Abstract
The human genome provides a reference sequence, which is a template for resequencing studies that aim to discover and interpret the record of common ancestry that exists in extant genomes. To understand the nature and pattern of variation and linkage disequilibrium comprising this history, we present a study of ∼31 kb spanning an ∼70 kb region of FMR1, sequenced in a sample of 20 humans (worldwide sample) and four great apes (chimp, bonobo, and gorilla). Twenty-five polymorphic sites and two insertion/deletions, distributed in 11 unique haplotypes, were identified among humans. Africans are the only geographic group that do not share any haplotypes with other groups. Parsimony analysis reveals two main clades and suggests that the four major human geographic groups are distributed throughout the phylogenetic tree and within each major clade. An African sample appears to be most closely related to the common ancestor shared with the three other geographic groups. Nucleotide diversity, π, for this sample is 2.63 ± 6.28 × 10−4. The mutation rate, μ, is 6.48 × 10−10 per base pair per year, giving an ancestral population size of ∼6200 and a time to the most recent common ancestor of ∼320,000 ± 72,000 per base pair per year. Linkage disequilibrium (LD) at the FMR1 locus, evaluated by conventional LD analysis and by the length of segment shared between any two chromosomes, is extensive across the region.
With the completion of the reference human genome sequence, we now have information available that is fundamental to understanding human genetic disease. Importantly, it will also provide the basis for new studies of human population history and genomic evolution and their impact on human disease. The human reference sequence is the template for large-scale resequencing studies that aim to discover and interpret the record of common ancestry that exists in modern genomes. This history is written in the form of human variation and its patterns. And, it is this variation that is a large determinant of complex human diseases.
Most recent studies of human evolution have been based on relatively short segments of DNA, often including only coding sequence (e.g., Dorit et al. 1995; Harding et al. 1997; Rana et al. 1999). Different classes of DNA, however, can have very different histories despite their proximity to one another (Wang et al. 1999). Furthermore, whereas short segments of sequence are useful for generating gene genealogies because of the decreased probability of recombination, large regions are necessary if we wish to understand genomic patterns of evolution. First, it is only through larger regions that we are likely to capture the effects of intragenic recombination, an important feature of protein evolution. Second, the contiguity of the data is paramount if we are to begin to understand how these patterns of variation relate to concepts such as linkage disequilibrium (LD). Lastly, with the larger data sets that come with larger regions, we will better be able to test competing hypotheses. Therefore, if we are to reconstruct a more complete view of human history, we must study the full complement of DNA, including coding and noncoding DNA, regions of high and low recombination, and regions with high and low mutation rates, as well as sequences from the autosomes, sex chromosomes, and mitochondrial DNA. In other words, the human genome is nonhomogeneous (Consortium 2001; Venter et al. 2001) and we must attempt to study it as such.
Because of the different histories of mitochondrial and Y chromosome DNA, each of which is a single locus, and nuclear DNA, the history evidenced in either of the single loci is not reflective of our collective history. Furthermore, DNA from different genomic locations has different time depths, as, on average, the mitochondrial genome and the Y chromosome have histories that span ∼Ne (the effective population size of the ancestral population) generations, the X chromosome, ∼3 Ne generations, and the autosomes, ∼4 Ne generations. By analyzing data from multiple loci, we are more likely to get a representative sample of the variation present in the genome — over several time periods — revealing a broader view of human and genomic history. To study this variation, we have chosen to look at the most common variable site, single nucleotide polymorphisms (SNPs).
In contrast to microsatellites, it is expected that SNPs are present in the genome at high enough density to be useful for a wide range of genetics studies (Chakravarti 1999). Furthermore, as with DNA from different locations, markers of different types also have different time depths, or different spans of history for which they hold information. Because of their relatively low mutation rate, SNP alleles remain in the population without sustaining recurrent mutation much longer than do microsatellite alleles, and therefore, have a greater time depth than microsatellites, revealing features of ancient history.
To uncover a piece of this history, we have completed a study of a large contiguous region of the nuclear genome, sequenced in multiple individuals from a variety of human populations. Here we present a study of an ∼70 kb region of the X chromosome, sequenced in a sample of 20 human males and four male great apes. We have cataloged the variation in the region, measured the level of LD and examined what these data can tell us about the history of the region and the alleles sampled.
We studied de novo SNP variation in the fragile X (FMR1) region of the X chromosome by resequencing; we also examined three well-studied microsatellite markers (DXS548, FRAXAC1, and FRAXAC2) in an effort not only to characterize the region in our sample, but also to compare what these two types of markers tell us about the history of the locus. We used these data to assess recombination and the level and pattern of LD in the region.
Several analyses were performed to determine if the evolutionary patterns present in the local variation is obscured by recombination. We looked at both conventional measures of linkage disequilibrium and a new statistic, S, length of segment shared between two sequences, around a variant site. Finally, we used phylogenetic analysis to investigate the evolutionary history of the FMR1 region and the alleles sampled in this study.
The data presented here also have important implications for association studies. If one is to create a high-density marker map and scan the entire genome for associations with a given phenotype, one must first know the density of polymorphisms needed to conduct such studies efficiently (Kruglyak 1999). The requisite density of polymorphisms, in turn, depends on the nature and pattern of background linkage disequilibrium in the human genome, a question to which studies such as this one may be applied.
