Abstract
A specialized nucleosome is a component of all eukaryotic kinetochores. The core of this nucleosome contains a centromere-specific histone, CENP-A (the Cse4 gene product in budding yeast), instead of the usual H3. Assembly of a centromeric nucleosome depends on a specific chaperone, called Scm3 in yeast and HJURP in higher eukaryotes. We describe here the structure of a complex formed by an N-terminal fragment of Scm3 with the histone-fold domains of Cse4, and H4, all prepared as recombinant proteins derived from the budding yeast Kluyveromyces lactis. The contacts of Scm3 with Cse4 explain its selectivity for the centromere-specific histone; key residues at the interface are conserved in HJURP, indicating a common mechanism for centromeric-histone deposition. We also report the structure of a (Cse4 : H4)2 heterotetramer; comparison with the structure of the Scm3:Cse4:H4 complex shows that tetramer formation and DNA-binding require displacement of Scm3 from the nucleosome core. The two structures together suggest that specific contacts between the chaperone and Cse4, rather than an altered overall structure of the nucleosome core, determine the selective presence of Cse4 at centromeres.
Faithful transfer of genetic information from a mother cell is crucial for the survival of its daughters. During mitosis, an assembly of protein complexes, the kinetochore, connects each centromere with spindle microtubules and monitors bipolar attachment (1). A hallmark of kinetochores in all eukaryotes is a centromere-specific nucleosome, in which a centromere-specific H3 variant, CENP-A (sometimes designated CenH3 and known as Cse4 in budding yeast), replaces the canonical H3 (2–5). CENP-A/Cse4 is very well conserved, despite the divergence of centromeric DNA from budding yeast (which have short “point centromeres,” approximately 150–220 bp in length) to higher eukaryotes (with much longer, “regional centromeres”) (6).
In point-centromere yeasts, a kinetochore-associated protein, Scm3, targets Cse4 nucleosomes to centromeres (7–9). Scm3, which associates specifically with Cse4 and not with H3, has orthologs in fission-yeast (Scm3SP) and in higher eukaryotes (HJURP) (10–15). Centromeric localization of Scm3 is determined by Ndc10, a component of centromere-binding-factor 3 (CBF3) (8); elimination of CBF3 blocks deposition of the centromeric nucleosome (16). In organisms with regional centromeres, CENP-A deposition appears to be epigenetically directed. Localization of the Scm3 homolog depends on a set of proteins known as the Mis16–Mis18 complex (13, 17), as well as on the presence of CENP-A in neighboring nucleosomes and on defined H3 modifications in the interspersed chromatin (18).
The so-called “CENP-A targeting domain” (CATD)—loop 1 and helix II of the CENP-A/Cse4 histone-fold domain (HFD)—is crucial for centromeric-histone function (19). Substitution of several CATD residues with their H3 counterparts disrupts CENP-A localization (20), and a chimeric H3-CATD histone functionally replaces CENP-A in vivo (19). Evidence that Scm3 and HJURP are assembly chaperones for Cse4 and CENP-A, respectively, therefore suggests that these proteins recognize features of the CATD (7–14).
We report here the crystal structure (at 2.3-Å resolution) of a complex containing Scm3, Cse4, and H4, all from the budding yeast Kluyveromyces lactis. The structure shows that Scm3 interacts with Cse4 helix II and that its contacts explain selectivity for Cse4. Comparison of this structure with those of a (Cse4 : H4)2 heterotetramer, also reported here, and of a conventional nucleosome (21) shows that tetramer formation and DNA-binding will displace Scm3. Conservation in HJURP and Scm3SP of key residues at the Scm3:Cse4 interface indicates a common mechanism by which these chaperones recognize CENP-A/Cse4 and deposit it at centromeres. Our structure thus suggests that the principal difference between point and regional centromeres is in recruitment of the centromeric-histone–chaperone and that the structure and higher-order interactions of the centromere-specific nucleosome itself are essentially the same in all eukaryotes.
Results
Cse4-Interacting Segment of Scm3.
