpsH | 2162 | 30S ribosomal protein S8 |
| rpsI | 2347 | 30S ribosomal protein S9 |
| rpsJ | 2170 | 30S ribosomal protein S10 |
| rpsK | 2153 | 30S ribosomal protein S11 |
| rpsL | 2355 | 30S ribosomal protein S12 |
| rpsM | 2154 | 30S ribosomal protein S13 |
| rpsN | 2164 | 30S ribosomal protein S14 |
| rpsN2 | 911 | 30S ribosomal protein S14 |
| rpsO | 1955 | 30S ribosomal protein S15 |
| rpsP | 1611 | 30S ribosomal protein S16 |
| rpsQ | 2165 | 30S ribosomal protein S17 |
| rpsR | 2274 | 30S ribosomal protein S18 |
| rpsS | 2167 | 30S ribosomal protein S19 |
| rpsT | 1797 | 30S ribosomal protein S20 |
| rpsU | 237 | 30S ribosomal protein S21 |
| ycjC | 293 | acetyltransferase |
| ycjD | 293 | acetyltransferase |
| yhdC | 740 | acetyl transferase |
| yhjG | 798 | acetyl transferase |
| ylxQ | 776 | probable ribosomal protein |
| fmt | 1962 | methyonyl-tRNA formyltransferase |
| gatA | 166 | Glu-tRNA amidotransferase subunit A |
| gatB | 168 | Glu-tRNA amidotransferase subunit B |
| gatC | 165 | Glu-tRNA amidotransferase subunit C |
| ksgA | 690 | kasugamycin dimethyltransferase |
| miaA | 615 | tRNA isopentenyltransferase |
| pth | 17 | peptidyl-tRNA hydrolase |
| rnpA | 132 | ribonuclease P protein component |
| tgt | 156 | queuine tRNA-ribosyltransferase |
| trmH | 1942 | tRNA-guanosine methyltransferase |
| trmU | 853 | tRNA-methyltransferase |
| truA | 485 | tRNA pseudouridine synthase A |
| truB | 1141 | tRNA pseudouridine synthase B |
| Translation factors | ||
| efp | 692 | elongation factor P |
| frr | 2087 | ribosome recycling factor |
| fusA | 2353 | elongation factor G |
| infA | 2154 | translation initiation factor IF-1 |
| infB | 777 | translation initiation factor IF-2 |
| infC | 1912 | translation initiation factor IF-3 |
| lepA | 1118 | GTP-binding protein LepA |
| prfA | 586 | peptide chain release factor 1 |
| prfB | 999 | peptide chain release factor 2 |
| prfC | 352 | peptide chain release factor 3 |
| rbfA | 779 | ribosome-binding factor A |
| tsf | 2227 | elongation factor Ts |
| tuf | 1930 | elongation factor Tu |
| TRANSPORT AND BINDING PROTEINS | ||
| General | ||
| ecsA | 2075 | ABC transporter ATP binding protein |
| ecsB | 2074 | ABC transporter permease protein |
| mscL | 2171 | large conductance mechanosensitive channel protein |
| yabE | 16 | ABC transporter ATP-binding protein |
| yahG | 74 | ABC transporter ATP binding protein |
| yaiE | 87 | transporter |
| yajA | 90 | transporter |
| ybaB | 102 | ABC transporter ATP binding protein |
| ycfB | 251 | ABC transporter ATP binding protein |
| ycfC | 252 | ABC transporter permease protein |
| ycfI | 260 | ABC transporter ATP binding protein |
| ycgA | 261 | ABC transporter ATP binding protein |
| ycgB | 262 | ABC transporter ATP binding protein |
| ychD | 276 | ABC transporter ATP-binding protein |
| ychE | 277 | ABC transporter ATP-binding protein |
| ychF | 278 | ABC transporter permease protein |
| ydaG | 310 | ABC transporter ATP binding and permease protein |
| ydbA | 312 | ABC transporter ATP binding and permease protein |
| ydcE | 325 | ABC transporter ATP binding protein |
| ydcF | 326 | ABC transporter permease protein |
| ydiA | 382 | permease |
| yfcA | 520 | ABC transporter ATP binding protein |
| yfcB | 521 | ABC transporter permease protein |
| yfgE | 563 | ABC transporter ATP binding protein |
| yfgF | 564 | ABC transporter permease protein |
| ygfA | 652 | ABC transporter ATP-binding protein |
| ygfB | 653 | ABC transporter permease protein |
| yhcA | 721 | ABC transporter ATP-binding and permease protein |
| yiiF | 886 | transporter |
| yijC | 894 | ABC transporter permease protein |
| yijD | 895 | ABC transporter ATP binding protein |
| yjcA | 921 | ABC transporter ATP binding protein |
| yjjC | 993 | ABC transporter ATP-binding protein |
| yjjD | 994 | ABC transporter permease protein |
| yjjF | 996 | transporter |
| ykhF | 1074 | ABC transporter ATP binding protein |
| ylbA | 1111 | ABC transporter ATP-binding protein |
| ylbB | 1113 | ABC transporter permease protein |
| yljI | 1199 | permease |
| ymeB | 1245 | ABC transporter ATP binding protein |
| ynaC | 1304 | ABC transporter ABC binding and permease protein |
| ynaD | 1306 | ABC transporter ABC binding and permease protein |
| yngB | 1364 | fibronectin-binding protein |
| ypgD | 1564 | ABC transporter ATP binding and permease protein |
| ypjG | 1597 | ABC transporter ATP binding protein |
| yrjE | 1794 | transport permease |
| ysaB | 1808 | ABC transporter permease and substrate binding protein |
| ysaC | 1809 | ABC transporter ATP-binding protein |
| ysdA | 1834 | ABC transporter permease protein |
| ysdB | 1835 | ABC transporter ATP binding protein |
| ysfB | 1852 | ABC transporter ATP-binding protein |
| ysiA | 1882 | transport protein |
| ysiB | 1883 | permease |
| ytaB | 1902 | transport protein |
| yteD | 1945 | transmembrane efflux protein |
| yudA | 2031 | transport protein |
| yujD | 2097 | ABC transporter ATP binding protein |
| ywiG | 2285 | ABC transporter ATP binding protein |
| ywiH | 2286 | ABC transporter permease protein |
| yxaA | 2300 | permease |
| yxdG | 2340 | transporter |
| yxeB | 2349 | ABC transporter ATP-binding protein |
| yxfA | 2358 | transporter |
| Amino acids, peptides and amines | ||
| arcD1 | 2112 | arginine/ornitine antiporter |
| arcD2 | 2107 | arginine/ornitine antiporter |
| brnQ | 685 | branched chain amino acid permease |
| busAA | 1475 | betaine ABC transporter ATP binding protein |
| busAB | 1474 | betaine ABC transporter permease and substrate binding protein |
| choQ | 865 | choline ABC transporter ATP binding protein |
| choS | 867 | choline ABC transporter permease and substrate binding protein |
| ctrA | 113 | cationic amino acid transporter |
| dtpT | 705 | di-/tripeptide transporter |
| gadC | 1326 | glutamate-gamma-aminobutyrate antiporter |
| glnP | 1818 | glutamine ABC transporter permease and substrate binding protein |
| glnQ | 1819 | glutamine ABC transporter ATP-binding protein |
| gltP | 1856 | glutamate ABC transporter permease protein |
| gltQ | 1855 | glutamate ABC transporter ATP-binding protein |
| gltS | 559 | glutamate or arginine ABC transporter substrate binding protein |
| lysP | 2277 | lysine specific permease |
| lysQ | 370 | lysine specific permease |
| oppA | 1906 | oligopeptide ABC trasporter substrate binding protein |
| oppB | 1908 | oligopeptide ABC trasporter permease protein |
| oppC | 1907 | oligopeptide ABC trasporter permease protein |
| oppD | 1910 | oligopeptide ABC trasporter ATP binding protein |
| oppF | 1909 | oligopeptide ABC trasporter ATP binding protein |
| optA | 346 | oligopeptide ABC transporter substrate binding protein |
| optB | 347 | oligopeptide ABC trasporter permease protein |
| optC | 348 | oligopeptide ABC trasporter permease protein |
| optD | 349 | oligopeptide ABC trasporter ATP binding protein |
| optF | 350 | oligopeptide ABC trasporter ATP binding protein |
| optS | 344 | oligopeptide ABC transporter substrate binding protein |
| potA | 1176 | spermidine/putrescine ABC transporter ATP-binding protein |
| potB | 1177 | spermidine/putrescine ABC transporter permease protein |
| potC | 1178 | spermidine/putrescine ABC transporter permease protein |
| potD | 1179 | spermidine/putrescine ABC transporter substrate binding protein |
| yagE | 70 | amino acid permease |
| ydcB | 322 | amino acid ABC transporter ATP binding protein |
| ydcC | 323 | amino acid ABC trasporter permease protein |
| ydgB | 361 | amino acid permease |
| ydgC | 362 | amino acid permease |
| yfcG | 525 | peptide-binding protein |
| yibG | 819 | amino acid permease |
| yjgC | 963 | amino acid ABC transporter substrate binding protein |
| yjgD | 964 | amino acid ABC transporter permease protein |
| yjgE | 965 | amino acid ABC transporter ATP binding protein |
| ylcA | 1121 | amino acid permease |
| yqfD | 1655 | amino acid permease |
| yrfD | 1756 | amino acid antiporter |
| yshA | 1876 | amino acid permease |
| ysjA | 1891 | amino acid permease |
| yvdF | 2138 | amino acid ABC transporter substrate binding protein |
| Anions | ||
| phnA | 2332 | alkylphosphonate uptake protein |
| phnB | 299 | phosphonate ABC transporter permease protein |
| phnC | 298 | phosphonate ABC transporter ATP-binding protein |
| phnE | 299 | phosphonate ABC transporter permease protein |
| pstA | 1772 | phosphate ABC transporter ATP binding protein |
| pstB | 1772 | phosphate ABC transporter ATP binding protein |
| pstC | 1773 | phosphate ABC transporter permease protein |
| pstD | 1774 | phosphate ABC transporter permease protein |
| pstE | 1775 | phosphate ABC transporter substrate binding protein |