RESULTS
Variation
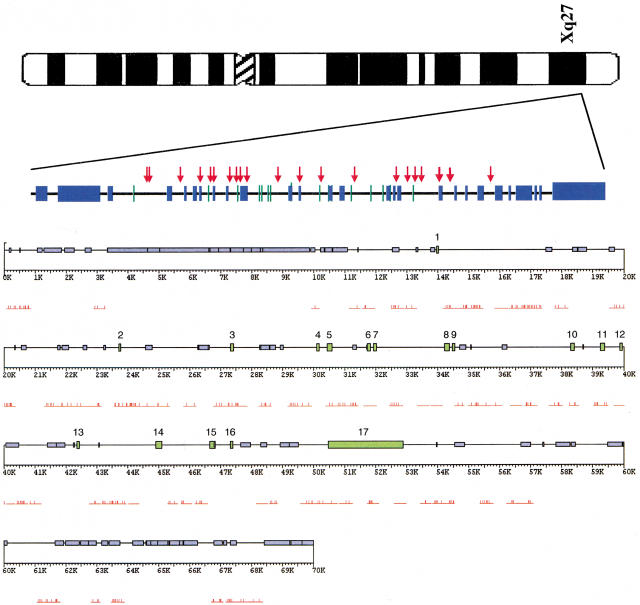
The 70-kb region encompassing the FMR1 gene was masked for repetitive sequences (e.g., LINEs, SINEs, and Alus), resulting in 47.7 kb (68.1%) of unique sequence, 22 kb of common repeats, and <0.5 kb of internal repeats (Fig. 1). The repeats are spaced fairly uniformly across the region containing the FMR1-coding sequence, with large blocks of repeats flanking the first and last exons. The overall GC content for this locus is 38.2%. Six of the 17 exons of the FMR1 gene were included in this analysis, as was ∼14 kb of sequence upstream of exon 1 and ∼17 kb of sequence downstream of exon 17. Twenty-five polymorphic sites and two insertion/deletions were found across an average of 30,946 bases compared between the sample and the GenBank record, spanning the first ∼70 kb of the FMR1 locus, among 20 human males (Fig. 2). No polymorphisms were identified within the human exonic sequences surveyed. Four polymorphic sites were identified in exons in the great apes (exons 3, 5, 11, and 17), but none resulted in a nonsynonymous change.
Figure 1.
The genomic structure of the FMR1 region. The ideogram of the X chromosome shows the location of the FMR1 region, subsequently magnified to show the region we studied. The green bars identify FMR1 exons, numbered accordingly. The blue bars represent regions masked by RepeatMasker. The red arrows locate the SNPs found among the 20 humans sampled. This region is further magnified below, with the modification that the red bars are the segments screened for variation and the thin red hash marks identify all differences from the reference sequence.
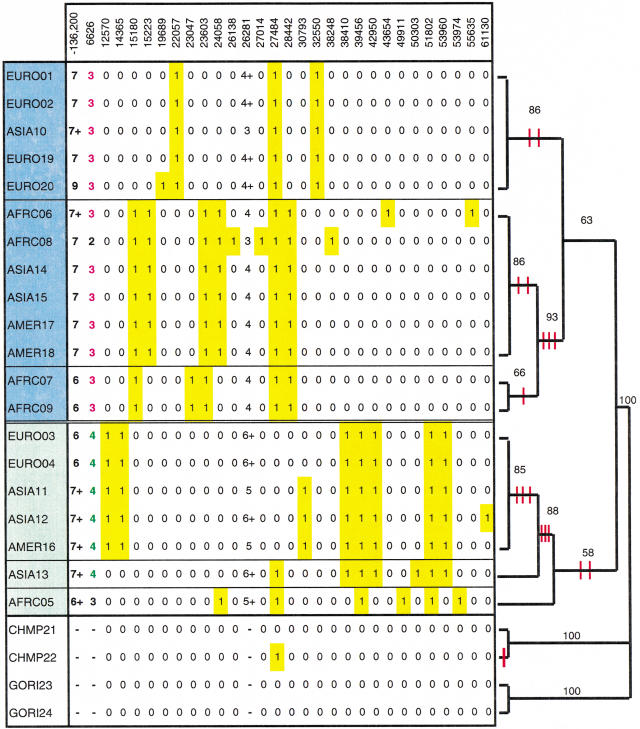
Figure 2.
FMR1 haplotypes grouped according to a branch and bound genealogy based on SNP haplotypes. All derived alleles are highlighted in yellow; microsatellite alleles are listed using the nomenclature in Macpherson et al. (1994) (alleles are given a number correlating to the number of base pairs in the microsatellite repeat tract; a whole number difference indicates a 2-bp increment, whereas a +'sign indicates a 1-bp increment). The sample names shaded in blue and green comprise Clades A and B, respectively. The red hash marks indicate the introduction of single nucleotide changes. The branch and bound algorithm generated five most parsimonious trees, with a tree length of 398, consistency index (CI) of 0.98 (Swofford 1998).
Among humans, 19 transitions, six transversions and two insertion/deletions were found (Table 1), none of which are located in coding sequence. Watterson's θ (Watterson 1975), for this sample (including indels in the analysis) is 2.42 ± .97 × 10−4, with no significant variation among geographic groups. Furthermore, π (the average number of nucleotide differences per site between two randomly chosen individuals from a population) and θ for this sample are not significantly different (π = 2.63 ± 6.28 × 10−4), with a value for Tajima's D of 0.277 (95% confidence interval of −1.800∼2.001). The mutation rate, μ, is 6.48 × 10−10 per base pair per year, giving an ancestral population size of ∼6200 breeding individuals and a time to most recent common ancestor (TMRCA) of 319,389 ± 72,583 years (see Appendix A, available as an on-line supplement at http://www.genome.org).
