Abstract
A detailed bioinformatics analysis of six glycopeptide biosynthetic gene clusters isolated from soil environmental DNA (eDNA) mega-libraries indicates that a subset of these gene clusters contains collections of tailoring enzymes that are predicted to result in the production of new glycopeptide congeners. In particular, sulfotransferases appear in eDNA derived gene clusters at a much higher frequency than would be predicted from the characterization of glycopeptides from cultured Actinomycetes. Enzymes found on tailoring enzyme-rich eDNA clones associated with these six gene clusters were used to produce a series of new sulfated glycopeptide derivatives in both in vitro and in vivo derivatization studies. The derivatization of known natural products with eDNA derived tailoring enzymes is likely to be a broadly applicable strategy for generating libraries of new natural product variants.
Introduction
In many cases, bacterial natural products can be classified into large families based on the presence of conserved core structures with shared biosynthetic origins.1 The structural diversity seen within a family of natural products is often attributable almost exclusively to differences in the constellation of functional groups that are appended around the conserved core. Differences in functionalization patterns arise from the action of tailoring enzymes found within the biosynthetic gene clusters that are associated with a given natural product family. Tailoring enzymes often exhibit broad substrate promiscuity, and therefore, enzymes that have evolved to be used in the biosynthesis of one member of a family will likely have the ability to carryout the same reaction on many related structural variants.2 Tailoring enzymes associated with a particular family of natural products, either alone or in combination, are therefore potentially useful tools for creating libraries of natural products with new functionalization patterns. The utility of this approach for generating collections of novel bioactive metabolites is limited mainly by the size and diversity of the set of tailoring enzymes associated with a specific natural product family. The emerging field of metagenomics provides an unprecedented opportunity to access new collections of tailoring enzymes that could be used to generate additional derivatives of many natural products.
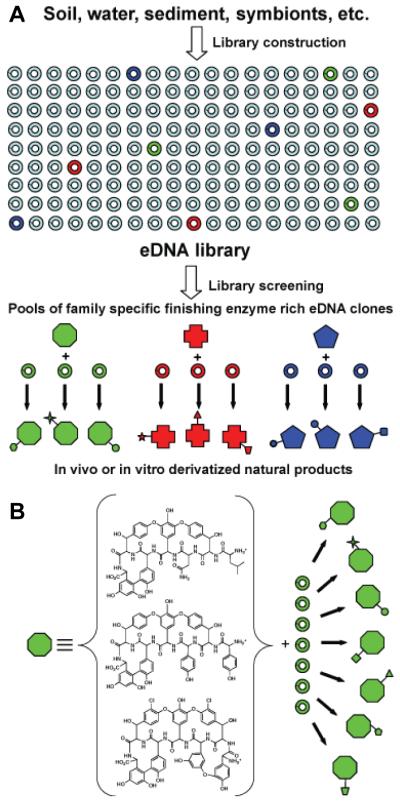
Environmental samples typically contain two to three orders of magnitude more bacterial species than is easily cultured in the laboratory.3-5 Metagenomics relies on the cloning of DNA directly from naturally occurring microbial populations (environmental DNA, eDNA) to circumvent difficulties associated with culturing bacteria and then the subsequent screening of the resulting clone libraries to identify new genes and gene products of interest.6-8 All of the genes required for the biosynthesis of a natural product, including genes that encode biosynthetic, regulatory, and self-immunity enzymes, are typically clustered on bacterial chromosomes. In addition to containing completely novel biosynthetic systems, eDNA samples are also likely to contain numerous gene clusters that resemble known systems, but that differ in their pathway-specific collections of tailoring enzymes. The construction of 35-40 kb insert cosmid based eDNA libraries is now routine, and a number of examples exist in the literature where large natural product gene clusters have been recovered on a series of overlapping eDNA clones.9-12 The construction of larger insert eDNA libraries capable of capturing intact natural product gene clusters that might differ in their pathway-specific collections of tailoring enzymes on individual eDNA clones remains a major challenge for the field. While it is still difficult to regularly clone large intact gene cluster from environmental samples, the systematic examination of eDNA cosmid libraries with probes designed to recognize conserved sequences found in known natural product gene clusters has the potential to uncover individual clones with unique collections of tailoring enzymes that could be used to generate metabolites with new functionalization patterns (Figure 1A).
Figure 1.
A.) Environmental DNA libraries are predicted to contain thousands of unique biosynthetic gene clusters. Many of these previously inaccessible clusters likely encode the biosynthesis of new members of previously characterized natural product families. Tailoring enzyme-rich eDNA clones from these clusters will encode collections of enzymes that have evolved to specifically interact with a conserved natural product core structure. These enzymes should therefore be useful for generating new collections of unnatural natural products using either in vitro or in vivo biosynthetic strategies. B.) Glycopeptides are derived from a small number of oxidatively cross-linked heptapeptides. In this manuscript we describe the use of eDNA derived glycopeptide tailoring enzymes to generate a family of new glycopeptide congeners.
Glycopeptides are one example of a large family of natural products where the individual members differ by the number and type of functional groups that are arranged around a set of closely related core structures (Figure 1B).13 Several members of this group of oxidatively cross-linked heptapeptides, including vancomycin and teicoplainin, are the preferred course of treatment for nosocomial antibiotic-resistant Gram-positive bacterial infections.14 Although cultured bacteria have been an important source of glycopeptides, there undoubtedly remain numerous previously undetected glycopeptide biosynthetic gene clusters in nature. Environmental samples likely contain both glycopeptide-producing bacteria that have remained recalcitrant to standard culture methods, as well as bacteria that possess cryptic glycopeptide gene clusters that have evaded detection in standard activity-based assays. Here we report the isolation of new glycopeptide biosynthetic gene clusters from eDNA libraries constructed from multiple soil samples and the use of the tailoring enzymes found in these gene clusters to produce a series of new sulfated glycopeptide derivatives. The tailoring of known natural product scaffolds with collections of eDNA derived tailoring enzymes is likely to be a broadly applicable strategy for generating libraries of new natural product variants.
Results and discussion
Isolation of glycopeptide tailoring enzyme-rich clones
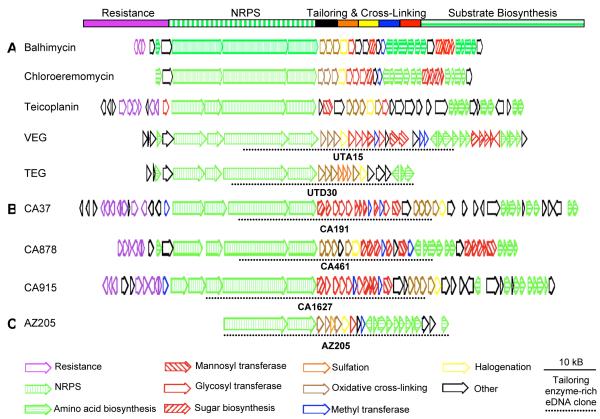
Five of the seven sequenced glycopeptide gene clusters show a very similar overall gene organization (Figure 2A).11,15-19 In these clusters, the position of the genes that encode each general class of enzyme required for the biosynthesis of a glycopeptide is conserved; resistance genes are grouped together at the beginning of the cluster, followed by nonribosomal peptide synthetase genes, unique collections of genes encoding tailoring enzymes and ultimately genes that encode the biosynthesis of essential substrates (amino acids and sugars) (Figure 2A).20 An interesting feature of this conserved architecture is that genes encoding tailoring enzymes tend to be grouped tightly together in the gene cluster. With such an arrangement, we postulated that it would be possible to capture large portions of glycopeptide tailoring enzyme systems on single eDNA derived cosmid clones, and that the genes found on these clones would be useful for generating glycopeptides with novel functionalization patterns.