| pstF | 1776 | phosphate ABC transporter substrate binding protein |
| yafB | 52 | sulfate transporter |
| Carbohydrates, organic alcohols and acids | ||
| glpF1 | 248 | glycerol uptake facilitator |
| glpF2 | 1270 | glycerol uptake facilitator |
| glpT | 549 | glycerol-3-phosphatase transporter |
| gntP | 2266 | gluconate permease |
| lacS | 2063 | lactose permease |
| maeP | 1205 | malate permease |
| malE | 1740 | maltose ABC transporter substrate binding protein |
| malF | 1741 | maltose ABC transporter permease protein |
| malG | 1742 | maltose ABC transporter permease protein |
| mleP | 924 | malate transporter |
| msmK | 428 | multiple sugar ABC transporter ATP-binding protein |
| rbsA | 1685 | ribose ABC transporter ATP binding protein |
| rbsB | 1683 | ribose ABC transporter substrate binding protein |
| rbsC | 1684 | ribose ABC transporter permease protein |
| rbsD | 1686 | ribose ABC transporter permease protein |
| rgpC | 204 | polysaccharide ABC transporter permease protein |
| rgpD | 205 | polysaccharide ABC transporter ATP-binding protein |
| tagG | 939 | teichoic acid ABC transporter permease protein |
| tagH | 938 | teichoic acid ABC transporter ATP binding protein |
| uxuT | 1676 | Na-galactoside symporter |
| xylT | 1542 | D-xylose proton-symporter |
| xynT | 1546 | xyloside transporter |
| yngE | 1366 | sugar ABC transporter ATP binding protein |
| yngF | 1368 | sugar ABC transporter permease protein |
| yngG | 1369 | sugar ABC transporter permease protein |
| ypbD | 1515 | sugar transport symporter |
| ypcG | 1528 | sugar ABC transporter substrate binding protein |
| ypcH | 1529 | sugar ABC transporter permease protein |
| ypdA | 1530 | sugar ABC transporter substrate binding protein |
| yqgE | 1668 | transporter |
| yvdD | 2134 | transporter |
| Cations | ||
| amtB | 1636 | ammonium transporter |
| cadA | 97 | cadmium efflux ATPase |
| copA | 847 | copper/potassium-transporting ATPase |
| copB | 872 | copper-potassium transporting ATPase B |
| feoA | 192 | ferrous ion transport protein A |
| feoB | 191 | ferrous ion transport protein B |
| fhuB | 328 | ferrichrome ABC transporter permease protein |
| fhuC | 327 | ferrichrome ABC transporter ATB binding protein |
| fhuD | 330 | ferrichrome ABC transporter substrate binding protein |
| fhuG | 329 | ferrichrome ABC transporter permease protein |
| kupA | 610 | potassium uptake protein |
| kupB | 613 | potassium uptake protein |
| mgtA | 1287 | cation-transporting P-ATPase |
| mtsA | 1350 | manganese ABC transporter substrate binding protein |
| mtsB | 1351 | manganese ABC transporter ATP binding protein |
| mtsC | 1351 | manganese ABC transporter permease protein |
| nah | 1994 | Na+/H+ antiporter |
| pacL | 677 | cation-transporting ATPase |
| ydaE | 308 | cation transporter |
| yddA | 332 | transporter |
| ydiF | 388 | Na+/H+ antiporter |
| yfgQ | 570 | cation-transporting ATPase |
| ygfE | 657 | divalent cation transport-related protein |
| yieF | 846 | mercuric reductase |
| yjdJ | 937 | potassium channel protein |
| yliI | 1190 | cation-transporting ATPase |
| yndG | 1337 | metal ABC transporter substrate binding protein |
| yoaB | 1404 | cation-transporting ATPase |
| yogJ | 1468 | cation transporter |
| ypbB | 1512 | cationic transporter |
| yqeI | 1650 | cation transport protein |
| yqgG | 1664 | cation transport ATPase |
| ysdE | 1838 | cation transporter |
| ytjB | 1990 | manganese transporter |
| yuiA | 2081 | metal transporting ATPase |
| yxdC | 2336 | cation-transporting ATPase |
| zitP | 2183 | zinc ABC transporter permease protein |
| zitQ | 2183 | zinc ABC transporter ATP binding protein |
| zitS | 2184 | zinc ABC transporter substrate binding protein |
| Nucleosides, purines and pyrimidines | ||
| pbuX | 1161 | xanthine permease |
| pnuC1 | 856 | nicotinamide mononucleotide transporter |
| pnuC2 | 901 | nicotinamide mononucleotide transporter |
| pyrP | 1647 | uracil permease |
| PTS system | ||
| celB | 178 | cellobiose-specific PTS system IIC component |
| fruA | 986 | fructose-specific PTS system enzyme IIBC component |
| mtlA | 29 | mannitol-specific PTS system IIBC component |
| mtlF | 33 | mannitol-specific PTS system IIA component |
| ptbA | 1492 | beta-glucoside-specific PTS system IIABC component |
| ptcA | 419 | cellobiose-specific PTS system IIA component |
| ptcB | 419 | cellobiose-specific PTS system IIB component |
| ptcC | 421 | cellobiose-specific PTS system IIC component |
| ptnAB | 1763 | mannose-specific PTS system component IIAB |
| ptnC | 1764 | mannose-specific PTS system component IIC |
| ptnD | 1765 | mannose-specific PTS system component IID |
| ptsH | 120 | phosphocarrier protein Hpr |
| ptsI | 122 | phosphoenolpyruvate-protein phosphotransferase |
| ptsK | 605 | Hpr(Ser) kinase |
| yedF | 439 | beta-glucoside-specific PTS system IIABC component |
| yidB | 832 | cellobiose-specific PTS system IIC component |
| yleD | 1146 | sucrose-specific PTS system IIBC component |
| yleE | 1147 | beta-glucoside-specific PTS system IIABC component |
| Multidrug resistance | ||
| blt | 126 | multidrug efflux transporter |
| cydC | 711 | cytochrome D ABC transporter ATP binding and permease protein |
| cydD | 713 | cytochrome D ABC transporter ATP binding and permease protein |
| lcnC | 84 | lactococcin A ABC transporter ATP binding and permease protein |
| lcnD | 85 | lactococcin A ABC transporter permease protein |
| lmrA | 717 | multidrug resistance ABC transporter ATP binding and permease protein |
| lmrP | 2242 | integral membrane protein LmrP |
| napC | 306 | multidrug-efflux transporter |
| pmrA | 661 | multidrug resistance efflux pump |
| pmrB | 130 | multidrug resistance efflux pump |
| ybfD | 158 | transporter |
| ycdH | 236 | transporter |
| ydiC | 385 | efflux pump antibiotic resistance protein |
| yfjF | 599 | membrane-bound transport protein |
| yjdE | 933 | multidrug resistance protein |
| yniG | 1386 | drug-export protein |
| ypfE | 1557 | transport protein |
| ypiB | 1582 | transporter |
| yqiA | 1682 | multidrug transporter |
| yweA | 2240 | membrane protein |
| yxbD | 2319 | transporter |
| OTHER CATEGORIES | ||
| Adaptations and atypical conditions | ||
| arsC | 1412 | arsenate reductase |
| clpB | 1568 | ClpB protein |
| clpC | 631 | ATP-dependent protease ATP-binding subunit |
| clpE | 552 | ATP-dependent protease ATP-binding subunit |
| clpP | 673 | ATP-dependent Clp protease proteolytic subunit |
| clpX | 1164 | ATP dependent Clp protease |
| cpo | 835 | non-heme chloride peroxidase |
| cspD | 517 | cold shock protein D |
| cspE | 173 | cold shock protein E |
| cstA | 414 | carbon starvation protein |
| ctsR | 630 | transctiptional regulator CtsR |
| dinF | 172 | damage-inducible protein DinF |
| dinP | 2105 | DNA-damage-inducible protein P |
| dpsA | 2102 | non-heme iron-binding ferritin |
| grpE | 978 | stress responce protein GrpE |
| hrcA | 977 | heat-inducible transcription repressor HrcA |
| osmC | 69 | osmotically inducible protein |
| phoL | 1094 | phosphate starvation inducible protein |
| tpx | 302 | thiol peroxidase |
| ybjA | 193 | reductase |
| yjbE | 915 | general stress protein GSP13 |
| Drug and analog sensitivity | ||
| bacA | 2276 | undecaprenol kinase |
| bar | 1837 | acyltransferase |
| pacA | 1150 | penicillin acylase |
| pacB | 1904 | penicillin acylase |
| ymdC | 1234 | kanamycin kinase |
| Phage related functions and prophages | ||
| pi101 | 448 | prophage pi1 protein 01, integrase |
| pi102 | 449 | prophage pi1 protein 02 |
| pi103 | 450 | prophage pi1 protein 03, transcriptional regulator |
| pi104 | 450 | prophage pi1 protein 04, transcriptional regulator |
| pi105 | 451 | prophage pi1 protein 05 |
| pi106 | 451 | prophage pi1 protein 06 |
| pi107 | 451 | prophage pi1 protein 07 |
| pi108 | 452 | prophage pi1 protein 08 |
| pi109 | 452 | prophage pi1 protein 09 |
| pi110 | 453 | prophage pi1 protein 10, transcriptional regulator |
| pi111 | 453 | prophage pi1 protein 11, recombinase |
| pi112 | 454 | prophage pi1 protein 12 |
| pi113 | 455 | prophage pi1 protein 13, replisome organiser |
| pi114 | 456 | prophage pi1 protein 14, DNA replication protein |
| pi115 | 457 | prophage pi1 protein 15 |
| pi116 | 457 | prophage pi1 protein 16 |
| pi117 | 458 | prophage pi1 protein 17 |
| pi118 | 458 | prophage pi1 protein 18 |
| pi119 | 459 | prophage pi1 protein 19 |
| pi120 | 459 | prophage pi1 protein 16, deoxyuridine 5'-triphosphate nucleotidohydrolase |
| pi121 | 459 | prophage pi1 protein 21 |
| pi122 | 460 | prophage pi1 protein 22 |
| pi123 | 460 | prophage pi1 protein 23 |
| pi124 | 460 | prophage pi1 protein 24 |
| pi125 | 461 | prophage pi1 protein 25 |