Table 1.
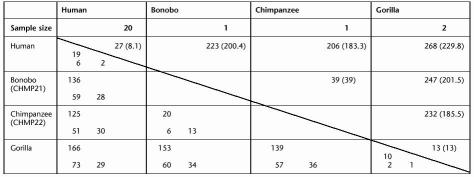
Sequence Variation Within and Between the Great Apes and Humans
The total (mean) number of variants for a given comparison are shown above the diagonal. The number of transitions (upper left corner), transversions (lower left), and insertion/deletions (lower right) are shown below the diagonal.
Within humans, 11 unique haplotypes were identified (Fig. 2). Three haplotypes are present among the six European males, four among the five Africans, five among the six Asians (one of which is shared with Europeans and two of which are shared with Amerindians). Among the three Amerindians, two haplotypes were identified, both of which are shared with Asians, as noted above. A total of 11 polymorphic sites were found among Europeans, 15 among Africans, 18 among Asians, and 14 among Americans. There are no fixed differences between any of the four geographic groups. The frequency spectrum reveals a relative paucity of singletons (10/27), given the rapid population growth during human history (in rapidly expanding populations, one expects to see more rare sites) (Donnelly and Tavare 1995).
To quantify the evolutionary history of the region, four non-human primates were also examined (Fig. 2). For 26/27 sites, one allele is fixed in the apes. This fixed allele is assumed to be the ancestral state, so that the derived allele in the human sample can be inferred (Table 2). One in 27 sites (27484) is polymorphic in both humans and chimps; here, the base that is fixed in the gorillas was assumed to be the ancestral state. Additionally, 39 sites differ between the two sub-species of chimp and 13 sites differ between the two gorilla samples. There are 74 fixed differences between the humans and the four great apes and ∼10-fold as much variation between humans and the great apes as among humans (Table 1). A modified McDonald-Kreitman test (McDonald and Kreitman 1991) was unable to reject the null hypothesis of neutral evolution at the FMR1 locus, for all possible comparisons between human and gorilla (P values of 0.24–1.0). Furthermore, the relative rate test (Li 1997) failed to reject the molecular clock hypothesis.
Table 2.
FMR1 Sequence Polymorphisms
| Position | Ancestral allele | Derived allele | Frequency of derived allele |
|---|---|---|---|
| 12570 | C | G | 0.25 |
| 14365 | C | A | 0.25 |
| 15180 | A | G | 0.40 |
| 15223 | G | C | 0.30 |
| 19689 | A | G | 0.05 |
| 22057 | G | A | 0.25 |
| 23047 | T | C | 0.10 |
| 23603 | A | G | 0.40 |
| 24058 | T | C | 0.35 |
| 26138 | A | G | 0.05 |
| 27014 | T | C | 0.05 |
| 27484 | A | T | 0.75 |
| 28442 | A | G | 0.40 |
| 30793 | G | A | 0.15 |
| 32550 | C | T | 0.25 |
| 38248 | T | C | 0.05 |
| 38410 | C | T | 0.30 |
| 39456 | G | A | 0.35 |
| 42950 | G | A | 0.30 |
| 43654 | C | *(del) | 0.05 |
| 49911 | C | T | 0.05 |
| 50303 | A | G | 0.05 |
| 51802 | T | C | 0.35 |
| 53960 | A | C | 0.30 |
| 53974 | * | C(ins) | 0.05 |
| 55635 | C | G | 0.05 |
| 61130 | C | T | 0.05 |
FMR1 sequence polymorphisms (relative to GenBank record L29074, 5′ to 3′), the ancestral and derived states, and the frequency of the derived allele. The derived allele was determined relative to the bonobo sample.
The 20 humans were also genotyped for the microsatellite markers DXS548, FRAXAC1, and FRAXAC2 (Fig. 2). We found a total of 11 microsatellite haplotypes in this sample. Three haplotypes were found in the six Europeans, four in the five Africans, four in the six Asians and two in the three Amerindians. Three unique (“singleton”) haplotypes were found in Africa, one in Europe, and one in Asia. The most common haplotype (4/20) is 7-3-4 (alleles at DXS548, FRAXAC1, and FRAXAC2, respectively) followed by 7-3-4+ (3/20). The FRAXAC1 alleles are the least diverse, with 19/20 individuals carrying either the 3 or 4 allele at this locus.
In contrast to the SNP data, the relationships among the microsatellite alleles, with only three markers (versus 27 SNPs) and just as many haplotypes as the SNPs, are much less obvious. These microsatellite data, however, provide additional information by virtue of previous work on the association of microsatellite alleles with fragile X disease (e.g., Macpherson et al. 1994; Eichler et al. 1996; Gunter et al. 1998). The most common haplotype in this sample, 7-3-4, is the second most common haplotype (12%) among chromosomes from the general (normal/control) population typed by Eichler et al. (1996), and has a significant association (P<0.02) with the 13+9 interspersion pattern (where the numbers are the counts of CGG repeats and the “+” represents an AGG interruption) (Eichler et al. 1996). The second most common haplotype here (7-3-4+) is the most common haplotype among normal chromosomes (44%) and is the only haplotype found to have a significant negative association with an interspersion pattern (9 + 9 + 9) (Eichler et al. 1996). The greatest diversity in haplotypes was again found in Africa (four haplotypes among five individuals), which also contains most (3/5) of the unique haplotypes.