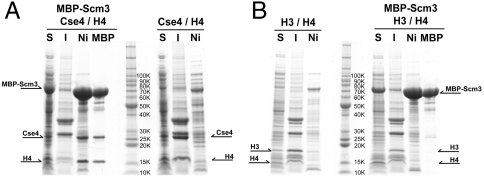
We have coexpressed yeast (K. lactis) Cse4, H4, and Scm3 in bacterial cells and isolated a soluble complex of the three proteins (Fig. 1A). Omission of Scm3 results in insoluble, aggregated histones. This folding-chaperone activity of Scm3 is specific for Cse4; coexpression of Scm3 with H3 and H4 does not yield a soluble product (Fig. 1B). The Cse4:H4:Scm3 complex that results from bacterial coexpression is a 1∶1∶1 heterotrimer as determined by sedimentation equilibrium centrifugation (expected mass = 56.2 kDa; calculated mass = 56 ± 1 kDa; Fig. S1A). Limited proteolysis, followed by mass spectrometric analysis of the products, identified the minimal fragment of each subunit necessary and sufficient to maintain a soluble heterotrimer. Those components are residues 53–115 of Scm3 and the HFDs of Cse4 and H4 (Fig. S1B). The Scm3 segment is moderately well conserved in mammalian HJURP as well as in other budding yeast. The corresponding bacterial coexpression experiment with Scm3, Cse4, and H4 from Saccharomyces cerevisiae did not yield a soluble product, but refolding of Cse4:H4 mixtures in vitro showed that addition of Scm3 led to formation of soluble heterotrimer, with properties similar to those for the K. lactis product (Fig. S2 A–D).
Fig. 1.
Scm3 as a Cse4-specific chaperone. (A) Polycistronic expression of K. lactis Cse4 and H4 in the presence or absence of Scm3. Scm3 has tandem N-terminal His6 tag and MBP tag; Cse4 and H4 have N-terminal His6 tag. S, soluble fraction; I, insoluble fraction; Ni, Ni-NTA eluate; MBP, amylose eluate. The soluble fraction of cell lysates was applied to a Ni-NTA affinity column, and the eluate was then applied to an amylose affinity column. The Ni-NTA eluate contains all soluble, His-tagged proteins; the amylose eluate identifies proteins that form stable complexes with MBP–Scm3. (B) Polycistronic expression of K. lactis H3 and H4 in the presence or absence of Scm3.
Structure of Scm3:Cse4:H4.
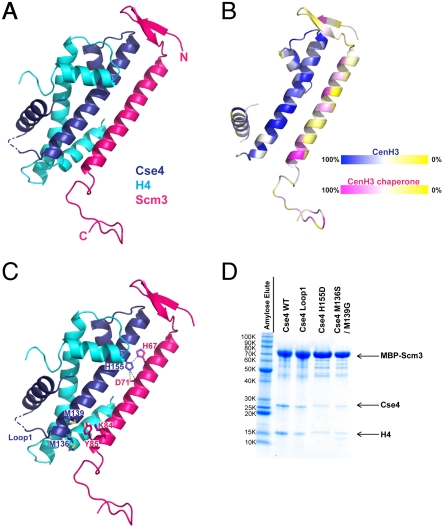
We crystallized the complex obtained by coexpression of Scm3 (41–115), Cse4 (103–184), and H4 (22–103), recorded diffraction to 2.3-Å resolution (Table S1 and Fig. S1C and D), and determined the structure by molecular replacement as outlined in Methods. The Cse4:H4 heterodimer resembles closely the H3:H4 heterodimer in a canonical nucleosome (Fig. 2A). The Scm3 fragment folds into a short β-ribbon, a long α-helix, and a C-terminal loop; the loop is probably flexible, and its observed conformation may depend in part on crystal contacts. The N- and C-terminal ends of the Scm3 fragment (residues 41–43 and 104–115) are disordered.
Fig. 2.
Structure of the Scm3:Cse4:H4 heterotrimer. (A) Ribbon diagram of the Scm3:Cse4:H4 heterotrimer. Residues 44–103 of Scm3, residues 108–180 of Cse4, and residues 24–97 of H4 are in red, dark blue, and cyan, respectively. (B) Sequence conservation in Scm3 and Cse4. Colors ramped from red and blue, respectively, to yellow, corresponding to degree of conservation in multiple sequence alignments (Fig. S4 A and B) of Scm3 and Cse4 from K. lactis, S. cerevisiae, Schizosaccharomyces pombe, bovine, mouse, and human. (C) Some key interacting residues of Cse4 and Scm3. (D) Amylose pull-down with MBP–Scm3 (the bait) coexpressed with H4 and mutants of Cse4. Mutants of Cse4 with substitutions at various positions of their H3 counterparts were coexpressed with MBP–Scm3 and H4. Substitutions in Cse4 loop1 are residues 76–88 of H3 (AQDFKTDLRFQSS) for residues 150–165 of Cse4 (TDQFTTESEPLRWQSM). The volume of sample used for SDS-PAGE was adjusted based on the amount of MBP–Scm3.