| pi126 | 461 | prophage pi1 protein 26 |
| pi127 | 461 | prophage pi1 protein 27 |
| pi128 | 462 | prophage pi1 protein 28 |
| pi129 | 462 | prophage pi1 protein 29 |
| pi130 | 462 | prophage pi1 protein 30 |
| pi131 | 463 | prophage pi1 protein 31 |
| pi132 | 464 | prophage pi1 protein 32 |
| pi133 | 464 | prophage pi1 protein 33, terminase small subunit |
| pi134 | 465 | prophage pi1 protein 34, terminase large subunit |
| pi135 | 467 | prophage pi1 protein 35 |
| pi136 | 468 | prophage pi1 protein 36, prohead protease |
| pi137 | 469 | prophage pi1 protein 37, capsid protein |
| pi138 | 470 | prophage pi1 protein 38 |
| pi139 | 470 | prophage pi1 protein 39 |
| pi140 | 470 | prophage pi1 protein 40, tail component |
| pi141 | 471 | prophage pi1 protein 41, tail component |
| pi142 | 471 | prophage pi1 protein 42, small major structural protein |
| pi143 | 472 | prophage pi1 protein 43 |
| pi144 | 474 | prophage pi1 protein 44, tail component |
| pi145 | 477 | prophage pi1 protein 45, tail component |
| pi146 | 480 | prophage pi1 protein 46, tail component |
| pi147 | 482 | prophage pi1 protein 47 |
| pi148 | 482 | prophage pi1 protein 48, holin |
| pi149 | 483 | prophage pi1 protein 49, muramidase |
| pi201 | 1037 | prophage pi2 protein 01, integrase |
| pi202 | 1038 | prophage pi2 protein 02 |
| pi203 | 1039 | prophage pi2 protein 03 |
| pi204 | 1039 | prophage pi2 protein 04 hypothetical protein |
| pi205 | 1040 | prophage pi2 protein 05 |
| pi206 | 1040 | prophage pi2 protein 06 |
| pi207 | 1041 | prophage pi2 protein 07 |
| pi208 | 1041 | prophage pi2 protein 08 |
| pi209 | 1042 | prophage pi2 protein 09 |
| pi210 | 1042 | prophage pi2 protein 10 |
| pi211 | 1043 | prophage pi2 protein 11, topoisomerase |
| pi212 | 1043 | prophage pi2 protein 12, single strand binding protein |
| pi213 | 1044 | prophage pi2 protein 13, replisome organiser |
| pi214 | 1045 | prophage pi2 protein 14 |
| pi215 | 1045 | prophage pi2 protein 15 |
| pi216 | 1046 | prophage pi2 protein 16 |
| pi217 | 1046 | prophage pi2 protein 17 |
| pi218 | 1047 | prophage pi2 protein 18 |
| pi219 | 1048 | prophage pi2 protein 19 |
| pi220 | 1048 | prophage pi2 protein 20 hypothetical protein |
| pi221 | 1049 | prophage pi2 protein 21, deoxyuridine 5'-triphosphate nucleotidohydrolase |
| pi222 | 1049 | prophage pi2 protein 22 |
| pi223 | 1049 | prophage pi2 protein 23 |
| pi224 | 1050 | prophage pi2 protein 24 |
| pi225 | 1051 | prophage pi2 protein 25 |
| pi226 | 1051 | prophage pi2 protein 26 |
| pi227 | 1052 | prophage pi2 protein 27 |
| pi228 | 1052 | prophage pi2 protein 28 |
| pi229 | 1053 | prophage pi2 protein 29 |
| pi230 | 1054 | prophage pi2 protein 30, terminase |
| pi231 | 1055 | prophage pi2 protein 31 |
| pi232 | 1056 | prophage pi2 protein 32 |
| pi233 | 1057 | prophage pi2 protein 33, capsid protein |
| pi234 | 1058 | prophage pi2 protein 34 |
| pi235 | 1059 | prophage pi2 protein 35 |
| pi236 | 1059 | prophage pi2 protein 36 |
| pi237 | 1059 | prophage pi2 protein 37 |
| pi238 | 1060 | prophage pi2 protein 38 |
| pi239 | 1060 | prophage pi2 protein 39 |
| pi240 | 1061 | prophage pi2 protein 40 |
| pi241 | 1061 | prophage pi2 protein 41 |
| pi242 | 1062 | prophage pi2 protein 42 |
| pi243 | 1064 | prophage pi2 protein 43 |
| pi244 | 1065 | prophage pi2 protein 44 |
| pi245 | 1068 | prophage pi2 protein 45 |
| pi246 | 1069 | prophage pi2 protein 46 |
| pi247 | 1069 | prophage pi2 protein 47 |
| pi248 | 1070 | prophage pi2 protein 48 |
| pi249 | 1070 | prophage pi2 protein 49 |
| pi250 | 1070 | prophage pi2 protein 50 |
| pi251 | 1071 | prophage pi2 protein 51, holin |
| pi252 | 1071 | prophage pi2 protein 52, muramidase |
| pi301 | 1414 | prophage pi3 protein 01 |
| pi302 | 1415 | prophage pi3 protein 02 |
| pi303 | 1415 | prophage pi3 protein 03 |
| pi304 | 1416 | prophage pi3 protein 04 |
| pi305 | 1416 | prophage pi3 protein 05, muramidase |
| pi306 | 1417 | prophage pi3 protein 06, holin |
| pi307 | 1418 | prophage pi3 protein 07 |
| pi308 | 1419 | prophage pi3 protein 08 |
| pi309 | 1420 | prophage pi3 protein 09 |
| pi310 | 1421 | prophage pi3 protein 10 |
| pi311 | 1422 | prophage pi3 protein 11 |
| pi312 | 1424 | prophage pi3 protein 12 |
| pi313 | 1425 | prophage pi3 protein 13, tail component |
| pi314 | 1428 | prophage pi3 protein 14 |
| pi315 | 1431 | prophage pi3 protein 15 |
| pi316 | 1431 | prophage pi3 protein 16, tail component |
| pi317 | 1432 | prophage pi3 protein 17, major tail protein |
| pi318 | 1433 | prophage pi3 protein 18, tail component |
| pi319 | 1433 | prophage pi3 protein 19, tail component |
| pi320 | 1433 | prophage pi3 protein 20, head-tail joining protein |
| pi321 | 1434 | prophage pi3 protein 21 |
| pi322 | 1435 | prophage pi3 protein 22, major head protein precursor |
| pi323 | 1436 | prophage pi3 protein 23, ATP dependent Clp protease |
| pi324 | 1436 | prophage pi3 protein 24 |
| pi325 | 1437 | prophage pi3 protein 25, head-tail joining protein |
| pi326 | 1438 | prophage pi3 protein 26, terminase large subunit |
| pi327 | 1439 | prophage pi3 protein 27, terminase small subunit |
| pi328 | 1440 | prophage pi3 protein 28 |
| pi329 | 1440 | prophage pi3 protein 29 |
| pi330 | 1441 | prophage pi3 protein 30 |
| pi331 | 1441 | prophage pi3 protein 31 |
| pi332 | 1442 | prophage pi3 protein 32 |
| pi333 | 1443 | prophage pi3 protein 33 |
| pi334 | 1443 | prophage pi3 protein 34 |
| pi335 | 1443 | prophage pi3 protein 35, deoxyuridine 5'-triphosphate nucleotidohydrolase |
| pi336 | 1444 | prophage pi3 protein 36 |
| pi337 | 1444 | prophage pi3 protein 37 |
| pi338 | 1445 | prophage pi3 protein 38 |
| pi339 | 1445 | prophage pi3 protein 39 |
| pi340 | 1446 | prophage pi3 protein 40 |
| pi341 | 1446 | prophage pi3 protein 41 |
| pi342 | 1446 | prophage pi3 protein 42 |
| pi343 | 1447 | prophage pi3 protein 43 |
| pi344 | 1447 | prophage pi3 protein 44 |
| pi345 | 1447 | prophage pi3 protein 45 |
| pi346 | 1448 | prophage pi3 protein 46, DNA replication protein |
| pi347 | 1449 | prophage pi3 protein 47, replisome organiser |
| pi348 | 1450 | prophage pi3 protein 48, single strand binding helix destabilising protein |
| pi349 | 1450 | prophage pi3 protein 49 |
| pi350 | 1451 | prophage pi3 protein 50 |
| pi351 | 1451 | prophage pi3 protein 51 |
| pi352 | 1452 | prophage pi3 protein 52 |
| pi353 | 1452 | prophage pi3 protein 53 |
| pi354 | 1452 | prophage pi3 protein 54 |
| pi355 | 1453 | prophage pi3 protein 55, antirepressor |
| pi356 | 1453 | prophage pi3 protein 56, cro-like repressor |
| pi357 | 1454 | prophage pi3 protein 57, cI-like repressor |
| pi358 | 1455 | prophage pi3 protein 58 |
| pi359 | 1455 | prophage pi3 protein 59 |
| pi360 | 1456 | prophage pi3 protein 60, integrase |
| pip | 1720 | phage infection protein |
| ps101 | 36 | prophage ps1 protein 01, hypothetical regulator |
| ps102 | 36 | prophage ps1 protein 02 |
| ps103 | 37 | prophage ps1 protein 03, terminase subunit |
| ps104 | 37 | prophage ps1 protein 04 |
| ps105 | 38 | prophage ps1 protein 05, DNA primase |
| ps106 | 40 | prophage ps1 protein 06 |
| ps107 | 40 | prophage ps1 protein 07 |
| ps108 | 41 | prophage ps1 protein 08 |
| ps109 | 41 | prophage ps1 protein 09 |
| ps110 | 41 | prophage ps1 protein 10 |
| ps111 | 42 | prophage ps1 protein 11, transcriptional regulator |
| ps112 | 42 | prophage ps1 protein 12 |
| ps113 | 42 | prophage ps1 protein 13 |
| ps114 | 43 | prophage ps1 protein 14 |
| ps115 | 44 | prophage ps1 protein 15, transcriptional regulator |
| ps116 | 44 | prophage ps1 protein 16 |
| ps117 | 45 | prophage ps1 protein 17 |
| ps118 | 45 | prophage ps1 protein 18 |
| ps119 | 45 | prophage ps1 protein 19 |
| ps120 | 46 | prophage ps1 protein 20 |
| ps121 | 47 | prophage ps1 protein 21 |
| ps122 | 48 | prophage ps1 protein 22 |
| ps123 | 49 | prophage ps1 protein 23, integrase |
| ps201 | 503 | prophage ps2 protein 01, integrase |
| ps202 | 504 | prophage ps2 protein 02 |
| ps203 | 505 | prophage ps2 protein 03 |
| ps204 | 505 | prophage ps2 protein 04 |
| ps205 | 506 | prophage ps2 protein 05, transcriptional repressor |
| ps206 | 506 | prophage ps2 protein 06 |
| ps207 | 506 | prophage ps2 protein 07, excisionase |
| ps208 | 507 | prophage ps2 protein 08 |
| ps209 | 507 | prophage ps2 protein 09 |
| ps210 | 507 | prophage ps2 protein 10 |
| ps211 | 508 | prophage ps2 protein 11 |
| ps212 | 508 | prophage ps2 protein 12 |
| ps213 | 508 | prophage ps2 protein 13 |
| ps214 | 509 | prophage ps2 protein 14 |
| ps215 | 510 | prophage ps2 protein 15 |
| ps216 | 511 | prophage ps2 protein 16 |
| ps218 | 512 | prophage ps2 protein 18 |
| ps219 | 512 | prophage ps2 protein 19 |
| ps220 | 513 | prophage ps2 protein 20 |