Finally, given the recent publication of the human genome (Consortium 2001; Venter et al. 2001), a comparison was made between the SNP data presented here and those that are currently publicly available (Sherry et al. 2001). Of the 59 SNPs reported within the sequences analyzed in this study, 27 are presented here and 24 were identified by us in a larger sample (of which the present sample is a subset), allowing independent confirmation that 23/24 are not found in the sample used in this study. The remaining 8/32 were not found in either of our samples.
Recombination and Linkage Disequilibrium
The four-gamete test revealed limited evidence of recombination, as only 5/300 pairs of sites have all four possible gametes. On further inspection, all five instances of four gametes involve site 24058 — which is neither a CpG site nor within a poly-nucleotide run — suggesting that this site might be homoplasic through recurrent mutation. To evaluate the nature of LD at the FMR1 locus, we used three measures: two traditional measures of LD, D2 and D‘ (Lewontin 1988), between pairs of markers and a new statistic, S, which measures the length of segment shared between any two chromosomes.
Conventional LD analysis using D2reveals moderate levels of LD (Fig. 3), though we have limited power to detect significant LD, given our sample size. Given the marginal frequencies of each of the 300 possible pairs pair of sites (indels were excluded), 97 could have been significant, based on a Fisher’s Exact test (Weir 1996), and of those, 41 exhibit significant LD (P <0.001). The maximum distance across which LD was detected is 45 kb, with 72% of the distances falling below 20 kb. Furthermore, the majority of D' values (295/300) are equal to 1.
Figure 3.
Tests of association (D2 statistic) for all pairwise comparisons between all variants. Statistical significance is based on P-values calculated by a Fisher's exact test. (Black) No power to detect LD; (white) no significant LD present; (grey) significant LD present (P < 0.001).
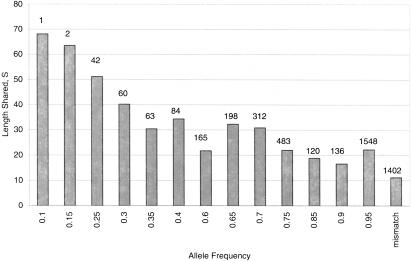
Another way to look at the genomic structure of a region is to look directly at the amount of sequence shared, identical-by-descent (IBD), between two chromosomes around a given focal site. This measure, S, reveals long-range shared haplotypes at the FMR1 locus (Fig. 4), and may be viewed as an alternative to traditional measures of LD. Our data show that the amount of sharing IBD between two chromosomes, around a focal site, decreases with increasing allele frequency, as expected, based on computer simulations (D. Mathews, R. Hudson, A. Chakravarti, unpubl.). The mean S values range from ∼70,000 bases (i.e., the entire region analyzed) around the allele present at the lowest frequency for which calculation of S is possible (i.e., a doubleton), to ∼20,000 bases around the most common (highest frequency) alleles. The standard deviations for these mean values range from 6%–125% of the mean. In contrast, mismatch comparisons (i.e., calculation of S around alternate alleles at a focal site) have a mean S value of only ∼11kb, assumed to be identity by state given their disparate histories.
Figure 4.
Sequence shared, S (in number of kilobases), as a function of derived allele frequency. Each bar represents the average of all alleles of the listed frequency class (classes listed represent only those present in our sample). The number of comparisons for each frequency class is listed above each bar. The “mismatch” bin represents all pairwise comparisons between alternative alleles at each variant site.
Phylogenetic Analysis
Phylogenetic analysis was conducted using the program PAUP (Swofford 1998). All trees reveal two major clades, referred to here as A and B (Fig. 2), but with weak statistical support. Among the five trees, Clade A is constant, whereas the relationship between five of the terminal branches of Clade B (EURO03, EURO04, ASIA11, ASIA12, and AMER16) varies. Furthermore, it is only among these five taxa that the majority rule and strict consensus trees differ. Bootstrap support for the strict consensus tree ranges from 58–100, with eight of the 11 nodes having bootstrap support of ≥85%.
The majority rule consensus tree suggests that the human geographic groups are distributed throughout the tree for this sample. Furthermore, all methods used produce trees in which individuals are likewise mixed with respect to geographic origin. AFRC05, a Biaka pygmy, appears to be most closely related to the inferred ancestral sequence. The Neighbor-Joining tree suggests the same two clades (A and B) as the parsimony tree.
The two most common microsatellite haplotypes identified in this study (7-3-4 and 7-3-4+) are both found in Clade A, but not among the African samples in this group. There is no overlap of FRAXAC2 alleles between clades A and B and AFRC05 is the only individual in Clade B with a 3 allele at FRAXAC1.
DISCUSSION
The human genome sequencing projects have demonstrated that our genome is remarkably nonhomogeneous (Consortium 2001; Venter et al. 2001), that is, different regions have different features. Therefore, only by studying variation in long segments of the genome, such as in this study, will properties of this heterogeneity become evident.
The FMR1 locus was chosen for analysis for several reasons: (1) the genomic sequence of the locus was known; (2) FMR1 is associated with fragile X and has an unusual pattern of disease linkage disequilibrium (LD); and (3) accurate sequence on complete haplotypes could be obtained. Our study shows that we have identified the major common polymorphisms in the region. Although no sample of 20 human males will ascertain all extant human variation, the comparison of our SNP data with those in the public resource (Sherry et al. 2001) demonstrates that we are able to confirm our variant sequences for 50/59 (85%) SNPs in regions of known overlap. Of the nine remaining SNPs, two are known to be at low frequency in humans (ATL3 ƒ <0.005, ATL4 ƒ <0.01) (Gunter et al. 1998).