Figure 2.
A. Many sequenced glycopeptide gene clusters show a similar overall gene organization. The balhimycin, chloroeremomycin and teicoplanin gene clusters were cloned from cultured soil bacteria while the VEG and TEG gene clusters were cloned directly from soil. Open reading frames have been color coded according their predicted functions. B. Glycopeptide gene clusters recovered from the CA eDNA mega-library are shown. C. AZ205, a tailoring enzyme-rich eDNA cosmid clone, recovered from the AZ library is shown. Dashed lines are used to indicate the region from each gene cluster that corresponds to the tailoring enzyme-rich clone used in this study.
The long history of culturing glycopeptide-producing bacteria from soil, coupled with the enormous unexplored bacterial diversity present in most soil environments, makes soil a particularly attractive starting point for the potential discovery of additional glycopeptide biosynthetic diversity using culture independent strategies. In this study, eDNA libraries were constructed from soil collected in California (CA) and Arizona (AZ) and these libraries were screened for genes encoding glycopeptide biosynthetic machinery. Previous studies have shown that much of the biosynthetic capacity present in a soil sample can be captured in a library containing at least 1.0 × 107 cosmid clones and therefore each of these libraries was expanded until it contained between 1.0 × 107 to 1.5 × 107 unique cosmid clones.21 We have termed these large libraries eDNA mega-libraries.
In a previous study, we were able to recover two glycopeptide gene clusters (TEG and VEG) from a Utah (UT) soil eDNA mega-library by initially screening the library for cosmids containing oxyC genes (Figure 2A).11 The oxyC gene product catalyzes the direct C-C coupling of amino acids 5 and 7 within the heptapeptide backbone of a glycopeptide.22 In known gene clusters, OxyC is highly conserved, yet has sufficiently diverged from other monooxygenases to permit the design of oxyC gene specific degenerate PCR primers. In most sequenced gene clusters, the oxyC gene is positioned directly adjacent to the tailoring enzyme cluster and therefore we felt it would be a good probe to use in our search for tailoring enzyme-rich eDNA clones. PCR screening of the AZ and CA mega-libraries with two sets of oxyC gene specific primers led to the identification of four oxyC gene sequences, three from the CA library and one from the AZ library. eDNA cosmid clones containing each unique oxyC gene sequence were recovered by serial dilution of the individual library aliquots from which the oxyC amplicon was originally generated and then fully sequenced using 454 pyrosequencing.
The annotation of the four oxyC containing clones recovered from the CA and AZ libraries, together with two oxyC containing clones associated with the VEG and TEG gene clusters found in the UT library revealed that each eDNA clone contained genes encoding unique collections of tailoring enzymes (Figure 2 A, B, and C). The three clones isolated from the CA library as well as the tailoring enzyme-rich clone associated with the VEG pathway contain different assortments of genes that encode glycosyl (Figure 2, hollow red) and methyl (Figure 2, blue) transferases. Since the bulk of the structural diversity seen in known glycopeptides arises from differences in the arrangement of sugar, methyl and acyl moieties positioned around the heptapeptide core, the isolation of multiple clones with numerous glycosyl and methyl transferase genes is not unexpected. The tailoring enzyme-rich clone associated with the TEG pathway, as well as the clone AZ205 isolated from the AZ library contain sulfotransferase genes (Figure 2, orange). Only two sulfated glycopeptides have been characterized from cultured bacteria.23,24 Two of the six eDNA derived tailoring enzyme systems contain genes that encode predicted sulfotransferases, enzymes that were previously thought to be rare in glycopeptide biosynthesis. The direct cloning of microbial DNA from environmental samples appears to have provided access to a pool of biosynthetic gene clusters that has not been functionally examined using traditional pure culture-based techniques.
In vivo and in vitro production of glycopeptide derivatives using eDNA derived tailoring enzymes
Telavancin, a semi-synthetic derivative of vancomycin recently approved for use by the FDA, differs from its natural counterpart by the addition of an anionic phosphono group and a long chain hydrophobic moiety.25 The phosphono group, which we believe likely mimics the naturally occurring sulfate functionality that has been seen in a small number of known glycopeptide congeners, is reported to markedly improve the ADME (absorption, distribution, metabolism, and excretion) characteristics of telavancin. Due to the dearth of naturally occurring anionic glycopeptides characterized to date, we chose to explore the possibility of using the genes found on the six eDNA derived tailoring enzyme-rich clones to generate additional sulfated glycopeptides using both in vivo and in vitro derivatization strategies.
For our in vitro derivatization studies each tailoring enzyme-rich clone was initially retrofitted with the genetic elements necessary for transfer and integration into Streptomyces and then transferred by conjugation into Streptomyces toyocaensis.26S. toyocaensis is the natural producer of the monosulfated glycopeptide congener A47934 (1).24 With the exception of the sulfate found on the N-terminal hydroxyphenylglycine (Hpg), A47934 is devoid of tailoring enzyme-derived functionality. Thus, A47934 provides numerous sites where new functionality could be added by eDNA derived tailoring enzyme genes introduced into S. toyocaensis. While it is unlikely that all of the tailoring enzymes from any one clone would be functionally expressed using this strategy, the fact that each clone contains genes for multiple potential modification enzymes significantly increases the likelihood of new functionality being introduced onto A47934.