| ps221 | 514 | prophage ps2 protein 21 |
| ps301 | 2014 | prophage ps3 protein 01 |
| ps302 | 2015 | prophage ps3 protein 02 |
| ps303 | 2015 | prophage ps3 protein 03 |
| ps304 | 2016 | prophage ps3 protein 04 |
| ps305 | 2016 | prophage ps3 protein 05 |
| ps306 | 2018 | prophage ps3 protein 06 |
| ps307 | 2019 | prophage ps3 protein 07 |
| ps308 | 2019 | prophage ps3 protein 08 |
| ps309 | 2019 | prophage ps3 protein 09 |
| ps310 | 2020 | prophage ps3 protein 10 |
| ps311 | 2021 | prophage ps3 protein 11 |
| ps312 | 2022 | prophage ps3 protein 12 |
| ps313 | 2023 | prophage ps3 protein 13 |
| ps314 | 2023 | prophage ps3 protein 14, transcriptional regulator |
| ps315 | 2024 | prophage ps3 protein 15 |
| ps316 | 2025 | prophage ps3 protein 16, integrase |
| Transposon related functions | ||
| tra1077A | 53 | transposase of IS1077A |
| tra1077B | 140 | transposase of IS1077B |
| tra1077C | 375 | transposase of IS1077C |
| tra1077D | 628 | transposase of IS1077D |
| tra1077E | 838 | transposase of IS1077E |
| tra1077F | 2156 | transposase of IS1077F |
| tra1077G | 2217 | transposase of IS1077G |
| tra904A | 54 | transposase of IS904A |
| tra904B | 138 | transposase of IS904B |
| tra904C | 40 | transposase of IS904C |
| tra904D | 374 | transposase of IS904D |
| tra904E | 627 | transposase of IS904E |
| tra904F | 836 | transposase of IS904F |
| tra904G | 839 | transposase of IS904G |
| tra904H | 2155 | transposase of IS904H |
| tra904I | 2215 | transposase of IS904I |
| tra905 | 1225 | transposase of IS905 |
| tra981A | 92 | transposase of IS981A |
| tra981B | 93 | transposase of IS981B |
| tra981C | 651 | transposase of IS981C |
| tra981D | 729 | transposase of IS981D |
| tra981E | 1217 | transposase of IS981E |
| tra981F | 1222 | transposase of IS981F |
| tra981G | 1276 | transposase of IS981G |
| tra981H | 1586 | transposase of IS981H |
| tra981I | 1748 | transposase of IS981I |
| tra981J | 2103 | transposase of IS981J |
| tra982 | 640 | transposase of IS982 |
| tra983A | 682 | transposase of IS983A |
| tra983B | 707 | transposase of IS983B |
| tra983C | 958 | transposase of IS983C |
| tra983D | 1338 | transposase of IS983D |
| tra983E | 1396 | transposase of IS983E |
| tra983F | 1556 | transposase of IS983F |
| tra983G | 1755 | transposase of IS983G |
| tra983H | 1954 | transposase of IS983H |
| tra983I | 1978 | transposase of IS983I |
| tra983J | 2012 | transposase of IS983J |
| tra983K | 2017 | transposase of IS983K |
| tra983L | 2084 | transposase of IS983L |
| tra983M | 2148 | transposase of IS983M |
| tra983N | 2203 | transposase of IS983N |
| tra983O | 2268 | transposase of IS983O |
| yafG | 53 | hypothetical protein |
| yafI | 55 | hypothetical protein |
| yajE | 92 | transposase |
| yajG | 94 | transposase |
| ybdK | 138 | hypothetical protein |
| ybdL | 139 | hypothetical protein |
| ybeG | 141 | hypothetical protein |
| ydhD | 373 | hypothetical protein |
| ydhE | 375 | hypothetical protein |
| yfjB | 593 | transposon-related protein |
| ygcD | 628 | hypothetical protein |
| ygcE | 629 | hypothetical protein |
| ygfF | 651 | transposase |
| yhcJ | 729 | transposase |
| yidF | 837 | hypothetical protein |
| yidG | 838 | hypothetical protein |
| yidH | 839 | hypothetical protein |
| ymbA | 1212 | integrase |
| ymbI | 1217 | transposase |
| ymcD | 1222 | transposase |
| ymfD | 1259 | integrase-recombinase |
| ymhB | 1276 | transposase |
| ypiI | 1587 | transposase |
| yrdA | 1748 | transposase |
| yuiI | 2104 | transposase |
| yvfC | 2157 | hypothetical protein |
| yvfD | 2156 | hypothetical protein |
| yvjF | 2216 | hypothetical protein |
| ywbC | 2217 | hypothetical protein |
| Other | ||
| crtK | 574 | carotenoid biosynthetic protein CrtK |
| yebB | 412 | carotenoid biosynthetic protein |
#Gene symbols, coordinates in kb, and definitions are shown. Bold italic symbols correspond to gene functions experimentally confirmed in L. lactis.
Origin and Terminus of Replication
Approximate position of the replication origin and terminus of the L. lactis chromosome was determined previously, using the GC and AT skews (Fig. 1; Bolotin et al. 1999). It should be noted that the precision of the origin mapping is greater than that of the terminus, as there are conserved elements (dnaA and dnaN genes, DnaA boxes) in the vicinity of the former but not of the latter (rtp gene was not found). We choose as the coordinate 1 of the genome the middle of a HindIII site localized near the replication origin (Fig 1).
Figure 1.
Distribution of IS elements and prophages in the IL1403 chromosome. Outer circle shows the scale in basepairs. IS981, IS983, and IS1077 are shown by yellow, red, and blue squares, respectively (enlarged for clarity). Red and blue arcs show the areas of IS983 and IS1077 insertions. pi prophages are shown in violet, and ps in black. Green circle shows GC skews (C−G/C+G) distribution (window 20 kb, step 5 kb), which indicates the origin (near bp 1) and terminus (near bp 1,260,000) of replication.
RNA, IS Elements, and Prophages
Location of six rRNA operons, 62 tRNA genes, the RNA component of RNAse P gene (rnpB), and the 10S RNA (ssrA) were determined earlier from the diagnostic sequence (Bolotin et al. 1999). There are six different IS elements in the IL1403 chromosome: IS981, IS982, IS983, IS904, IS905, and IS1077, present in 10, 1, 15, 9, 1, and 7 copies, respectively (Fig. 1) and totaling 42 kb. It is remarkable that one or two copies of IS904 always accompany IS1077 and that the relative orientation of the two is generally not the same. The former element might be a satellite of the latter. Another remarkable feature is that three of the IS elements are not randomly distributed over the chromosome (Fig. 1). Seven copies of IS1077 (and the associated IS904) occupy the region between 2150 and 840 kb, encompassing the replication origin, whereas 15 copies of IS983 occupy a different region, between 680 and 2270 kb. The two regions overlap by only ∼150 kb. As the 10 copies of IS981 are distributed over the whole genome, the uneven distribution of three other IS elements is not caused by a particular property of the L. lactis cell. We suggest that this distribution indicates a lateral transfer of a large portion of the genome from a lactococcus donor, carrying one type of IS, to a recipient, carrying the other type. Two lines of evidence lend support to this hypothesis. First, IS1076, which corresponds to the association of IS1077 and IS904 described above, is distributed over the whole genome of the strain L. lactis ssp. cremoris MG1363 (Le Bourgeois et al. 1995) rather than being restricted to one region of the genome, as is the case in IL1403. This transposon has, therefore, no particular hot region for insertion in the lactococcal genome. Second, the restriction map of another strain, L. lactis ssp. lactis DL11, coincides with that of IL1403 in the area between rrnF (550 kb) and rrnE (1980 kb), while it is divergent elsewhere (Le Bourgeois et al. 1992). We suggest that DL11 may be close to one of the putative parental strains of IL1403. Investigations of the distribution of IS1077 and IS983 among different lactococci might allow identification of both putative parents of the IL1403 strain.
Three potential prophages, designated pi1, pi2, and pi3, were detected at positions near 460, 1050, and 1460 kb (Fig. 1). They are large (35–44 kb), encode 49–60 proteins, and are related to known temperate phages of L. lactis. Another three prophages, designated ps1, ps2, and ps3, are localized near 42, 509, and 2020 kb (Fig. 1). They are small (11–15 kb), encode only 16–23 proteins and might be satellites of the other phages, as they lack most of the genes that code for phage structural elements. A copy of IS983 is present in ps3, which might, thus, be defective. The six prophages comprise a total of 175 kb of DNA and 221 protein coding genes. Recently, Chopin et al. (2001) characterized five phages, which can be found in the supernatant of IL1403 after mitomycin C treatment, and demonstrated the correspondence between the phage DNA extracted from the supernatant and the chromosome sequence. Phage bIL285 from the supernatant corresponds to pi2, bIL286 to pi3, bIL309 to pi1, bIL310 to ps1, and bIL312 to ps2. ps3, designated also as bIL311 (Chopin et al. 2001), cannot be induced, probably because of the IS983 element present in its genome. Detecting the circular forms of DNA of these phages allowed precise determination of the integration sites. About 9.2% of the L. lactis genome is thus formed by IS elements and prophages, suggesting that they may be important for horizontal gene transfer in these bacteria.