The SNP data presented here are useful for addressing the original striking finding of LD across 170 kb at the FMR1 locus (Eichler et al. 1996), which was surprising given that fragile X disease is caused by recurrent lethal mutations. To explain this result, it was suggested that there are two mutational pathways leading to fragile X disease alleles, one involving recurrent loss of AGG interspersions on multiple haplotypes and the other involving a founder effect of a haplotype that is refractory to the loss of AGG interspersions (Eichler et al. 1996). This study, which also demonstrates extensive LD in the region, supports the hypothesis of recurrent mutation on a limited number of haplotypes. Such a high level of LD results from the apparent lack of recombination in the region and/or ancient population subdivision followed by admixture (Wall 2000).
We analyzed linkage disequilibrium at the FMR1 locus by two methods — conventional LD analysis, using D2 and segment sharing around each SNP. D2 analysis reveals moderate levels of LD, with a maximum length of 45 kb and with 72% of the distances falling below 20 kb. It is important to keep in mind, however, that our sample size limits the utility of this analysis, as traditional measures of LD depend on allele frequencies. This difference between our value and that of Eichler et al. (1996) — 45 kb versus 170 kb — results from the markers used to detect LD in these studies and from the complex mutational and recombinational history of the FMR1 locus (Macpherson et al. 1994; Eichler et al. 1996; Gunter et al. 1998;). Nevertheless, at 45 kb, the level of LD found in this study is higher than predicted by early simulation studies (Kruglyak 1999), and is in sharp contrast to the findings of other similar studies on autosomal genes (Clark et al. 1998).
To study the relationships among alleles further, we searched for sequences shared IBD. Each SNP arises in the population at a specific point in time, and therefore has a unique history. The sharing statistic, S, takes advantage of this fact to study the relationship between all copies of a specified allele. The average amount of sharing between two chromosomes is directly proportional to their length of time in the population. Consequently, this measure of the amount of sequence shared at any site in the genome, between two haplotypes — be they from families, patient and control populations, or different geographic groups — can be used to localize the position of a disease gene, as originally done for cystic fibrosis (Kerem et al. 1989).
The S statistic is a direct measure of the relationship between two alleles; as such, the value of S varies from zero to the target length, unlike the traditional measure of LD, D, which is meant to vary between zero and one, but does not because of its dependence on allele frequencies. Furthermore, S takes into account not only the recombinational history of a region as do traditional measures of LD, but also mutational history, allowing it to capture more of the evolutionary signal and making it more powerful. Therefore, S is expected to depend on the density of variants in a region, the local rate of recombination, and age (or, frequency as a surrogate) of each variant allele; hence the importance of inferring ancestral alleles.
At the FMR1 locus, the pattern of sharing is as expected based on simulation studies (data not shown) in which rarer (younger) alleles have larger S values and common (older) alleles have smaller values (Fig. 4). This pattern is the record of the history of a population and results from generations of mutation and recombination whittling away at the amount of shared sequence associated with a given variant allele, along with the effects of selection, hitch-hiking, etc. In general, rarer alleles entered the population relatively recently; therefore, there have been fewer generations during which the sequence they share IBD with other chromosomes has degraded. In contrast, older alleles exist in the population for many generations, leaving only a small amount of shared sequence as a record of the common ancestry between alleles. Our data, both from the FMR1 locus and from simulation studies, show that SNPs of frequency ≥20% will have ∼20–50 kb of sequence associated with them. This suggests that a marker map of density ∼1/10 kb may be adequate to facilitate association studies.
The SNP haplotype diversity we observed corresponds well, but not precisely, to microsatellite haplotype diversity at the FMR1 locus. SNPs, by virtue of their low mutation rate, reflect a much deeper history than do microsatellites, which carry a higher probability of recurrent mutation. Whereas their relatively high mutation rate makes microsatellite markers useful for studying diseases such as fragile X, in which the mutation rate of normal to unstable alleles is likewise high (∼0.8×10−4; Chiurazzi et al. 1996), and therefore, the alleles young, they are not appropriate for investigating older population and evolutionary events. In particular, for reconstructing the evolutionary history of a region, many types of variants are needed. Additionally, not only the mutational history, but also the recombinational history must be considered.
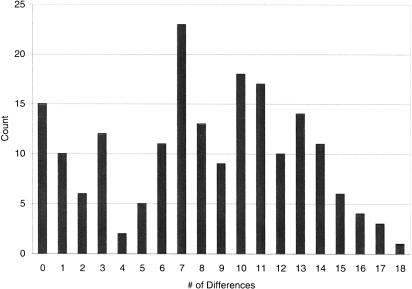
Although the four-gamete test reveals evidence of recombination at FMR1, on further inspection, all instances of four gametes involve site 24058. This, along with the plot of D' (data not shown), which fails to provide any evidence of recombination, suggests to us that site 24058 is more likely the result of recurrent mutation than recombination. Given this fact, and the above analysis of LD, our data reveal little or no recombination in the FMR1 region examined. As such, the history of the locus can be explained largely by the mutational history. Because of decreased recombination, a deep historical record is preserved, as reflected by the mismatch distribution (Fig. 5), which suggests multiple deep branches in this sample.
Figure 5.
The mismatch distribution in base pairs among 20 human samples.