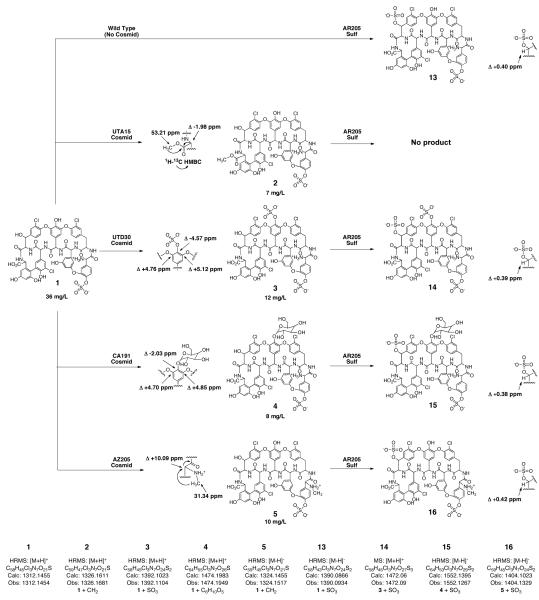
Cultures of both wild type S. toyocaensis as well as S. toyocaensis transformed with each of the six retrofitted eDNA cosmids were grown in Streptomyces antibiotic medium for 7 days and then crude glycopeptide extracts were generated from the resulting culture broths.27 Reversed-phase HPLC analysis of the crude extracts indicated that five out the six S. toyocaensis transformants produced new A47934 derivatives. Each new compound was purified by HPLC and then analyzed by HRMS. Based on the molecular formulas predicted by HRMS, the introduction of either clone UTA15 (VEG pathway) or clone AZ205 leads to the addition of a new methyl substituent onto A47934, the introduction of clone UTD30 (TEG pathway) leads to the addition of a second sulfate onto A47934 and the introduction of either clone CA191 (CA37 pathway) or clone CA1627 (CA915 pathway) leads to the addition of a sugar onto A47934 (Figure 3). The position of each new functional group was determined by comparing 1D and 2D NMR spectra obtained from A47934 with those obtained from each derivative.
Figure 3.
New glycopeptide congeners produced using eDNA derived tailoring enzymes. Compounds 2-5 were produced in vivo by S. toyocaensis transformed with tailoring enzyme-rich eDNA clones. Compounds 13-16 were generated in vitro using recombinantly expressed, eDNA-derived sulfotransferases.
Compound 2, the C-terminal methyl ester of A47934, is produced by S. toyocaensis transformed with clone UTA15. The new oxygen substituted methyl (53.21 ppm in 1H-13C HMQC spectra) is visible as a deshielded singlet (3.85 ppm) in the 1H spectra and an 1H-13C HMBC correlation from this new methyl to the C-terminal carbonyl carbon at 172.32 ppm, allowed us to position the methyl at the C-terminus of the heptapeptide. Clone UTA15 contains three predicted methyltransferase genes (Figure 2A, blue), two of which are predicted to encode N-methyltransferases. The methyltransferase encoded by the third methyltransferase gene found on this clone is therefore likely responsible for the addition of the methyl group to A47934.
The position of the second sulfate found on the disulfated derivative of A47934 (compound 3) that is produced by S. toyocaensis transformed with clone UTD30, was elucidated based on changes in 13C chemical shifts observed for the Hpg at position 4 in the heptapeptide. In compound 3, the chemical shift for carbon 4d shifts up field by 4.70 ppm and the chemical shift for the carbons ortho to this carbon (4c and 4e) shift downfield by 5.12 and 4.76 ppm, compared to the chemical shifts for these same carbons in A47934. This pattern of chemical shift changes is predicted to arise from the sulfation of an aromatic hydroxyl and mimics the changes we observed when the Teg14 sulfotransferase was used to sulfate the teicoplanin aglycone in vitro.28 The teg14 gene is found on clone UTD30 and this sulfotransferase is likely responsible for this modification in vivo as well.
Cultures of S. toyocaensis transformed with either cosmid CA191 or cosmid CA1627 produce spectroscopically identical glycosylated derivatives of A47934 (4). Based on the new chemical shifts and observed coupling constants seen in 1D and 2D NMR experiments this new sugar substituent was determined to be a β-linked D-glucose (see supplementary material).28 As is the case with the disulfated derivative of A47934 described above, changes in 13C chemical shifts for the carbons found in the Hpg at position 4 in the heptapeptide allowed us to place the glucosyl-group on this amino acid (Figure 3). Both cosmid CA191 and cosmid CA1627 contain multiple genes encoding glycosyltransferases (Figure 2B, hollow red) that could be responsible for appending the glucose moiety onto A47934.
Compound 5, the N-terminal methyl amine of A47934, is produced by S. toyocaensis transformed with clone AZ205. As seen with compound 2, the 1H and 1H-13C HMQC NMR spectra from compound 5 indicate the presence of a new heteroatom-substituted methyl substituent (1H 2.48 ppm, 13C 31.34 ppm) (Figure 3). In this case, however, the chemical shifts suggest that the methyl is bound to a nitrogen atom instead of an oxygen atom. An 1H-13C HMBC correlation from the new methyl to the alpha carbon (64.25 ppm) of the Hpg at position one in the heptapeptide confirmed the N-terminal methylation in compound 5. ORF10 in clone AZ205 is predicted to encode an N-methyl transferase (Figure 2C, blue) that is likely responsible for the addition of the methyl to the N-terminus of A47934.
Many of the tailoring enzymes encoded on eDNA clones will not be expressed in a particular heterologous system and the activity of others is undoubtedly thwarted either by incompatibly with the available substrate or by the occupation or absence of the site to be functionalized. In A47934, for example, the hydroxyl on Hpg1 is blocked by a sulfate and Tyr2 is missing the beta carbon oxidation that is often seen in glycopeptides. Even with these limitations five out of six eDNA-derived tailoring enzyme-rich clones resulted in the production of new A47934 derivatives. The addition of clones in a combinatorial fashion or the cloning of individual eDNA derived tailoring enzymes under the control of model promoters would likely provide access to even more highly functionalized derivatives, as would the introduction of these same clones into other genetically tractable glycopeptide producers.
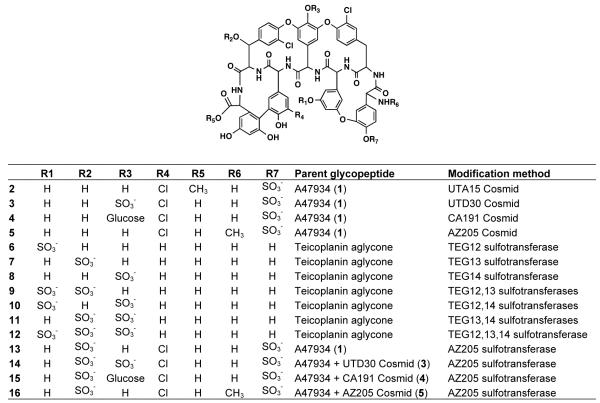
In some instances where tailoring enzymes are not expressed in vivo it should be possible to gain access to the functionality of these enzymes using an in vitro approach. Previously we showed that the three sulfotransferases (Teg12, 13 and 14) from the TEG gene cluster (Figure 2A, orange) could be used in vitro to produce new sulfated teicoplanin aglycone derivatives (6 - 12).11 By coupling the in vivo approach used to generate compounds 2 - 5 with the in vitro approach used to generate compounds 6 - 12, we were able to produce an additional set of sulfated A47934 analogs.11 In the presence of the co-substrate 3′-phosphoadenosine-5′-phosphosulfate (PAPS) and the teicoplanin aglycone, recombinant AZ205 sulfotransferase expressed as a His-tagged protein in E. coli was found to produce a monosulfated teicoplanin aglycone that is spectroscopically identical to compound 7, the sulfated product generated by Teg13. Both sulfotransferases from clone AZ205 and Teg13 therefore specifically sulfate the beta carbon hydroxyl on the beta-hydroxytyrosine (Bht) found at position six in glycopeptides. Sulfonation reactions using recombinant AR205 sulfotransferase were subsequently carried out on A47934 as well as on each of the A47934 derivatives that were generated in vivo. For compounds 1, 3, 4, and 5, we observed complete conversion of each metabolite to a new polysulfated derivative (13, 14, 15, and 16) after 16 h. In each case HRMS confirmed that one additional sulfate had been added to the glycopeptide starting material. As expected from the addition of a sulfate, each beta carbon proton at position six is deshielded by ~0.4 ppm compared to the chemical shift for this same proton in the starting material (Figure 3). No reaction was observed when compound 2 was used as a substrate. Methylation of the C-terminal carboxylic acid must disrupt a key interaction that is required for catalysis or substrate recognition by the AR205 sulfotransferase.