Residues 44–103 of Scm3 have extensive contact with both Cse4 and H4; the interface residues, with a mixture of polar and nonpolar side chains, are very well conserved among point-centromere yeasts (Fig. S3A). We illustrate the distribution of conserved residues from a broader alignment with orthologs from yeast to humans (Fig. 2B and Fig. S4 A and B). Most of the conserved positions in both partners are at the chaperone–histone interface, suggesting that the CENP-A:HJURP complex has the same structure and selectivity mechanism. The chaperone–histone interface in the Asf1:H3:H4 heterotrimer (22), the only other histone–chaperone complex for which a structure has been determined, is quite different from the interface in Scm3:Cse4:H4, with only slight overlap if H3 and Cse4 are superposed.
Scm3 Selectivity.
To determine the basis of the selectivity for the kinetochore-specific H3 ortholog, we made the following Cse4 variants (Fig. 2C). (i) Substitution of loop 1 from H3 into Cse4. This loop is the most divergent segment in the two orthologs, but it has no contact with Scm3. (ii) The double mutation M136S/M139G, in which two methionine residues at the Scm3–Cse4 interface are replaced by their H3 counterparts. The methionines contact Lys84 and Tyr85, two residues conserved in nearly all Scm3 orthologs (Fig. S4B). (iii) The point mutation H155D. The histidine in Cse4 bridges between His67 and Asp71 of Scm3; the Asp155 of H3 should change an attractive electrostatic configuration into a repulsive one. We coexpressed the three Cse4 variants with maltose-binding protein (MBP)–Scm3 and H4 and monitored the efficiency of soluble heterotrimer formation by pull-down assays (Fig. 2D). Both mutations in helix II of the HFD (the double methionine mutation and the histidine to aspartate mutation) decreased the yield of folded heterotrimer, while the loop 1 substitution had a minor effect. These results are consistent with previous work showing that Cse4 substitution mutations that included H155D led to severe chromosome loss in vivo (23). The extended interface between Cse4 helix II and the long helix in Scm3 thus appears to be a critical determinant of Scm3:Cse4 specificity.
After this work was complete and under review, two related papers appeared. One showed a solution NMR structure of a single-chain construct that included the S. cerevisiae Cse4 HFD, Scm3 (residues 93–169, corresponding to 64–140 in K. lactis), and H4 (24). The construct deleted helix I of H4, however, and this deletion, together with omission of about 20 residues in the N-terminal part of the conserved region of Scm3, appears to have generated severe distortions in about half the structure, leading to incorrect conclusions about histone conformational differences and about a number of the Scm3–Cse4 contacts (Fig. S5 A and B). The undistorted C-terminal end of the long Scm3 helix has contacts with Cse4 identical to those in our structure. The other recent paper reported a structure of an N-terminal fragment of human HJURP in complex with the HFDs of CENP-A and H4 (25). That structure is in substantial agreement with ours, reinforcing the conclusions we draw here about conservation from yeast to human (Fig. S5A). The HJURP fragment caps, with a small β-sheet, the N-terminal end of the long CENP-A helix—the opposite end from the one capped by a β-hairpin in the Scm3–Cse4–H4 complex described here. In both cases, however, the interaction is expected to block DNA binding.
(Cse4 : H4)2 Heterotetramer Structure.