Because recombination is not a major feature at this locus, and as our data suggest that we can assume a molecular clock, we can estimate several aspects of the history. Our population parameter estimates from these data (see Appendix A, available as an on-line supplement at http://www.genome.org) are in line with what have been seen in other studies of the X (e.g., Huang et al. 1998; Nachman et al. 1998); although, the number of singletons identified in this study is less than expected for an expanding population, which may be a property of the region or the result of our sequencing strategy. The range of the parameter estimates in the literature, however, is very broad (see , Appendix A, available as an on-line supplement at http://www.genome.org). The scatter in these data, and even in the data from the X chromosome alone, may be caused by differences in estimation methods (especially in TMRCA), differences in experimental design (e.g., sampling strategy), and/or locus-to-locus variation. As noted above, the human genome is remarkably heterogeneous, so it is not surprising that the variance in these parameter estimates is high, attributable to biological factors. A further problem may be that the data are used to make inferences about the history of peoples, rather than solely about the genomic region under study that they represent. The mutational and recombinational history of the region must be understood before making a judgement as to the appropriateness of the data for making inferences about human history.
Given our data on the mutational and recombinational history of the FMR1 locus, we can further address the evolutionary history of the alleles in our sample. The phylogenetic analysis of these data is remarkable for its inability to parse out the different population groups (Fig. 2). Whereas both parsimony and distance trees separate the individuals into two major clades (a division that is also supported by FRAXAC1 alleles), Africans are dispersed throughout the tree and members of each of the four major geographic groups are in each of the two clades. It appears that the oldest human sequence is found in Africa (AFRC05). Additionally, 70% of the singletons found in this study are found in Africa, and Africans are the only group that does not share at least one haplotype with another geographic group. Lastly, although these findings and our variation data for this sample are consistent with the Out of Africa Hypothesis, the sequences in the Asians and Americans are as, if not more, diverse than the African sequences, although sample sizes are small. The diversity found in Asia, however, is supported by other studies (Foley 1998; Kaessmann et al. 1999).
Rather than generating neat clusters for each geographic group, we suspect that large contiguous regions of DNA are likely to produce commingled genealogies attributable to the disparate histories of neighboring regions of the genome (e.g., Wang et al. 1999). Furthermore, because of the high mutation rate of microsatellites, these markers are more likely to result in identity-by-state than by descent, suggesting spurious conclusions about common history. In contrast, SNP alleles have distinct ages and shared sequence lengths, and allow us to delve more deeply into human and genomic history. That said, it is important to keep in mind that not all SNPs are created equal, as evidenced by the apparent recurrent mutation identified in this study, which has important implications for haplotype studies.
METHODS
Sequence and Microsatellite Data
Approximately 31 kb from the fragile X locus (FMR1) was sequenced from 24 males, including six Europeans (four Utah Mormons, two French), five African Pygmies (two Biaka, three Mbuti), six Asians (one Cambodian, two Melanesians, three Chinese), three Amerindians, two chimpanzees (one Pan troglodytes and one Pan paniscus), and two gorillas (Gorilla gorilla gorilla). This ∼31 kb spans ∼70 kb of the FMR1 region, from base positions 49 to 68,395 of GenBank record L29074. L29074 contains 185,775 bp of sequence, including ∼14 kb upstream of exon 1 and all 17 exons of the FMR1 gene.
Unique sequence was identified by analyzing the first 70 kb of L29074 with the program RepeatMasker2 (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker). The processed sequence was then analyzed by the program Miropeats (http://genome.wustl.edu/gsc/programs/finishing/repeats/miropeats.html) to check for internal repeats. This output was then analyzed by BLAST (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-newblast?Jform=1) (Altschul et al. 1990), against the nonredundant and high- throughput genomic sequence databases for uncharacterized repeat elements. Repeats were largely excluded from analysis as these sequences are expected to have significantly different properties and mutational histories than do unique sequences. The distribution of repeats in this region is fairly uniform, with the exception of two large blocks just upstream of exon 1 and downstream of exon 17. Likewise, the unique sequences are fairly evenly distributed across the region, suggesting that our data provide a representative view of variation at this locus.
PCR primers were designed to amplify products of 350–500 bases, in a tiling pattern across unique regions and small repeats, and covering only six of the 17 exons of FMR1. PCR amplification used genomic DNA from the Coriell Institute (20 humans) and from the Center for Reproduction of Endangered Species (CRES) at the San Diego Zoo (four nonhuman primates). PCR products were sequenced directly using the forward primer. Variant sites in the humans were confirmed by sequencing of the opposite strand from an independent PCR amplification. Sequencing was performed on ABI 377 Automated Sequencers and the data were collected and analyzed using ABI sequencing software.
ABI trace files were analyzed using a suite of programs that includes PHRED, PHRAP, and CONSED (Ewing and Green 1998; Ewing et al. 1998; Gordon et al. 1998) to align the sequences and allow visual identification of variant sites. Sites flagged as variant in the CONSED view were evaluated based on the PHRED score, with a minimum score of 20 and the nature (poly-nucleotide tracts, proximity to primer sequence, etc.) of the flanking sequence. Furthermore, it should be noted that neither ATL1 (Gunter et al. 1998) nor FMRb (Kunst and Warren 1994), SNPs in the FMR region studied previously, were within the regions sequenced in this study.
The exonic changes in the apes were evaluated by analyzing the four ape and two human nucleotide sequences using the program Genscan (Burge and Karlin 1997), aligning the resulting amino acid sequences and visually inspecting the alignments for changes. At site 27484, the allele present in three out of four apes was inferred to be the ancestral allele.
The microsatellite typing was performed as described in Murray et al. (1996) and Richards et al. (1991).