Paralogous Gene Families
We define here as a paralogous protein family a group of proteins within which each protein shares at least one homologous domain with another protein of the group. By this criterion, there are 370 paralogous families, comprising 1189 gene products, in the L. lactis genome. Among the smaller families (<10 members) there are 208 of two members, 80 of three, 36 of four, 13 of five, 13 of six, 8 of seven, 4 of eight, and 2 of nine. The larger families contain 10, 11, 15, 18, 26, and 60 members, the last corresponding to ATP-binding proteins of ABC transporters, as is the case in many bacteria. In the four smallest families, distribution of the number of proteins resembles that of B. subtilis (Kunst et al. 1997). It decreases, very approximately, twofold when the family member count increases by one (568:273:168:100 in B. subtilis and 416:240:144:65 in L. lactis for doublets, triplets, quadruplets, and quintuplets, respectively).
Information Processing and Gene Regulation
Information processing refers to the genes constituting replication, transcription, and translation machinery. In L. lactis, it is overall very similar to that of B. subtilis, the best characterized AT-rich gram-positive bacterium (Kunst et al. 1997). There are 67 genes involved in DNA metabolism in L. lactis. All the genes involved in DNA replication in B. subtilis are present in L. lactis, including counterparts of dnaB, dnaD, and dnaI, genes essential for initiation of replication in B. subtilis and absent in gram-negative bacteria. Two DNA-polymerase III α-chain genes, one corresponding to polC and another to dnaE of B. subtilis, were also detected in L. lactis. In contrast, E. coli has only the dnaE gene.
Transcription machinery in both L. lactis and B. subtilis comprises some 30 genes other than the ς-factors. However, the number of ς-factors differs greatly, as there are only three in L. lactis, while there are 18 in B. subtilis, pointing to a considerable difference in the mode of gene-expression regulation in the two organisms. Translation machinery comprises 119 genes in L. lactis and 131 genes in B. subtilis. There are no duplicated aminoacyl-tRNA synthetase genes in L. lactis, while there are three (for threonine, tyrosine, and histidine) in B. subtilis. Posttranslational protein modification genes mostly differ, as there are 27 such genes in B. subtilis and only 10 in L. lactis. A particular regulation of translation might also operate in L. lactis. As discussed more fully below, all the late competence genes of L. lactis seem to be controlled by a mechanism relaying on leaderless mRNAs and, thus, on a particular mode of translation. Recent evidence shows that the involvement of translation initiation factor 3, present in all bacteria, in start codon recognition is important for restriction of translation in such systems (Tedin et al. 1999). This provides a link between regulation of translation and competence in L. lactis. Such interaction has not been detected previously.
Analysis of homology allowed us to assign regulatory functions to 138 genes, half of which were classified further by their similarity to regulatory proteins of known families. The overall number of regulatory systems is about twofold lower in L. lactis than in B. subtilis, but the proportion of these genes is similar in the two organisms. Among the interesting differences is a much lower number of the two-component signal transducers in L. lactis than in B. subtilis (eight instead of 34) and of ς-factors (three instead of 18), both of which regulate complex responses to changing environmental conditions.
Energy Metabolism and Transporters
The most important industrial applications of L. lactis are based on its energy metabolism, which leads mainly to the production of high amounts of lactic acid (homolactic fermentation). Anaerobic glycolysis is the principal energy-generating process in L. lactis, and very little of the fermented sugar (∼5%) is used for synthetic reactions (Poolman 1993). All the genes required for the conversion of the glucose to pyruvate are present in the genome. The pyruvate is converted into lactic acid, thus allowing the oxidation of reduced NAD, and the lactate dehydrogenase gene ldh, essential for this process, was studied intensely (Griffin et al. 1992). Three other genes, highly similar to ldh, (ldhB, ldhX and hicD) are present in the genome, but their role is not known. The product of the last gene has a high similarity (42% identity) to hydroxyisocaproate dehydrogenase and may, therefore, be involved in the catabolism of branched-chain amino acids. Lactate is transported into the growth medium, causing the efflux of protons and, thus, providing transmembrane potential indispensable for growth and energy recycling (Ten Brink et al. 1985).
Genome analysis indicates that the full citric acid cycle, gluconeogenesis enzymes, and many anaplerotic reactions do not exist in L. lactis. Unexpectedly, the functions necessary for aerobic respiration are encoded in the genome. L. lactis has men and cytABCD operons, encoding proteins required for menaquinone synthesis and cytochrome d biogenesis. It also has three genes involved in the late steps of heme synthesis (hemH, hemK, and hemN, required for oxidation of porphyrinogen and attachment of iron to heme) but not the genes required for the early steps. L. lactis may thus be able to carry out oxidative phosphorylation if the protoporphyrinogen is provided. Indeed, improved growth properties in media containing hemin were observed for certain Streptococci (Sijpesteijn 1970; Mickelson 1972). The genome analysis thus suggests the existence of aerobic respiration in this bacterium, generally considered an exclusively fermentative microorganism.
Use of L. lactis in the food industry also exploits its ability to form fermentation products other than lactate (mixed acid fermentation). The balance of products depends on activities of enzymes that act on the key metabolite generated by glycolysis, the pyruvate. A number of genes encoding such enzymes (pyruvate dehydrogenase, pdhABCD; α-acetolactate synthase, als; pyruvate-formate lyase, pfl; and lactate dehydrogenase, ldh) have been identified previously in L. lactis and confirmed by genome analysis. We detected a novel gene, poxL, encoding pyruvate oxidase, which also acts on pyruvate and might, therefore, play a role in switching between different fermentation modes.
Besides gene activity, the availability of cofactors, such as NADH and FAD, also affects the balance of different fermentation products. Artificial changing of NADH/NAD ratio in L. lactis can redirect carbon flow from lactic acid to acetoin and diacetyl (Lopez de Felipe et al. 1998). There are more than five NADH dehydrogenase genes in the L. lactis genome, which may affect the type of fermentation products. Some NADH dehydrogenases generate hydrogen peroxide, which is toxic for the cells. L. lactis has no gene encoding catalase, which can remove the toxic H2O2. However, there is a gene encoding thiol peroxidase (tpx) and two genes (ahpC and ahpF) encoding alkyl hyperoxide reductases. These proteins could possibly act on H2O2. Active sodA, encoding superoxid dismutase, which converts oxygen radicals to H2O2, was shown to be important for the oxidative stress response (Sanders et al. 1995). Also, the gshR gene encoding glutathion reductase may be involved in response of L. lactis to the aerobic growth conditions.
The heterofermentative metabolism takes place in L. lactis when pentoso-phosphate pathway is active, as in this case, glycolysis generates not only a three-carbon compound that can be converted to lactate but also a two-carbon compound. We detected glucose-6P dehydrogenase (zwf), phosphogluconate dehydrogenase (gnd), and ribuloso-5P epimerase (rpe), which can lead to the formation of xyluloso-5P. Phosphoketolase, encoded by ptk gene, can catalyze formation of glyceraldehyde-3P and acetyl-P, which enter the fermentation pathways that yield lactate and ethanol, respectively.
Understanding the molecular basis of the switch between different fermentation types is of interest not only for standard uses of L. lactis but also for the metabolic engineering in this organism, aiming to enhance synthesis of certain metabolites to industrially useful levels. We detected a correlation between the presence of the phosphoenolpyruvate dependent transport system (PTS) and the fermentation profile for a given carbon source. PTS systems for fructose, mannose, sucrose or trehalose, mannitol, and cellobiose are present in the genome, and the homolactic fermentation profiles were reported for growth on fructose, mannose, glucose (which uses mannose or mannitol PTS) and sucrose (Cocaign-Bousquet et al. 1996). In contrast, mixed acid or heterofermentation profiles were observed for growth on galactose, xylose, maltose, gluconate, ribose, and lactose, which are not imported by a PTS system. When L. lactis cells harbor a plasmid encoding lactose-specific PTS system, lactose fermentation becomes homolactic (Gasson 1983). Our genome analysis thus strengthens the proposal that sugar consumption rate, which is the highest when PTS system is available, determines the ability for efficient homolactic fermentation (Cocaign-Bousquet et al. 1996). The correlation of information derived from genome analysis with experimental data on fermentation product distribution indicates that critical parameters regulating the final product balance may be found by a thorough analysis of the carbon source use and transport systems.
Proteases and Amino Acid Catabolism Genes
Proteases and peptidases provide a selective advantage for bacteria growing in milk, as this medium is rich in caseins and relatively poor in free amino acids. Amino acid catabolism has an impact on fermentation regulation and on the flavor of dairy products.
Genome sequence revealed 19 protease-encoding genes (Table 1). These include the membrane protease HtrA, which is responsible for degradation of the precursors of foreign exported proteins (Pouquet et al. 2000). Some 16 peptidases from LAB were characterized previously, including the products of 13 genes detected in L. lactis (Christensen et al. 1999).
Catabolism of amino acids usually starts by deamination. Arginine catabolic genes, organized in an operon near 2110 kb, encode the enzymes for the deaminase pathway as well as the arginine tRNA synthetase, suggesting complex regulation. Another operon for arginine catabolism, near 1755 kb, contains genes arcC3 and otcA. It could have a regulatory function, as it also contains the genes llrH and yrfE, representing a signal transduction system of a new type. Aspartate aminotransferase (aspC) and asparaginase (ansB) are involved in aspartate and asparagine catabolism. No genes for aspartate decarboxylase or aspartase were detected, although such enzymatic activities were identified in Lactobacillus, another prominent group of LAB (Rollan et al. 1985). Recent studies on catabolism and biosynthesis of glutamate in L. lactis identified the existence of a pathway leading to the production of γ-aminobutyrate (GABA; Sanders et al. 1998). We identified gadRCB operon for GABA production, gltBD genes for glutamate synthase, and an operon involved in citric acid metabolism: pycA, gltA, citB, and icd. Under appropriate physiological conditions, products of some of these genes might carry out glutamate catabolism, rather than biosynthesis. Serine can be directly converted to pyruvate by serine dehydratase encoded by the sdaAB operon.