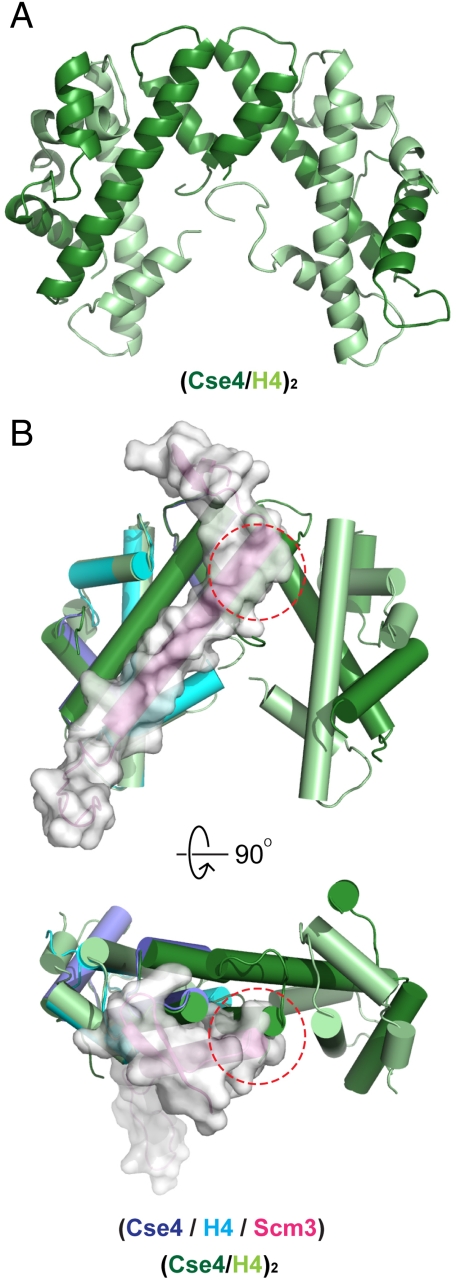
In the course of screening the Scm3:Cse4:H4 complex for crystallization, we also obtained crystals of the (Cse4 : H4)2 heterotetramer, which presumably had formed because of dissociation of Scm3 under the crystal growth conditions (Fig. 3A). We determined the structure at a resolution of 2.6 Å, as outlined in Methods. There are two heterotetramers in the asymmetric unit; they overlay well on each other, showing that the Cse4:Cse4 interface that generates them is robust and insensitive to molecular packing differences between the two crystallographic environments (Fig. S6). Superposition of the Scm3:Cse4:H4 complex and the (Cse4 : H4)2 heterotetramer shows that Scm3 overlaps the Cse4:Cse4 interface in the latter, explaining why the complex with Scm3 is a stable, 1∶1∶1 heterotrimer (Fig. 3B). The N-terminal β-ribbon of Scm3 that covers loop 2 in the Cse4 HFD occupies space that would contain DNA backbone in a nucleosome. The presence of Scm3 is therefore incompatible both with Cse4:H4 tetramerization and DNA-binding.
Fig. 3.
Structure of the (Cse4 : H4)2 heterotetramer and structural alignment with the Scm3:Cse4:H4 heterotrimer. (A) Ribbon diagram of the (Cse4 : H4)2 heterotetramer. Cse4 is in dark green; H4 is in light green. (B) Superposition of the Scm3:Cse4:H4 and (Cse4 : H4)2 heterotetramers. The structures of Cse4 and H4 in the heterotrimer align well with those in the heterotetramer. In the superposition, the central helix of Scm3 clashes with helix III of neighboring Cse4 in the tetramer (red dashed circle), illustrating that Scm3 binding and formation of a Cse4:H4 tetramer are mutually exclusive.
In a recently published crystallographic analysis of a (CENP-A : H4)2 heterotetramer (26), three structural features were cited that appeared to distinguish it from the canonical (H3 : H4)2 heterotetramer: (i) a small rotation of the tetramer interface, generating a somewhat more compact overall structure; (ii) a bulge, with enhanced positive charge, on the molecular surface at the top of loop 1; (iii) a set of hydrophobic contacts that appears to restrict conformational flexibility. These differences are all within the so-called CATD, the region critical for centromeric localization of CENP-A. Comparison with both (CENP-A : H4)2 [Protein Data Bank (PDB) ID code 3NQJ] and (H3 : H4)2 (PDB ID code 1ID3) shows that the (Cse4 : H4)2 heterotetramer is intermediate in compactness between the other two. The largest local displacements are about 5 Å at the positions most distant from the tetramer interface (Fig. S7 A and B). We note, however, that the reference (H3 : H4)2 structure is in the context of an intact nucleosome, in which either binding of DNA or association with the H2A:H2B heterodimers may influence the conformation of the central (H3 : H4)2 heterotetramer. We therefore cannot judge whether the differences are significant. Moreover, the surface bulge in the CATD of Cse4 has an overall negative charge, as in H3, rather than the positive charge seen in CENP-A (Fig. S7C). Finally, inspection of yeast H3 sequences suggests that the “hydrophobic stitching” that appears to characterize the (CENP-A : H4)2 structure does not carry over to the yeast orthologs (Fig. S7D). Thus, none of the observed differences between (CENP-A : H4)2 and (H3 : H4)2 are conserved in budding yeast. These results suggest that the selective presence of CENP-A/Cse4 at centromeres derives from the specificity of its chaperone, HJURP or Scm3, as determined by the molecular interactions outlined above, rather than by an altered global geometry of the nucleosome core.