Database Comparison
Two approaches were used to collect all publicly available SNPs that map to the FMR1 locus. First, the masked sequence from L29074 was BLASTed against dbSNP. Second, a LocusLink (Pruitt et al. 2000) search was performed for FMR1, providing the list of 105 SNPs (there is considerable, although not complete, overlap between the two sets of results). Of these SNPs, 52/105 were already mapped to L29074, with the positions reported in LocusLink. For the remainder, BLAST2 Sequences (Tatusova and Madden 1999) was used to align the 5′ flanking sequence reported with the SNP to L29074, providing the positions of these SNPs relative to the reference sequence.
Statistical Analysis
θ was calculated based both on numbers of segregating sites (Watterson 1975) and by pairwise differences (π) (Tajima 1983). All calculations, except those used for the application of the HKA test, include indels such that each insertion/deletion event was counted as a single polymorphism. μ was calculated assuming a human–chimp divergence time of 5 × 106 years, using the number of differences with CHMP22 (common chimpanzee) (Zuckerkandl and Pauling 1965). Effective population size was calculated from Ne =θ/3μg, where g= 20 yr (Watterson 1975). TMRCA was calculated using the program Genetree, written by R.C. Griffiths and available at http://www.maths.monash.edu.au/∼mbahlo/mpg/gtree.html. The estimates calculated by this program are based on the coalescent model and an assumption of an infinite sites mutational model. To meet the infinite sites assumption, site 24058 (the putative recurrent mutation) was trimmed from the data set for the estimation of TMRCA.
Tajima's D was calculated as described in Tajima (1989). To calculate the confidence interval for this statistic, the absence of recombination is assumed.
The McDonald-Kreitman test of neutrality asks whether two classes of mutations exhibit the same ratio of polymorphism to divergence (McDonald and Kreitman 1991). McDonald and Kreitman used silent and replacement sites in their original tests, whereas we used the proportions of transitions/transversions, transitions/indels, and transversions/indels between the 20 humans and two gorillas. The relative rate test was performed as described by Li (1997).
The four-gamete test makes all pairwise comparisons between sites and counts the number of the four possible gametes that are present for each pair of sites. The presence of all four gametes for a given pair of sites is evidence of recombination or recurrent mutation.
Conventional linkage disequilibrium measures, D2 (Lewontin 1988) and D‘ (Lewontin 1964), were calculated from a set of aligned FASTA files. D is a measure of disequilibrium, based on the frequencies of double heterozygotes. D2 is that value (D) normalized by allele frequencies and D‘ is that value (D) divided by the maximum possible value of D for a given pair of sites. The significance values were calculated using a two-tailed Fisher’s exact test on gametic counts (Weir 1996).
The tests of sharing were performed for all pairwise comparisons among a set of haplotypes to calculate the shared length around the majority and minority alleles at each polymorphic site (focal site). The mean shared length for each allele, within each frequency class (i.e., majority and minority classes), was then calculated. For example, for any polymorphic site x, all pairwise comparisons were made between haplotypes with the majority allele at that focal site, and likewise for all haplotypes carrying the minority allele. The mean and standard deviation of the shared lengths were then calculated within the majority and minority allele class, respectively. The mean shared length was calculated as the distance from the focal site to the base before the first site that is different between any two haplotypes, on either side of the focal site.
Phylogenetic Analysis
The parsimony tree was constructed using the software package PAUP (Phylogenetic Analysis Using Parsimony) (Swofford 1998) and the branch and bound algorithm (Hendy and Penny 1982). Support for the individual nodes was estimated from 1000 bootstrap replications, using the heuristic algorithm. For these analyses, insertions and deletions were treated as a fifth character state and sites for which there were missing data were dropped from the analysis for all comparisons affected. PAUP was also used to calculate Kimura two-parameter distances (Kimura 1980) and p-distances (Nei 1987).
A neighbor-joining tree (Saitou and Nei 1987) was constructed using uncorrected p-distances, mean character differences, and total number of differences; all three distance measures resulted in trees with the same two major clades and similar bootstrap values.
Acknowledgments
We thank Kimberley Bentley for DNA sequencing; Oliver Ryder and Leona Chemnick for chimpanzee and gorilla samples; James Macpherson and Catherine Lewis for their help with microsatellite genotyping; and Michael Zwick and David Cutler for comments and suggestions on this manuscript. This study was funded by National Institutes of Health grant HG01847.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL aravinda@jhmi.edu; FAX (410) 502-7544.