In total, fifteen new sulfated glycopeptides have been generated using the enzymes found on tailoring enzyme-rich clones recovered from the UT, CA and AZ eDNA mega-libraries (Figure 4). This represents a significant addition to the sulfated glycopeptides that have been characterized from cultured Actinomycetes. Minimum inhibitory concentrations (MIC) for each compound were determined using both glycopeptide sensitive (Staphylococcus aureus ATCC 6538P and Enterococcus faecalis ATCC 47077) and glycopeptide resistant (methicillin resistant Staphylococcus aureus USA30029 and VanA-type resistant Enterococcus faecalis EF1830) bacteria (Table 1). As has been seen with other glycopeptide congeners, the more highly functionalized derivatives tend to show higher MICs. While highly functionalized glycopeptide congeners often show increased in vitro MICs, they have proved useful as therapeutics because some exhibit better pharmacological properties in vivo. The systematic introduction of tailoring enzyme-rich clones to easily cultured glycopeptide producers should prove to be a simple yet effective strategy for generating collections of glycopeptides with new functionalization patterns that can be examined for improved biological activities.
Figure 4.
New sulfated glycopeptide derivatives produced by either in vivo or in vitro methods using eDNA derived tailoring enzymes.
Table 1.
MIC (μg/mL) for compounds 1-16 and the teicoplanin aglycone.
| Compound | S. aureus | E. faecalis | ||
|---|---|---|---|---|
| ATCC 6538P |
USA300 | ATCC 47077 |
EF18 | |
| 1 | 2 | 2 | 16 | >64 |
| 2 | 1 | 1 | 8 | >64 |
| 3 | 4 | 8 | 64 | >64 |
| 4 | 4 | 8 | 64 | >64 |
| 5 | 2 | 2 | 16 | >64 |
| 6 | 2 | 2 | 32 | >64 |
| 7 | 1 | 1 | 16 | >64 |
| 8 | 4 | 4 | 64 | >64 |
| 9 | 4 | 2 | 64 | >64 |
| 10 | 8 | 8 | >64 | >64 |
| 11 | 8 | 8 | >64 | >64 |
| 12 | 16 | 8 | >64 | >64 |
| 13 | 2 | 4 | 16 | >64 |
| 14 | 16 | 16 | >64 | >64 |
| 15 | 8 | 16 | 16 | >64 |
| 16 | 2 | 4 | 16 | >64 |
| teicoplanin aglycone |
1 | 1 | 8 | >64 |
Recovery of complete eDNA derived glycopeptide gene clusters
With so few sulfated glycopeptides having been characterized from cultured bacteria, it is clear that the two tailoring enzyme-rich clones that contain sulfotransferases must have arisen from gene clusters that contain the genetic capacity to generate novel glycopeptide congeners. However, in those cases where the tailoring enzyme-rich clones do not contain sulfotransferases, it is not immediately obvious from the information captured on these individual clones whether they arose from gene clusters that have the capacity to encode the production of novel congeners. A more complete picture of each CA gene cluster was therefore obtained from overlapping cosmid clones that were recovered from the CA mega-library in additional rounds of library screening. This re-screening process was continued iteratively until end sequencing of the newly recovered overlapping clones failed to identify additional glycopeptide biosynthetic machinery. After full sequencing and annotation, each set of overlapping cosmid clones could be assembled into a single 100-120 kb contig that appears to contain a full glycopeptide biosynthetic gene cluster (Figure 2B) (GenBank accession numbers: CA37 HM486074, CA878 HM486075, CA915 HM486076). All three gene clusters recovered from the CA library are predicted to contain a complete complement of glycopeptide biosynthetic genes and show the same conserved gene cluster architecture that is seen in most previously sequenced glycopepetide gene clusters (Figure 2A).
The structure of the congener(s) encoded by a glycopeptide gene cluster is determined by the specificity of the nonribosomal peptide synthetases (NRPSs) and the tailoring enzymes that are encoded by the gene cluster. A detailed accounting of these enzymes should therefore shed light onto the structure of the molecule that each CA gene cluster has the potential to encode, and thereby allow for a comparison of these predicted structures to known glycopeptide congeners (Figure 5). Each CA gene cluster contains seven NRPS modules distributed over either four or five open reading frames (Figure 2B, vertical striped green). In addition to the seven NRPS modules that are predicted to produce the heptapeptide, each CA gene cluster encodes four predicted monooxygenases that are homologs of OxyA-D (Figure 2B, brown). OxyA-D catalyze the four oxidative couplings that are needed to generate the four macrocycles seen in many glycopeptides.18
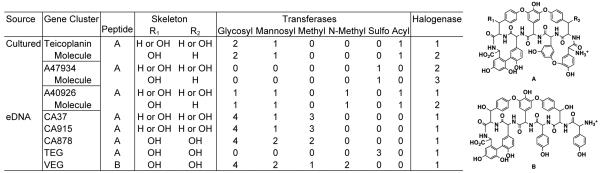
Figure 5.
Structures A and B correspond to the cross-linked core peptides that are predicted to arise from eDNA derived NRPS systems. An inventory of the number and type of tailoring enzymes (or functional group transferases) found in sequenced gene clusters from cultured soil bacteria as well as gene clusters cloned from eDNA is shown. For gene clusters associated with known metabolites, the number of tailoring enzyme derived functional groups found on the encoded metabolite is also shown.