Discussion
Conflicting experimental data have led to three quite different models for the composition of the centromeric nucleosome and even to suggestions that its structure is different in different organisms. (i) Addition of Scm3 displaces H2A:H2B in vitro from a Cse4-containing histone octamer, leading to the suggestion that the centromeric nucleosome core is a heterohexamer, (Scm3 : Cse4 : H4)2 (7). (ii) The thickness of centromeric nucleosomes from Drosophila and human cells, determined by atomic force microscopy, appears to be about half that of ordinary nucleosomes, leading to a proposal that the former contain a CenH3:H4:H2A:H2B “hemisome” (27, 28). (iii) Careful measurements of content and stoichiometry appear to be compatible with a conventional histone octamer, in which Cse4 replaces H3 (29). Our structural data exclude the heterohexamer model, by showing that Scm3 binding and (Cse4 : H4)2 tetramer formation are mutually exclusive (Fig. 3B). They are consistent with a recent observation that overexpression of Cse4 can rescue the lethality of an Scm3 deletion, which implies that Scm3 is not a structural component of the mature centromeric nucleosome (29). Our data also disfavor a hemisome model, because the tetramer interface in our (Cse4 : H4)2 crystals is stable and independent of the crystal-packing environment (Fig. S6). Moreover, mutations in the tetramer interface that would not interfere with Scm3 chaperone function prevent formation of Cse4 nucleosomes (29)—results best explained by assuming that the tetramer interface in our crystals is indeed present in the nucleosome. Our structures are fully compatible with an octamer model (although they cannot directly demonstrate its validity).
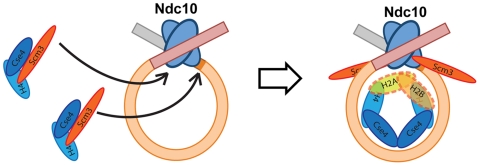
Our structural and biochemical experiments, along with the body of published data, are consistent with the picture of centromeric nucleosome assembly illustrated schematically in Fig. 4. The centromere-binding factors CBF3, Cbf1, and Mif2 (the yeast ortholog of CENP-C) (30) mark the location of Cse4 deposition (1, 31, 32). Ndc10, a dimeric component of CBF3, generates a DNA loop (U.-S.C. and S.C.H., manuscript in preparation) and recruits two Scm3:Cse4:H4 heterotrimers. Interactions of the histones with DNA and formation of the Cse4:Cse4 interface promote dissociation of Scm3 from Cse4:H4 and assembly of an octameric nucleosome core. Ndc10-dependent localization of Scm3 at centromeres (8) and tight association of Scm3 with Ndc10 (U-S. C. and S.C.H., manuscript in preparation) probably explain the continuing presence of Scm3 at yeast kinetochores after full assembly of the centromeric nucleosome (8). We suggest that CBF3, Cbf1, and/or Mif2 dictate the more limited extent of DNA winding around the Cse4-containing nucleosome core than around a conventional histone octamer.
Fig. 4.
Schematic model for Cse4:H4 incorporation at budding-yeast centromeres. Model for Cse4 nucleosome assembly. Scm3 stabilizes the Cse4:H4 heterodimer in solution by forming an Scm3:Cse4:H4 heterotrimeric complex, which is localized to a centromere through an interaction of Scm3 with Ndc10 (8). An Ndc10 dimer, as a part of the CBF3 complex, recognizes and associates with centromeric DNA and generates a DNA loop (U-S.C. and S.C.H., manuscript in preparation). Tetramer formation and DNA association detach Scm3 from Cse4:H4, but Scm3 remains at the centromere through its contact with Ndc10.
The presence of CENP-A/Cse4-containing nucleosomes at a centromere is essential for subsequent steps in kinetochore assembly. Because substitution into H3 of the CATD from CENP-A rescues cell viability (19), the CATD must provide a surface not only for chaperone recognition but also for association of one or more additional kinetochore subassemblies. The chaperone contact does not include the CATD loop I component—a somewhat negatively charged bulge in K. lactis Cse4. This bulge must therefore have some role in nucleating the next steps in the hierarchy of kinetochore construction. The helix II surface that contacts Scm3 could also have a role, after displacement of Scm3 by DNA and (probably) by H2A:H2B. Even the full CATD, of which there is only a pair in the single centromeric nucleosome of budding yeast, is a relatively small platform on which to build a complete kinetochore superstructure.