Article and publication are at http://www.genome.org/cgi/doi/10.1101 /gr.172601.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Chakravarti A. Population genetics–making sense out of sequence. Nat Genet. 1999;21:56–60. doi: 10.1038/4482. [DOI] [PubMed] [Google Scholar]
- Chiurazzi P, Macpherson J, Sherman S, Neri G. Significance of linkage disequilibrium between the fragile X locus and its flanking markers [editorial] Am J Med Genet. 1996;64:203–208. doi: 10.1002/(SICI)1096-8628(19960712)64:1<203::AID-AJMG37>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Clark AG, Weiss KM, Nickerson DA, Taylor SL, Buchanan A, Stengard J, Salomaa V, Vartiain E, Perola M, Boerwinkle E, et al. Haplotype structure and population genetic inferences from nucleotide- sequence variation in human lipoprotein lipase. Am J Hum Genet. 1998;63:595–612. doi: 10.1086/301977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TGIS. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Donnelly P, Tavare S. Coalescents and genealogical structure under neutrality. Annu Rev Genet. 1995;29:401–421. doi: 10.1146/annurev.ge.29.120195.002153. [DOI] [PubMed] [Google Scholar]
- Dorit RL, Akashi H, Gilbert W. Absence of polymorphism at the ZFY locus on the human Y chromosome. Science. 1995;268:1183–1185. doi: 10.1126/science.7761836. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Macpherson JN, Murray A, Jacobs PA, Chakravarti A, Nelson DL. Haplotype and interspersion analysis of the FMR1 CGG repeat identifies two different mutational pathways for the origin of the fragile X syndrome. Hum Mol Genet. 1996;5:319–330. doi: 10.1093/hmg/5.3.319. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Foley R. The context of human genetic evolution. Genome Res. 1998;8:339–347. doi: 10.1101/gr.8.4.339. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Gunter C, Paradee W, Crawford DC, Meadows KA, Newman J, Kunst CB, Nelson DL, et al. Re-examination of factors associated with expansion of CGG repeats using a single nucleotide polymorphism in FMR1. Hum Mol Genet. 1998;7:1935–1946. doi: 10.1093/hmg/7.12.1935. [DOI] [PubMed] [Google Scholar]
- Hammer MF. A recent common ancestry for human Y chromosomes. Nature. 1995;378:376–378. doi: 10.1038/378376a0. [DOI] [PubMed] [Google Scholar]
- Harding RM, Fullerton SM, Griffiths RC, Bond J, Cox MJ, Schneider JA, Moulin DS, Clegg JB. Archaic African and Asian lineages in the genetic ancestry of modern humans. Am J Hum Genet. 1997;60:772–789. [PMC free article] [PubMed] [Google Scholar]
- Harris EE, Hey J. X chromosome evidence for ancient human histories. Proc Natl Acad Sci. 1999;96:3320–3324. doi: 10.1073/pnas.96.6.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendy MD, Penny D. Branch and bound algorithms to determine minimal evolutionary trees. Math Biosci. 1982;59:277–290. [Google Scholar]
- Huang W, Fu YX, Chang BH, Gu X, Jorde LB, Li WH. Sequence variation in ZFX introns in human populations. Mol Biol Evol. 1998;15:138–142. doi: 10.1093/oxfordjournals.molbev.a025910. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Heissig F, von Haeseler A, Paabo S. DNA sequence variation in a non-coding region of low recombination on the human X chromosome. Nat Genet. 1999;22:78–81. doi: 10.1038/8785. [DOI] [PubMed] [Google Scholar]
- Kerem BS, Buchanan JA, Durie P, Corey ML, Levison H, Rommens JM, Buchwald M, Tsui LC. DNA marker haplotype association with pancreatic sufficiency in cystic fibrosis. Am J Hum Genet. 1989;44:827–834. [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet. 1999;22:139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- Kunst CB, Warren ST. Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell. 1994;77:853–861. doi: 10.1016/0092-8674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— On measures of gametic disequilibrium. Genetics. 1988;120:849–852. doi: 10.1093/genetics/120.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H. Molecular evolution. 1997. p. 80. , 216–218. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Macpherson JN, Bullman H, Youings SA, Jacobs PA. Insert size and flanking haplotype in fragile X and normal populations: possible multiple origins for the fragile X mutation. Hum Mol Genet. 1994;3:399–405. doi: 10.1093/hmg/3.3.399. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila [see Comments] Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Murray A, Youings S, Dennis N, Latsky L, Linehan P, McKechnie N, Macpherson J, Pound M, Jacobs P. Population screening at the FRAXA and FRAXE loci: Molecular analyses of boys with learning difficulties and their mothers. Hum Mol Genet. 1996;5:727–735. doi: 10.1093/hmg/5.6.727. [DOI] [PubMed] [Google Scholar]
- Nachman MW, Bauer VL, Crowell SL, Aquadro CF. DNA variability and recombination rates at X-linked loci in humans. Genetics. 1998;150:1133–1141. doi: 10.1093/genetics/150.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York, NY: Columbia University Press; 1987. p. 99. [Google Scholar]
- Pruitt KD, Katz KS, Sicotte H, Maglott DR. Introducing RefSeq and LocusLink: Curated human genome resources at the NCBI. Trends Genet. 2000;16:44–47. doi: 10.1016/s0168-9525(99)01882-x. [DOI] [PubMed] [Google Scholar]
- Rana BK, Hewett-Emmett D, Jin L, Chang BH, Sambuughin N, Lin M, Watkins S, Bamshad M, Jorde LB, Ramsay M, et al. High polymorphism at the human melanocortin 1 receptor locus. Genetics. 1999;151:1547–1557. doi: 10.1093/genetics/151.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards RI, Shen Y, Holman K, Kozman H, Hyland VJ, Mulley JC, Sutherland GR. Fragile X syndrome: Diagnosis using highly polymorphic microsatellite markers. Am J Hum Genet. 1991;48:1051–1057. [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. PAUP*: Phylogenetic analysis using parsimony (*and other methods) version 4. Sunderland, MA: Sinauer Associates; 1998. [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wall JD. Detecting ancient admixture in humans using sequence polymorphism data. Genetics. 2000;154:1271–1279. doi: 10.1093/genetics/154.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RL, Stec A, Hey J, Lukens L, Doebley J. The limits of selection during maize domestication. Nature. 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Weir BS. Genetic data analysis II: Methods for discrete population genetic data. 1996. pp xii, 445. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. New York, NY: Academic Press; 1965. pp. 97–166. [Google Scholar]