NRPS modules are predicted to carry out a single condensation reaction that extends the growing peptide by one amino acid.31 At a minimum, a module contains a condensation domain for condensing an incoming amino acid with the growing peptide, an adenylation domain for selecting and activating the incoming amino acid and a thiolation domain that carries the growing peptide. The amino acid incorporated by a module is determined by a small number of variable residues found in the adenylation domain.32,33 Table 2 shows these key amino acid specificity residues from each adenylation domain found in the three CA derived glycopeptide gene clusters. Based on a comparison of these residues to those seen in adenylation domains from sequenced glycopeptide NRPS modules, all three CA gene clusters are expected to encode the production of an Hpg-Bht(Tyr)-Dpg-Hpg-Hpg-Bht(Tyr)-Dpg-like heptapeptide (Table 2).34 Potential ambiguities in the predicted peptide arise from the mode of Bht incorporation, which has been reported to occur by two distinct mechanisms during glycopeptide biosynthesis. Bht can be biosynthesized and incorporated directly into the glycopeptide backbone through Bht-specific adenylation domains, or it can be produced by oxidizing tyrosine in a reaction that takes place after the selective incorporation of tyrosine into the growing peptide.35,36 Both CA37 and CA915 are predicted to generate Bht through the latter, tyrosine-oxidation strategy. Modules 2 and 6 from both gene clusters are predicted to incorporate Tyr into the growing peptide. However, as can be seen in a similar analysis of both the teicoplanin and A47934 NRPS modules, the final residues at these positions will be Bht if the tyrosines are subsequently oxidized.
Table 2.
Adenylation domain specificity predictions for all NPRS modules cloned from the CA megalibrary.
| CA37 | CA915 | CA878 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed |
Closest |
Closest Relative |
Amino Acid |
Observed |
Closest |
Closest Relative |
Amino Acid |
Observed |
Closest |
Closest Relative |
Amino Acid |
|
| Mod1 | DAFLLGLL | DAFLLGLL | Tcp-Mod1 | Hpg | DAFLLGLL | DAFLLGLL | Tcp-Mod1 | Hpg | DAFNLGLL | DAFLLGLL | Tcp-Mod1 | Hpg |
| Mod2 | DASTVAAV | DASTVAAV | A47-Mod2 | Tyr | DASTVAAV | DASTVAAV | A47-Mod2 | Tyr | DTSKVAAI | DTSKVAAI | Cep-Mod2 | BHT |
| Mod3 | DAYFSGSL | DAYFLGTL | Tcp-Mod3 | Dpg | DAYFLGTL | DAFLLGLL | A47-Mod1 | Hpg | DAYFQGTF | DAYFLGTL | Tcp-Mod3 | Dpg |
| Mod4 | DIFTLGLL | DIFHLGLL | Cep-Mod4 | Hpg | DIFHLGLL | DIFHLGLL | Cep-Mod4 | Hpg | DIFTLGLL | DIFHLGLL | Cep-Mod4 | Hpg |
| Mod5 | DALLLGLL | DALLLGVG | Tcp-Mod5 | Hpg | DALLLGVG | DALLLGVG | Tcp-Mod5 | Hpg | DAVLLGLL | DAVHLGLL | Cep-Mod5 | Hpg |
| Mod6 | DASTVAAV | DASTVAAV | A47-Mod2 | Tyr | DASTVAAV | DASTIAGV | Tcp-Mod6 | Tyr | DASTLGAI | DASTLGAI | Bal-Mod6 | BHT |
| Mod7 | DPYLGGTL | DPYLGGTL | Cep-Mod7 | Dpg | DPYLGGTL | DPYLGGTL | Cep-Mod7 | Dpg | DPYLGGTL | DPYLGGTL | Cep-Mod7 | Dpg |
Tcp= teicoplanin, A47=A47934, Cep, chloroeremomycin, Bal=balhimycin, Mod=module.
For sequenced gene clusters, the number and type of tailoring enzymes found in these clusters is a reliable predictor of the features that are seen in the most highly functionalized congener it produces (Figure 5).15-18,37 The inventory of tailoring enzymes found in cryptic eDNA-derived glycopeptide gene clusters should in turn be a reasonable predictor of the features found in the congeners produced by these gene clusters. This type of analysis does not allow for precise structural predictions to be made because it does not account for differences in regioselectivity, subtle differences in substrate specificities or the functional order of the tailoring enzymes. It can, however, provide an indication as to whether or not a cryptic gene cluster has the potential to generate a glycopeptide congener with a unique number or type of prosthetic groups.
The CA37 and CA915 gene clusters each contain genes encoding four glycosyltransferases, a mannosyltransferase, a halogenase, and three SAM-dependent methyltransferases. Both gene clusters also contain genes encoding enzymes used in the biosynthesis of a number of different rare glycopeptide-associated sugars, including vancosamine, acosamine or ristosamine.38,39,40 It is not possible from the DNA sequence alone to tell which of these sugars will actually be produced by the two gene clusters. Of the known glycopeptide congeners that contain five sugars, only actaplanin contains the same cross-linked heptapeptide that is predicted to arise from the CA37 and CA915 gene clusters.40 These gene clusters may therefore encode the biosynthesis of actaplanin itself or more highly glycosylated and methylated actaplanin-like glycopeptides.
The CA878 gene cluster contains genes encoding four glycosyltransferases, two mannosyltransferases, a halogenase, and two methyltransferases. This gene cluster also contains homologs of the same sugar biosynthetic genes that are seen in CA37 and CA915. Based on the collection of tailoring enzyme genes present in CA878, this gene cluster would appear to encode the biosynthesis of a ristocetin-like congener. Ristocetin contains the same heptapeptide as is predicted to arise from the CA878 gene cluster as well as six sugars, including a ristosamine, and two methyl substituents.41 The presence of a halogenase in CA878 suggests that this cluster probably does not produce ristocetin itself, but instead produces a halogenated ristocetin-like congener.
Conclusions
First generation glycopeptide antibiotics like vancomycin and teicoplainin are still used extensively in the clinic.14 However, their generally poor pharmacological properties and the appearance of resistant isolates have drawn attention to the increasing need for second-generation glycopeptides with improved ADME characteristics and broader spectra of activity.25,42-44 In response to this increased interest, both biosynthetic and synthetic approaches have been used to generate additional glycopeptide derivatives containing functional groups and functional group arrangements that differ from those seen in naturally occurring metabolites.27,45-47 The tailoring of glycopeptide scaffolds with new eDNA derived tailoring enzymes provides an additional means of generating collections of new glycopeptide derivatives that can be examined in future screening efforts.
With culture independent strategies, it is possible to explore the biosynthetic potential of thousands of unique soil associated bacterial species simultaneously. Large environmental DNA libraries are likely to be bountiful sources of tailoring enzyme-rich clones that could serve as toolboxes for either the in vivo or the in vitro derivatization of many, if not most, different classes of bacterially derived natural products. One key advantage to using this approach, over other enzyme based strategies for generating unnatural natural product variants, is that each tailoring enzyme used in a modification reaction will have evolved to interact with the specific family of natural products being derivatized.