Genome sequence provides inventory of 12 aminotransferases, of which some can initiate degradation of aromatic, branched-chain, and sulfur-containing amino acids, important for cheese flavor. The specificity of seven aminotransferases (aspC, serC, argD, glmS, hisC, aspB, and arcT) can be predicted from sequence comparisons, whereas those of other five (araT, nifZ, yeiG, bcaT, and ytjE) are less obvious. It was recently shown that araT and bcaT are involved in the degradation of aromatic and branched-chain amino acids, respectively (Yvon et al. 2000). The product of ytjE might be specific for methionine, as the gene is cotranscribed with the relevant biosynthesis genes. Degradation of tryptophane seems to proceed via indole aldehyde because of indole pyruvate decarboxylase gene ipd. It is not clear which pathways L. lactis uses to catabolize phenylalanine and tyrosine. It is possible that phenyl pyruvate and p-OH-phenyl pyruvate are degraded further by decarboxylation. This would depend on the specificity of the phenolic acid decarboxylase encoded by pdc.
Amino Acid, Vitamin, and Nucleotide Biosynthesis
L. lactis requires certain metabolites in the growth medium, although it has a genetic potential to synthesize some of them. Synthetic medium for L. lactis should contain at least six amino acids (isoleucine, valine, leucine, histidine, methionine, and glutamic acid) and seven vitamins (biotin, pyridoxal, folic acid, riboflavin, nicotinamide, thiamine, and pantothenic acid; Jensen and Hammer 1993). L. lactis has the genes to synthesize the 20 standard amino acids and at least four cofactors (folic acid, menaquinone, riboflavin, and thioredoxin). One reason for the requirement of the compounds that can potentially be synthesized is that some of the existing genes are not functional, as was reported previously for amino acid biosynthesis genes (Godon et al. 1993). We carefully checked sequencing tracks for the genes that could contain a frameshift mutation and could not rule out the presence of a mutation in 30 of them. This relatively high level of pseudogenes in IL1403 could possibly be, at least in part, caused by the treatments used to cure the parental strain of its plasmids (Chopin et al. 1984).
Milk does not contain sufficient levels of purine compounds to support growth of L. lactis and, therefore, de novo biosynthesis is necessary (Dickely et al. 1995). We detected 57 genes involved in this metabolism. Therefore, physiological and genomic evidence shows that L. lactis has sufficient and fairly active capacities for biosynthesis and also for salvage of nucleic acid compounds.
Cell Wall Metabolism
Many L. lactis properties that are important for applications, such as phage sensitivity, stress resistance, autolysis, and mucosal immunostimulation, depend on the structure of the cell wall. There are 29 genes encoding enzymes required for the synthesis of the main cell wall component, peptidoglycan. Among these, three encode amino acid racemases: dal for alanine, murI for glutamate, and racD for aspartate. D-alanine and D-glutamate are the components of linear peptide moiety of peptidoglycan, whereas D-aspartate forms cross-bridges. There are no genes for synthesis of modified peptidoglycan, containing D-lactate or D-serine instead of D-alanine, reported for several other LAB.
Cheese ripening can be accelerated by induction of enzymes that process peptidoglycan. There are six genes related to such processing in L. lactis: dacA and dacB, encoding alanine–alanine carboxypeptidase; and acmA, B, C, and D, encoding four lysozymes. Carboxypeptidases alone cannot cause the cell lysis, as their activity does not destabilize the wall. Modulation of the level of their production can, however, influence the action of lysozymes. acmA, responsible for separation of daughter cells, was used for artificial induction of autolysis (Buist et al. 1997).
Lipoteichoic acid is another main component of the L. lactis cell wall. Neither teichoic nor teichuronic acids were detected in this microorganism (Valyasevi et al. 1990). However, there is a cluster of seven tag genes near 950 kb. Only three genes from teichuronic acid biosynthesis pathway were found: ycbK, ycbF, and ycbH, corresponding to tuaB, tuaC, and tuaG of B. subtilis. dlt operon, encoding D-alanylation of lipoteichoic acid, is of crucial importance for properties of the cell wall and whole-cell physiology. A knockout mutation in dltD causes filamentous growth and UV sensitivity and facilitates penetrability of the cells (Duwat et al. 1997).
Synthesis of extracellular polysaccharides is important for the industrial use of many LAB, as these polymers affect the texture of the fermented products. There are >20 genes involved in the biosynthesis of such molecules in the region near 200 kb. They encode functions providing activated sugars and other components involved in production of surface or extracellular polysacharide. A plasmid that carries an operon involved in the formation of the repeating unit, linking activated sugar to the lipid carrier, export, and polymerization, was recently identified (Van Kranenburg et al. 1997). Conjunction of plasmid-carried and chromosomal functions presumably determine the amounts and the structure of extracellular polysaccharides.
Protein Secretion
L. lactis has only eight genes identified as implicated in protein secretion. Contrary to B. subtilis and E. coli, this bacterium does not have secDF genes, known to improve the secretion efficiency (Pogliano and Beckwith 1994; Bolhuis et al. 1998). There is only one membrane protease, HtrA, involved in degradation of hybrid exported proteins (Pouquet et al. 2000). Gene pmpA (protein maturation protein) encodes a homolog of PrsA from B. subtilis and might be involved in stabilization of secreted proteins by facilitating their folding. L. lactis was shown to secrete up to 20 mg/L of foreign protein with optimized gene constructs (Le Loir et al. 1998). This value could possibly be improved by manipulating the gene expression levels and supplying the missing components of the secretion machinery.
Competence to Genetic Transformation
Natural competence to DNA transformation was not demonstrated in L. lactis. We detected four operons (comE, comF, comC, and comG) containing genes similar to the late competence genes from B. subtilis and S. pneumoniae. In addition, we found a gene for ComX, which is similar to the S. pneumoniae ECF-type ς-factor required for transcription of the competence genes (Lee and Morrison 1999). The regions preceding the first ORF of the four operons resemble competence promoters from S. pneumoniae and might be transcribed by ComX. There are three common sequences in front of all competence operons, two of which, GTTACATT and TTTTCGTATA, are in the −35 and −10 domains of the promoter, while the third, AGTATG, includes the ATG start codon of the first gene in each operon. The relative position of the three conserved elements indicates that all mRNAs start at the ATG codon of the first gene and are, therefore, leaderless, lacking the canonical ribosome-binding site. Search for the consensus sequence over the whole genome, using PatScan (Dsouza et al. 1997), revealed six such promoters other than those of the late competence operons. The genes downstream of these promoters are radA, coiA, dprA, recQ, ssbA, and yqfG. Only the radA gene, encoding a DNA repair protein, has leaderless mRNA. Three of the genes, coiA, dprA, and recQ, affect DNA transformation in S. pneumoniae, H. influenzae, and B. subtilis, respectively (Karudapuram et al. 1995; Fernandes et al. 1998; Pestova and Morrison 1998). ssbA encodes single-strand DNA-binding protein and could be involved in the processing of transforming DNA, which enter gram-positive bacteria in the single-stranded form. yqfG encodes a protein of unknown function. The existence of the competence-related genes in L. lactis indicates that this bacterium might be naturally transformable by DNA. There are no genes homologous to those involved in early steps of competence development in S. pneumoniae, which indicates that, in L. lactis, the regulation cascade upstream of ComX ς-factor is very different from that in Streptococcae.
Another difference between L. lactis and S. pneumoniae competence systems is that the leaderless mRNAs are present in the former organism only. The translation of such mRNAs requires that they start precisely at the initiation codon of the gene (Kravchenko et al. 1988; Van Etten and Janssen 1998). Synthesis of competence-related proteins would, therefore, not take place on spurious transcription of the cognate genes by leakage from upstream operons. This might tighten the control of the competence development and does limit it to very strict environmental conditions.
Horizontal Gene Transfer between Lactococci and Gram-Negative Enteric Bacteria
We detected a gene of unknown function, designated ycdB, which appears to be present in all bacteria and some eukaryotes. The level of identity between the YcdB protein and a homolog from S. pyogenes or S. pneumococcus, phylogenetically close to L. lactis, is ∼80%, while the identity with the homologous genes from gram-negative bacteria is ∼40%. Very surprisingly, the E. coli and S. typhimurium genomes encode not only a protein that is 40% identical with YcdB but also a protein that is 94% identical to YcdB. We conclude that this second ycdB gene has been transferred from lactococci to enteric bacteria. The divergence of the synonymous nucleotide sites in L. lactis IL1403, compared with Salmonella and E. coli, is ∼10%. If the rate of nucleotide changes at such sites is ∼1% per million years (Ochman et al. 1999), the genes in Salmonella/E. coli and L. lactis IL1403 started to diverge 10 million years ago. However, comparison of the ycdB genes in different strains of lactococci and in gram-negative enteric bacteria may reveal even more closely related genes and allow us to better assess the time of the gene transfer, the species that may have been involved in the transfer, and the mechanism of the transfer. Nevertheless, anticipating that closer homologs will be found, it is tempting to speculate that the transfer may have taken place in the digestive tract of ruminants, if it involved wild-type lactococci, or of humans, if it involved the domesticated lactococci, massively introduced there by cheese consumption.
Analysis of completely sequenced genomes, available from the NCBI server, revealed that most bacteria have only one homolog to YcdB. Some (E. coli, S. typhimurium, B. subtilis, E. faecalis, and Shewanella putrefaciens), however, have two, indicating that the family might be undergoing an expansion where, at least for enteric bacteria, a lateral gene transfer from lactococci might be a driving force. As the function of this gene is unknown, the advantage that the second copy confers is not known. Elucidation of the gene function would help to answer this question.