Materials and Methods
Library construction
The eDNA cosmid libraries screened in this study were constructed using previously described methods.48 In brief: soil which had been passed through a 1/8″ screen was mixed with lysis buffer (100 mM Tris-HCl, 100 mM Na EDTA, 1.5 M NaCl, 1% (w/v) cetyl trimethyl ammonium bromide (CTAB), 2% (w/v) SDS, pH 8.0) and incubated at 70°C for 2 h. Soil particulates were removed by centrifugation (3500 × g, 30 min), and crude eDNA was precipitated from the centrifuge-clarified soil suspension with the addition of 0.7 volumes of isopropanol. The crude eDNA extract was collected by centrifugation (3500 × g, 30 min), washed with 70% ethanol and allowed to air dry. DNA was separated from the residual mud using preparative agarose gel electrophoresis (1.0 %), and after 16h at 20 V, the high molecular weight band (>25 kb) was excised from the gel. Purified eDNA was collected by electroelution (2 h at 100V) and concentrated using a centrifugal concentrator (30K MWCO, Vivascience). Blunt ended (End-It Kit, Epicentre) eDNA was ligated into either pWEB or pWEB-TNC, packaged into lambda phage and transfected into E. coli EC100 (Epicentre). 2,178 unique 4,000-5,000 membered libraries were constructed from the transfected E. coli. For each unique library, a matching DNA miniprep (Qiagen) and glycerol stock set was archived.
Clone recovery and sequencing
DNA from the individual 4,000-5,000 membered library pools was arrayed into 8X8 grids. Aliquots of DNA from each pool in a grid were then combined into unique sublibraries. DNA from each sublibrary pool was screened by PCR with two different sets of oxyC specific degenerate primers: PS1FWD: 5′-ATGCTSACSCCSGAGTTCACSGTVCGG-3′ and PS1REV: 5′-CGAGTRRTGGAYGCCGTGCCCGAA-3′; PS2FWD: 5′-CCGCAATTCASCMARKMGAARTCSG-3′ and PS2REV: 5′-TGCCKGGCRAKGAGGTTGTC-3′. PS1 was designed based on the oxyC gene sequences found in five cultured glycopeptide producers: (A. orientalis which produces chloroeremomycin, A. balhimyceticus which produces balhimycin, S. toycaensis which produces A47934, Nonomuraea sp. ATCC 39727 which produces A40926, and A. teichomyceticus which produces teicoplanin). PS2 was designed to encompass the five oxyC sequences used in the design of PS1, as well as the two oxyC sequences previously cloned from eDNA. Each 25 μL PCR reaction contained 0.5 μL of cosmid DNA, 2.5 μL of ThermoPol Buffer (New England Biolabs), 1.25 μL of DMSO, 0.625 μL of each 100mM oligonucleotide primer, 0.5 μL of 10 mM dNTPs mix and 1 unit of Taq DNA Polymerase. For the PS1 primer set, the cycling parameters were as follows: 7 touch down cycles of 95°C for 30 s, 70-62°C (−1°C/cycle) for 30 s and 72°C for 90 s followed by 20 cycles of 95°C for 30 s, 62°C for 30 s, 72°C for 90 s. For the PS2 primer set the following cycling parameters were used: 30 cycles of 95°C for 30 s, 62°C for 30 s, 72°C for 30 s. PCR amplicons of the correct predicted size (700-800 bp for PS1 and 380-410BP for the PS2) were gel purified, A/T cloned into PCR2.1 (Invitrogen) and sequenced. DNA from the individual pools that were used to create each large sublibrary that yielded an oxyC sequence of interest was then screened using the same set of PCR primers. Once an individual sublibrary pool containing that sequence was identified, the cosmid of interest was recovered from the corresponding glycerol stock using successive rounds of serial dilution and PCR screening. Recovered cosmid clones were end-sequenced using vector specific primers: T7 Promoter and M13 Universal Fwd −40. End-sequencing data was used to design new sets of PCR primers that could be used to recover clones containing overlapping DNA sequences. This PCR screening process was repeated iteratively as needed for each gene cluster.
All cosmid clones were 454 sequenced (Roche) by the Memorial Sloan Kettering Cancer Center Genomics Core Laboratory. Raw reads were processed with Newbler (Roche) and Velvet.49 Sequence manipulation and gene identification were carried out using MacVector and BLASTX, respectively. The specificity of each NRPS adenylation domain was determined using NRPS-PKS analysis Web-site.34
Cosmid retrofitting and conjugation into S. toyocaensis
Cosmids containing glycopeptide tailoring enzymes were each retrofitted to contain the genetic elements required for conjugation, integration and selection in Streptomyces. Each cosmid was initially digested with PsiI (New England Biolabs) and then ligated to the 6.7 kb DraI fragment from pOJ436.50 This DraI fragment contains the φC31 integration machinery, the RK2 oriT and the aac(3)IV apramycin resistance gene. Upon transformation into E. coli, retrofitted cosmids could be easily identified by selecting with both ampicillin and apramycin. Correctly retrofitted cosmids were transformed into E. coli S17.1 and conjugated into Streptomyces toyocaensis NRRL 15009 using previously described methods, with a few minor modifications.26 For each conjugation, a 50 mL culture of E. coli S17.1 transformed with an appropriately retrofitted cosmid clone was grown to mid-logarithmic phase (LB with 50 μg/mL apramycin and 10 μg/mL trimethoprim). Cells were then pelleted by centrifugation (3500 × g, 30 min) and the resulting cell pellet was washed twice with 50 mL of antibiotic free LB. Washed E. coli were resuspended in 1 mL SOC (per liter: 20 gm Bacto Tryptone 5 gm Bacto Yeast Extract 10 mM NaCl, 2.5 mM Kcl, 10 mM MgCl2 10 mM MgSO4 20 mM glucose). 20 μL of a heat treated (50°C for 10 min) spore slurry (OD450=40-80) was mixed with 50 μL of washed E. coli and this cell suspension was then plated onto R2 agar. After 16 hours at 30°C, the plates were flooded with 1 mL of sterile water containing 0.5 mg of apramycin and 0.5 mg of nalidixic acid and then returned to the incubator until exconjugants appeared (3-4 days). Exconjugants were restruck on modified Bennett’s agar containing 25 μg/mL apramycin and 25 μg/mL nalidixic acid.
Glycopeptide congener production
S. toyocaensis was initially grown on modified Bennett’s agar for 3 days at 30°C. Single colonies were inoculated into 50 mL of Streptomyces Vegetative Medium (SVM) and incubated at 30°C for 3 days (200 rpm orbital shaking). 50 mL of Streptomyces antibiotic medium containing 1 g/L yeast extract (SAM) was inoculated with a 1:1000 dilution of the 3-days-old starter culture. After 7 days (30°C, 200 rpm orbital shaking) the cell mass was collected by centrifugation (3500 × g, 10 min). The wet weight of the pellet was determined and the pellet was resuspended (vortex, 30s) in 0.75 mL of 2.5% (v/v) NH4OH per gram of wet cell pellet. This basic cell suspension (final pH >11) was centrifuged (3500 × g, 10 min) and the supernatant collected. The pH of the supernatant was adjusted to 7.5 with 1 M HCl, and analyzed directly by HPLC [C18 (4.6 μm × 150 mm), 20 mM ammonium acetate:CH3CN, from 95:5 to 70:30 over 20 min, 1.5 mL/min or C18 (10 μm × 150 mm), 20 mM ammonium acetate:CH3CN, from 95:5 to 70:30 over 20 min, 7 mL/min].
In vitro glycopeptide production
The sulfotransferase gene from AZ205 was amplified from cosmid DNA using the Failsafe system (Epicentre) and the following primers: AZ205SulfFWD(BamHI) GCGCGGATCCATGAACGGAATCCGATGG and AZ205SulfREV (HindIII) GCGCAAGCTTTCCCTAATCAGCGTACCCGTA (restriction sites added for cloning are shown in bold). PCR conditions were as follows: 30 rounds of 30 s at 98°C, 30 s at 65°C, and 1 min at 72°C. The amplicon was doubly digested (BamHI/HindIII) (New England Biolabs), ligated into similarly digested pET28a, and transformed into E. coli BL21(DE3) for protein expression. For protein expression, cultures were grown at 37°C to an OD600 of 0.6, induced with 0.5 mM IPTG and then allowed to shake at 20°C for 16h. The induced culture was then pelleted by centrifugation (3,200 × g for 30 min) and the cell pellet was resuspended in 40 mL of lysis buffer [50 mM Hepes, pH 7.5, 0.5 M NaCl, 5% (vol/vol) glycerol, 20 mM imidazole, pH 8, 10 mM β-mercaptoethanol and 0.5% (vol/vol) Triton X-100]. The resuspended cells were lysed by sonication. The cleared cell lysate (15,000 × g for 30 min) was incubated with 1 mL Ni-NTA resin for 15 min. The resulting slurry was loaded onto a column, washed with 40 mL of lysis buffer, followed by 40 mL of wash buffer [50 mM Hepes, pH 7.5, 0.5 M NaCl, 5% (vol/vol) glycerol, 20 mM imidazole, pH 8.0, and 10 mM β-mercaptoethanol] and the protein was then eluted with the addition of 15 mL of elution buffer (buffer [50 mM Hepes, pH 7.5, 0.5 M NaCl, 5% (vol/vol) glycerol, 125 mM imidazole, pH 8, and 10 mM β-mercaptoethanol]. Sulfation reactions were run at 30°C for 16h in a final volume of 6 mL [15 mM HEPES, pH 7.5, 1 mM 3′-phosphoadenosine-5′-phosphosulfate (PAPS), 0.1 mM DTT]. Each reaction contained 6 mg of purified recombinant protein and 0.5 mg of substrate (compound 1 – 5). Sulfated products (13 – 16) were then purified from heat-killed (99°C, 10min) reaction mixtures by preparative HPLC.
Glycopeptide congener analysis
Hi-Res ESI-MS experiments were carried out either on a Thermo-Fisher LTQ-Obritrap mass spectrometer interfaced with a Dionex U3000 capillary/nano-HPLC system, or by direct injection onto a Waters LCT Premier XE mass spectrometer. NMR data was collected for all samples in a 3:1 mixture of D2O:CD3CN on a Bruker 600 MHz spectrometer and was processed using the MestreNova Software Package (MestreLab Research).
MIC determination
Stationary phase cultures of Staphylococcus aureus and Enterococcus faecalis, grown in tryptic soy broth or brain heart infusion broth, respectively, were diluted 1:106. Individual wells in 96 well microtiter plates were then filled with 100 μL of these dilute cultures. A stock solution of an individual compound dissolved in dimethylsulfoxide was diluted 1:100 into culture media and 100 μL of this solution was added to the first well of a row in the filled microtiter plate. This well was then serially diluted 1:2 across the plate. Microtiter plates were incubated at 37°C with 100 rpm orbital shaking for 18 hours. MIC values are reported as the lowest concentration at which no bacterial growth was observed.
Supplementary Material
Acknowledgements
This work was supported by NIH GM077516, NIH MSTP GM07739 (J.W.C.), the Howard Hughes Medical Institute and the Beckman Foundation. We thank Dr. Alexander Tomasz for providing S. aureus USA300 and E. faecalis EF18.
References
- (1).Buckingham J. Dictionary of natural products. 1st ed Chapman & Hall; London; New York: 1994. [Google Scholar]
- (2).Firn RD, Jones CG. Nat. Prod. Rep. 2003;20:382–91. doi: 10.1039/b208815k. [DOI] [PubMed] [Google Scholar]
- (3).Rappé MS, Giovannoni SJ. Annu. Rev. Microbiol. 2003;57:369–94. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- (4).Torsvik V, Goksøyr J, Daae FL. Appl. Environ. Microbiol. 1990;56:782–7. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Torsvik V, Ovreas L, Thingstad TF. Science. 2002;296:1064–6. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- (6).Seow KT, Meurer G, Gerlitz M, Wendt-Pienkowski E, Hutchinson CR, Davies J. J. Bacteriol. 1997;179:7360–8. doi: 10.1128/jb.179.23.7360-7368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Chem.Biol. 1998;5:R245–9. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- (8).Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C, Tiong CL, Gilman M, Osburne MS, Clardy J, Handelsman J, Goodman RM. Appl. Environ. Microbiol. 2000;66:2541–7. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Piel J. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14002–7. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sudek S, Lopanik NB, Waggoner LE, Hildebrand M, Anderson C, Liu H, Patel A, Sherman DH, Haygood MG. J. Nat. Prod. 2006;70:67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- (11).Banik JJ, Brady SF. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17273–7. doi: 10.1073/pnas.0807564105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Fisch KM, Gurgui C, Heycke N, van der Sar SA, Anderson SA, Webb VL, Taudien S, Platzer M, Rubio BK, Robinson SJ, Crews P, Piel J. Nat. Chem. Biol. 2009;5:494–501. doi: 10.1038/nchembio.176. [DOI] [PubMed] [Google Scholar]
- (13).Nicolaou KC, Boddy CNC, Bräse S, Winssinger N. Angew. Chem. Int. Ed. 1999;38:2096–2152. doi: 10.1002/(sici)1521-3773(19990802)38:15<2096::aid-anie2096>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- (14).Pace JL, Yang G. Biochem. Pharmacol. 2006;71:968–80. doi: 10.1016/j.bcp.2005.12.005. [DOI] [PubMed] [Google Scholar]