METHODS
Genome Cloning, Sequencing, and Data Verification
The strain IL1403 is a plasmid-free derivative of the strain IL594, isolated from a cheese starter culture (Chopin et al. 1984). Diagnostic sequencing, involving 10,235 sequencing reactions and yielding a total of 4,687,630 bases, has been described previously (Bolotin et al. 1999). Further sequencing was carried out to assure us that each nucleotide in the genome was read at least four times and at least once on each strand. For this purpose, a collection of short insert clones was constructed. A total of 9,888,620 bases, covering 93% of the total genome, were produced by 15,578 more sequencing reactions. To reduce the error rate level to <0.01%, 978 more reactions, with average read length of 632 bases, were carried out using genome-specific primers. The redundancy of the final assembling is 6.44.
Informatics and Gene Nomenclature
Assembling manual corrections of sequencing errors and consensus generation were carried out concurrently with data accumulation, using the XBAP program (Dear and Staden 1991; version 14.0). To predict protein-coding regions, we used a conceptual translation of the whole genome in six possible coding frames. The predicted proteins >60 amino acids were checked for the statistical consistency with the output of the GENMARK program (Borodovsky and McIninch 1993) using parameters for Streptococcal genes. EBI server (http://www2.ebi.ac.uk/genemark) and pyogenes_3.xdr matrix dated November 14, 1996, were used for this analysis. The presence of a putative ribosome-binding site upstream of the 5′ end of the candidate was searched next. As a ribosome binding site, we considered the presence of initiator codon ATG, TTG, or GTG and a short sequence homologous to the 3′ end of 16S rRNA of L. lactis (5′…GGAUCACCUCCUUUCUAA 3′) upstream of it (Chiaruttini and Milet 1993). Genome annotation was done by using several homemade shell or Perl scripts, generating convenient html format tables linked to BLAST (Altschul et al. 1990) output files. NCBI server (http://www.ncbi.nlm.nih.gov/Entrez) was used to generate updated bacterial protein databases. Homology analysis of YcdB with the unpublished genome sequences was carried out by using the relevant NCBI server (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html). The functional classification of genes was done according to the list of categories presented earlier (Bolotin et al. 1999). Fully automatic computer-generated classification was used as the starting material. Each protein was then analyzed by an expert to improve the category assignment, which is presented in Table 1 and Figure 2. The expert usually used three means to confirm or to alter the automated function assignment and classification: first, phylogenetic or COGnitor (Tatusov et al. 1997) assisted scrutiny of BLAST or FASTA reslts (performed with different parameters); second, complete knowledge of particular biochemical pathways or biological systems, existing in other than L. lactis IL1403 organisms (such as protein secretion or the competence system). Phage-specific proteins were classified to those because of their clustering in the areas identified as prophages. Also, specialized databases (Quentin et al. 1999) were used by the expert to classify the ABC transporters; third, results of numerous experiments in L. Lactis, published previously (148 functional assignments). Although it is never absolutely explicit, the provided classification of gene functions in L. lactis IL1403 is biological, rather than biochemical.
Figure 2.
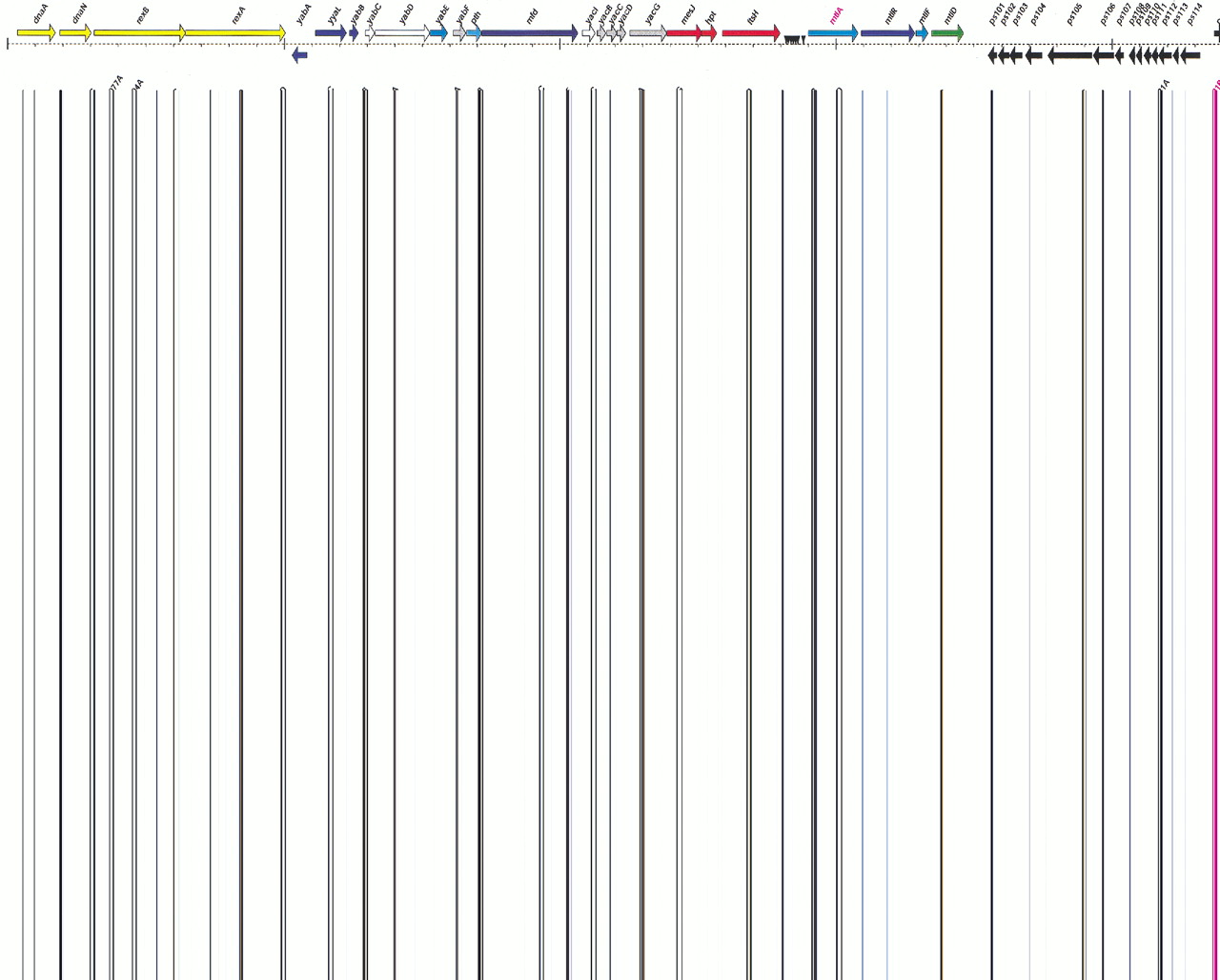
Linear map of the Lactococcus lactis ssp. lactis IL1403 chromosome. Coding regions are shown as arrows color-coded to the assigned functional categories. IS-elements and rRNA genes are shown as black arrows with white designation numbers inside. Symbols shown in pink identify genes in which frameshifts were detected.
L. lactis paralogous gene families were constructed by searching each predicted protein against all predicted proteins, using BLASTP with different parameters. Alignments of proteins in the identified families were then scrutinized to make a decision of how many proteins belong to a family. This decision was based either on the size of homologous domains or on the similarity levels. A protein was always assigned to only one family of paralogs.
We tried to keep the same gene symbols as proposed by the previous authors for ORFs with functions experimentally confirmed in L. lactis (148 genes). A y prefix with the gene symbol consistent with its position on the chromosome (Fig. 2) was kept for unascertained functions (1149 genes). Other gene symbols, consistent with those for homologs found in other bacteria, are proposed here (1017 genes).
Accessibility of Data
The nucleotide sequence of the L. lactis IL1403 genome is available from NCBI with accession no. AE005176. Updated annotations are supported at the Génétique Microbienne (INRA) server at http://spock.jouy.inra.fr. A PatScan of Ross Overbeek (Dsouza et al. 1997) for pattern searches in DNA sequence and proteins, implemented for IL1403, and peptide spectrum identification tool PeptOko for L. lactis proteome research are also available from this server.
Acknowledgments
We thank Jacek Bordovski and Saulius Kulakauskas for giving samples of the L. lactis IL1403 strain and Marie-Christine and Alain Chopin, Patrick Duwat, Emmanuel Jamet, Alexandra Gruss, Emmanuelle Maguin, Isabelle Poquet, Pierre Renault, and Catherine Robert for helpful discussions. We thank also the Genome Centers that contributed to the Unfinished Microbial Genome Database available for BLAST search through the NCBI server (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sorokine@biotec.jouy.inra.fr; FAX 33-1-34-65-25-21.