- (15).Pelzer S, Süssmuth R, Heckmann D, Recktenwald J, Huber P, Jung G, Wohlleben W. Antimicrob. Agents Chemother. 1999;43:1565–73. doi: 10.1128/aac.43.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).van Wageningen AM, Kirkpatrick PN, Williams DH, Harris BR, Kershaw JK, Lennard NJ, Jones M, Jones SJ, Solenberg PJ. Chem. Biol. 1998;5:155–62. doi: 10.1016/s1074-5521(98)90060-6. [DOI] [PubMed] [Google Scholar]
- (17).Sosio M, Kloosterman H, Bianchi A, de Vreugd P, Dijkhuizen L, Donadio S. Microbiology. 2004;150:95–102. doi: 10.1099/mic.0.26507-0. [DOI] [PubMed] [Google Scholar]
- (18).Pootoolal J, Thomas MG, Marshall CG, Neu JM, Hubbard BK, Walsh CT, Wright GD. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8962–7. doi: 10.1073/pnas.102285099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Sosio M, Stinchi S, Beltrametti F, Lazzarini A, Donadio S. Chem. Biol. 2003;10:541–9. doi: 10.1016/s1074-5521(03)00120-0. [DOI] [PubMed] [Google Scholar]
- (20).Donadio S, Sosio M, Stegmann E, Weber T, Wohlleben W. Mol. Genet. Genomics. 2005;274:40–50. doi: 10.1007/s00438-005-1156-3. [DOI] [PubMed] [Google Scholar]
- (21).Kim J, Feng Z, Bauer JD, Kallifidas D, Calle PY, Brady SF. Biopolymers. 2010;93:833–844. doi: 10.1002/bip.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Bischoff D, Pelzer S, Bister B, Nicholson GJ, Stockert S, Schirle M, Wohlleben W, Jung G, Süssmuth RD. Angew. Chem. Int. Ed. 2001;40:4688–4691. doi: 10.1002/1521-3773(20011217)40:24<4688::aid-anie4688>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- (23).Holdom KS, Maeda H, Ruddock JC, Tone J. Pfizer, Ltd.; 1988. p. 265143. EP-B. [Google Scholar]
- (24).Boeck LD, Mertz FP. J. Antibiot. (Tokyo) 1986;39:1533–40. doi: 10.7164/antibiotics.39.1533. [DOI] [PubMed] [Google Scholar]
- (25).Leadbetter MR, Adams SM, Bazzini B, Fatheree PR, Karr DE, Krause KM, Lam BM, Linsell MS, Nodwell MB, Pace JL, Quast K, Shaw JP, Soriano E, Trapp SG, Villena JD, Wu TX, Christensen BG, Judice JK. J. Antibiot. (Tokyo) 2004;57:326–36. doi: 10.7164/antibiotics.57.326. [DOI] [PubMed] [Google Scholar]
- (26).Matsushima P, Baltz RH. Microbiology. 1996;142(Pt 2):261–7. doi: 10.1099/13500872-142-2-261. [DOI] [PubMed] [Google Scholar]
- (27).Lamb SS, Patel T, Koteva KP, Wright GD. Chem. Biol. 2006;13:171–81. doi: 10.1016/j.chembiol.2005.12.003. [DOI] [PubMed] [Google Scholar]
- (28).Pretsch E. Structure determination of organic compounds : tables of spectral data. 4th ed Springer; New York: 2009. [Google Scholar]
- (29).Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- (30).Mato R, de Lencastre H, Roberts RB, Tomasz A. Microb. Drug. Resist. 1996;2:309–17. doi: 10.1089/mdr.1996.2.309. [DOI] [PubMed] [Google Scholar]
- (31).Marahiel MA, Stachelhaus T, Mootz HD. Chem. Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- (32).Stachelhaus T, Mootz HD, Marahiel MA. Chem. Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- (33).Challis GL, Ravel J, Townsend CA. Chem. Biol. 2000;7:211–24. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- (34).Ansari MZ, Yadav G, Gokhale RS, Mohanty D. Nucleic Acids Res. 2004;32:W405–13. doi: 10.1093/nar/gkh359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Puk O, Bischoff D, Kittel C, Pelzer S, Weist S, Stegmann E, Süssmuth RD, Wohlleben W. J. Bacteriol. 2004;186:6093–100. doi: 10.1128/JB.186.18.6093-6100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Stinchi S, Carrano L, Lazzarini A, Feroggio M, Grigoletto A, Sosio M, Donadio S. FEMS Microbiol. Lett. 2006;256:229–35. doi: 10.1111/j.1574-6968.2006.00120.x. [DOI] [PubMed] [Google Scholar]
- (37).Goldstein BP, Selva E, Gastaldo L, Berti M, Pallanza R, Ripamonti F, Ferrari P, Denaro M, Arioli V, Cassani G. Antimicrob. Agents Chemother. 1987;31:1961–6. doi: 10.1128/aac.31.12.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Chen H, Thomas MG, Hubbard BK, Losey HC, Walsh CT, Burkart MD. Proc. Natl. Acad. Sci. U. S. A. 2000;97:11942–7. doi: 10.1073/pnas.210395097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Heald SL, Mueller L, Jeffs PW. J. Antibiot. (Tokyo) 1987;40:630–45. doi: 10.7164/antibiotics.40.630. [DOI] [PubMed] [Google Scholar]
- (40).Debono M, Merkel KE, Molloy RM, Barnhart M, Presti E, Hunt AH, Hamill RL. J. Antibiot. (Tokyo) 1984;37:85–95. doi: 10.7164/antibiotics.37.85. [DOI] [PubMed] [Google Scholar]
- (41).Fehlner JR, Hutchinson RE, Tarbell DS, Schenck JR. Proc. Natl. Acad. Sci. U. S. A. 1972;69:2420–1. doi: 10.1073/pnas.69.9.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Lancet. 1997;350:1670–3. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- (43).Cetinkaya Y, Falk P, Mayhall CG. Clin. Microbiol. Rev. 2000;13:686–707. doi: 10.1128/cmr.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Ge M, Chen Z, Onishi HR, Kohler J, Silver LL, Kerns R, Fukuzawa S, Thompson C, Kahne D. Science. 1999;284:507–11. doi: 10.1126/science.284.5413.507. [DOI] [PubMed] [Google Scholar]
- (45).Solenberg PJ, Matsushima P, Stack DR, Wilkie SC, Thompson RC, Baltz RH. Chem. Biol. 1997;4:195–202. doi: 10.1016/s1074-5521(97)90288-x. [DOI] [PubMed] [Google Scholar]
- (46).Chen L, Walker D, Sun B, Hu Y, Walker S, Kahne D. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5658–63. doi: 10.1073/pnas.0931492100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Kahne D, Leimkuhler C, Lu W, Walsh C. Chem. Rev. 2005;105:425–48. doi: 10.1021/cr030103a. [DOI] [PubMed] [Google Scholar]
- (48).Brady SF. Nat. Protoc. 2007;2:1297–305. doi: 10.1038/nprot.2007.195. [DOI] [PubMed] [Google Scholar]
- (49).Zerbino DR, Birney E. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Matsushima P, Broughton MC, Turner JR, Baltz RH. Gene. 1994;146:39–45. doi: 10.1016/0378-1119(94)90831-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.