Article published on-line before print: Genome Res., 10.1101/gr.169701.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.169701.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bolhuis A, Broekhuizen CP, Sorokin A, van Roosmalen ML, Venema G, Bron S, Quax WJ, van Dijl JM. SecDF of Bacillus subtilis, a molecular Siamese twin required for the efficient secretion of proteins. J Biol Chem. 1998;273:21217–21224. doi: 10.1074/jbc.273.33.21217. [DOI] [PubMed] [Google Scholar]
- Bolotin A, Mauger S, Malarme K, Ehrlich SD, Sorokin A. Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome. Antonie Leeuwenhoek. 1999;76:27–76. [PubMed] [Google Scholar]
- Borodovsky M, McIninch J. GENMARK: A parallel gene recognition for both DNA strands. Comput Chem. 1993;17:123–133. [Google Scholar]
- Buist G, Karsens H, Nauta A, van Sinderen D, Venema G, Kok J. Autolysis of Lactococcus lactis caused by induced overproduction of its major autolysin, AcmA. Appl Environ Microbiol. 1997;63:2722–2728. doi: 10.1128/aem.63.7.2722-2728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini C, Milet M. Gene organization, primary structure and RNA processing analysis of a ribosomal RNA operon in Lactococcus lactis. J Mol Biol. 1993;230:57–76. doi: 10.1006/jmbi.1993.1126. [DOI] [PubMed] [Google Scholar]
- Chopin A, Chopin MC, Moillo-Batt A, Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984;11:260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- Chopin A, Bolotin A, Sorokin A, Ehrlich SD, Chopin MC. Analysis of six prophages in Lactococcus lactis IL1403: Different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 2001;29:644–651. doi: 10.1093/nar/29.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Dudley E, Pederson J, Steele J. Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Leeuwenhoek. 1999;76:217–246. [PubMed] [Google Scholar]
- Cocaign-Bousquet M, Garrigues C, Loubiere P, Lindley ND. Physiology of pyruvate metabolism in Lactococcus lactis. Antonie Leeuwenhoek. 1996;70:253–267. doi: 10.1007/BF00395936. [DOI] [PubMed] [Google Scholar]
- Cousineau B, Smith D, Lawrence-Cavanagh S, Mueller JE, Yang J, Mills D, Manias D, Dunny G, Lambowitz AM, Belfort M. Retrohoming of a bacterial group II intron: Mobility via complete reverse splicing, independent of homologous DNA recombination. Cell. 1998;94:451–462. doi: 10.1016/s0092-8674(00)81586-x. [DOI] [PubMed] [Google Scholar]
- Dear S, Staden R. A sequence assembly and editing program for efficient management of large projects. Nucl Acids Res. 1991;19:3907–3911. doi: 10.1093/nar/19.14.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickely F, Nilsson D, Hansen EB, Johansen E. Isolation of Lactococcus lactis nonsense suppressors and construction of a food-grade cloning vector. Mol Microbiol. 1995;15:839–847. doi: 10.1111/j.1365-2958.1995.tb02354.x. [DOI] [PubMed] [Google Scholar]
- Dsouza M, Larsen N, Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- Duwat P, Cochu A, Ehrlich SD, Gruss A. Characterization of Lactococcus lactis UV-sensitive mutants obtained by ISS1 transposition. J Bacteriol. 1997;179:4473–4479. doi: 10.1128/jb.179.14.4473-4479.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez S, Sorokin A, Alonso JC. Genetic recombination in Bacillus subtilis 168: Effects of recU and recS mutations on DNA repair and homologous recombination. J Bacteriol. 1998;180:3405–3409. doi: 10.1128/jb.180.13.3405-3409.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PF. Cheese: An overview. In: Fox PF, editor. Cheese: Chemistry, Physics and Microbiology. London: Chapman & Hall; 1989. pp. 1–36. [Google Scholar]
- Fraser CM, Fleischmann RD. Strategies for whole microbial genome sequencing and analysis. Electrophoresis. 1997;18:1207–1216. doi: 10.1002/elps.1150180803. [DOI] [PubMed] [Google Scholar]
- Gasson MJ. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon JJ, Delorme C, Bardowski J, Chopin MC, Ehrlich SD, Renault P. Gene inactivation in Lactococcus lactis: Branched-chain amino acid biosynthesis. J Bacteriol. 1993;175:4383–4390. doi: 10.1128/jb.175.14.4383-4390.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin HG, Swindell SR, Gasson MJ. Cloning and sequence analysis of the gene encoding L-lactate dehydrogenase from Lactococcus lactis: Evolutionary relationships between 21 different LDH enzymes. Gene. 1992;122:193–197. doi: 10.1016/0378-1119(92)90049-u. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Hammer K. Minimal requirements for exponential growth of Lactococcus lactis. Appl Environ Microbiol. 1993;59:4363–4366. doi: 10.1128/aem.59.12.4363-4366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karudapuram S, Zhao X, Barcak GJ. DNA sequence and characterization of Haemophilus influenzae dprA+, a gene required for chromosomal but not plasmid DNA transformation. J Bacteriol. 1995;177:3235–3240. doi: 10.1128/jb.177.11.3235-3240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchenko VV, Gileva IP, Dobrynin VN, Filipov SA, Korobko VG. Location of the initiation codon AUG in relation to the 5′-end of mRNA mediates the effectiveness of translation in E. coli cells. Bioorg Khim. 1988;14:1387–1392. [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Le Bourgeois P, Lautier M, Mata M, Ritzenthaler P. Physical and genetic map of the chromosome of Lactococcus lactis subsp. lactis IL1403. J Bacteriol. 1992;174:6752–6762. doi: 10.1128/jb.174.21.6752-6762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourgeois P, Lautier M, van den Berghe L, Gasson M, Ritzenthaler P. Physical and genetic map of the chromosome of Lactococcus lactis subsp. cremoris MG1363 chromosome: Comparison with that of Lactococcus lactis subsp. lactis IL1403 reveals a large genome inversion. J Bacteriol. 1995;177:2840–2850. doi: 10.1128/jb.177.10.2840-2850.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Morrison DA. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol. 1999;181:5004–5016. doi: 10.1128/jb.181.16.5004-5016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Loir Y, Gruss A, Ehrlich SD, Langella P. A nine-residue synthetic propeptide enhances secretion efficiency of heterologous proteins in Lactococcus lactis. J Bacteriol. 1998;180:1895–1903. doi: 10.1128/jb.180.7.1895-1903.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Felipe FL, Kleerebezem M, de Vos WM, Hugenholtz J. Cofactor engineering: A novel approach to metabolic engineering in Lactococcus lactis by controlled expression of NADH oxidase. J Bacteriol. 1998;180:3804–3808. doi: 10.1128/jb.180.15.3804-3808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelson MN. Glucose degradation, molar growth yields, and evidence for oxidative phosphorylation in Streptococcus agalactiae. J Bacteriol. 1972;109:96–105. doi: 10.1128/jb.109.1.96-105.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Elwyn S, Moran NA. Calibrating bacterial evolution. Proc Natl Acad Sci. 1999;96:12638–12643. doi: 10.1073/pnas.96.22.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova EV, Morrison DA. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J Bacteriol. 1998;180:2701–2710. doi: 10.1128/jb.180.10.2701-2710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano JA, Beckwith J. SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 1994;13:554–561. doi: 10.1002/j.1460-2075.1994.tb06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman B. Energy transduction in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:125–147. doi: 10.1111/j.1574-6976.1993.tb00015.x. [DOI] [PubMed] [Google Scholar]
- Pouquet I, Saint V, Seznec E, Simoes N, Bolotin A, Gruss A. HtrA in the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol Microbiol. 2000;35:1042–1051. doi: 10.1046/j.1365-2958.2000.01757.x. [DOI] [PubMed] [Google Scholar]
- Quentin Y, Fichant G, Denizot F. Inventory, assembly and analysis of Bacillus subtilus ABC transport systems. J Mol Biol. 1999;287:467–484. doi: 10.1006/jmbi.1999.2624. [DOI] [PubMed] [Google Scholar]
- Rollan G, de Nadra MCM, Holgado PR, Oliver G. Aspartate metabolism in Lactobacillus murinus CNRS 313. I. Aspartase. J Gen Appl Microbiol. 1985;31:403–409. [Google Scholar]
- Sanders JW, Leenhouts KJ, Haandrikman AJ, Venema G, Kok J. Stress response in Lactococcus lactis: Cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J Bacteriol. 1995;177:5254–5260. doi: 10.1128/jb.177.18.5254-5260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JW, Leenhouts K, Burghoorn J, Brands JR, Venema G, Kok J. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol. 1998;27:299–310. doi: 10.1046/j.1365-2958.1998.00676.x. [DOI] [PubMed] [Google Scholar]
- Schleifer KH, Kraus J, Dvorak C, Kilpper-Bälz R, Collins MD, Fischer W. Transfer of Streptococcus lactis and related streptococci to the genus Lactococcus gen. nov. Syst Appl Microbiol. 1985;6:183–195. [Google Scholar]
- Sijpesteijn AK. Induction of cytochrome formation and stimulation of oxidative dissimilation by hemin in Streptococcus lactis and Leuconostoc mesenteroides. Antonie Leeuwenhoek. 1970;36:335–348. doi: 10.1007/BF02069035. [DOI] [PubMed] [Google Scholar]
- Sorokin A, Lapidus A, Capuano V, Galleron N, Pujic P, Ehrlich SD. A new approach using multiplex long accurate PCR and yeast artificial chromosomes for bacterial chromosome mapping and sequencing. Genome Res. 1996;6:448–453. doi: 10.1101/gr.6.5.448. [DOI] [PubMed] [Google Scholar]
- Steidler L, Hans W, Schotte L, Neirynck S, Obermeier F, Falk W, Fiers W, Remaut E. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289:1352–1355. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tedin K, Moll I, Grill S, Resch A, Graschopf A, Gualerzi CO, Blasi U. Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol Microbiol. 1999;31:67–77. doi: 10.1046/j.1365-2958.1999.01147.x. [DOI] [PubMed] [Google Scholar]
- Ten Brink B, Otto R, Hansen UP, Konings WL. Energy recycling by lactate flux in growing and non-growing cells of Streptococcus cremoris. J Bacteriol. 1985;162:383–390. doi: 10.1128/jb.162.1.383-390.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE, Eisen JA, Ketchum KA, Hood DW, Peden JF, Dodson RJ, et al. Complete genome sequence of Naisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- Valyasevi R, Sandine WE, Geller BL. The bacteriophage kh receptor of Lactococcus lactis subsp. cremoris KH is the rhamnose of the extracellular wall polysaccharide. Appl Environ Microbiol. 1990;56:1882–1889. doi: 10.1128/aem.56.6.1882-1889.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten WJ, Janssen GR. An AUG initiation codon, not codon-anticodon complementarity, is required for the translation of unleadered mRNA in Escherichia coli. Mol Microbiol. 1998;27:987–1001. doi: 10.1046/j.1365-2958.1998.00744.x. [DOI] [PubMed] [Google Scholar]
- Van Kranenburg R, Marugg JD, van Swam II, Willem NJ, de Vos WM. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol Microbiol. 1997;24:387–397. doi: 10.1046/j.1365-2958.1997.3521720.x. [DOI] [PubMed] [Google Scholar]
- Yvon M, Chambellon E, Bolotin A, Roudot-Algaron F. Characterisation and role of the branched-chain aminotransferase (BcaT) isolated from Lactococcus lactis subsp. cremoris NCDO763. Appl Environ Microbiol. 2000;66:571–577. doi: 10.1128/aem.66.2.571-577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]