The results reported here show that Scm3, acting as a Cse4-specific histone–chaperone, can localize a Cse4 nucleosome to a centromere, by interacting in turn with Ndc10. Moreover, conservation of key residues in centromeric H3s and their chaperones points to a conserved recognition mechanism. Indeed, the mammalian HJURP:CENP-A:H4 complex does resemble closely the budding yeast Scm3:Cse4:H4 heterotrimer (25). This conclusion suggests that restriction of centromeric nucleosomes to centromeres depends primarily on the mechanism for localizing the chaperone complexes, rather than on structural properties of the nucleosome itself. Deposition of a single Cse4 nucleosome at a point centromere requires a specific, DNA-binding complex, CBF3, to identify the centromere after each replication cycle. At a regional centromere, multiple CENP-A nucleosomes and specific modifications (H3K4me2 and H3K36me2) of the interspersed H3-containing nucleosomes (18, 33) guarantee a persistent centromeric label, even when a subset of those nucleosomes has dissociated, thus creating the conditions for a self-renewing deposition mechanism. The link to HJURP recruitment may involve the Mis16–Mis18 complex or their homologs (13, 17) and noncoding transcription (18, 25). The structures of the resulting centromeric nucleosomes are probably very similar to those on point centromeres, deposited by CBF3–Scm3.
Methods
Cloning, Expression, and Protein Purification.
Codon-optimzed cDNAs of full-length K. lactis Cse4 and H4 were synthesized by Geneart and subcloned into a pET vector modified for ligation-independent cloning (LIC), which contains an N-terminal His6 tag and a tobacco etch virus (TEV) protease cleavage site. A clone of full-length K. lactis Scm3 was obtained by PCR amplification from a genomic library of K. lactis and subcloned into a pET vector modified for LIC, which has both an N-terminal His6 tag and MBP tag with a TEV protease cleavage site. Constructs of his-Cse4, his-H4, and his-MBP–Scm3 including a ribosome-binding site were PCR amplified and used to make a polycistronic construct with three genes. The same strategy was used to generate all truncated and mutated constructs of Scm3, Cse4, and H4. Polycistronic constructs were then transformed into Rosetta (DE3) pLysS Escherichia coli competent cells (Novagen) and used for protein expression. Cells were grown at 37 °C in ZYM-5052 autoinducible medium (34); the temperature was switched to 25 °C when the OD reached 1.0, and the incubation continued overnight at the lower temperature. Cells were harvested and resuspended in binding buffer (30 mM Tris-HCl, pH 8.0, 500 mM NaCl, 3 mM β-mercaptoethanol) with protease inhibitor cocktail tablets. Cells were sonicated, and the cell debris was removed by centrifugation at 20,000 × g for 1 h. Soluble fractions were then passed through a 0.45-μm filter and applied to a Ni-NTA column (Qiagen) preequilibrated with binding buffer. The column was washed with binding buffer and 30 mM Tris-HCl, pH 8.0, 500 mM NaCl, 30 mM imidazole, 3 mM β-mercaptoethanol and then eluted with 30 mM Tris-HCl. pH 8.0, 500 mM NaCl, 300 mM imidazole, 3 mM β-mercaptoethanol. The elution fraction of Ni-NTA was then mixed with amylose resins preequilibrated with binding buffer and incubated for 1 h at 4 °C with rotation. Unbound proteins were then washed out with binding buffer and eluted with 30 mM Tris-HCl, pH 8.0, 500 mM NaCl, 20 mM maltose, 3 mM β-mercaptoethanol. The elution fraction was dialyzed overnight at 4 °C in 30 mM Tris-HCl. pH 8.0, 500 mM NaCl, 1 mM EDTA, and 1 mM DTT with TEV protease, and applied to a second Ni-NTA column to remove his-MBP. The flow-through fraction from Ni-NTA was then applied into HiTrap SP column 5 mL (GE healthcare) preequilibrated with 30 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM DTT. Fractions containing target proteins were obtained by gradually increasing NaCl concentration up to 1 M, and fractions were then pooled, concentrated, and applied to a Superdex 200 size exclusion column (Prep grade 16/60: GE healthcare) preequilibrated with 30 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM TCEP [tris(2-carboxyethyl)phosphine]. Fractions containing target proteins were pooled and concentrated and used for biochemical and structural studies.
Sedimentation Equilibrium Analytical Ultracentrifugation.
To determine the oligomerization states of K. lactis Scm3:Cse4:H4, sedimentation equilibrium experiments were performed in a Beckman-Coulter ProteomeLab XL-I analytical ultracentrifuge equipped with 12-mm Epon double-sector cells in an An-60 Ti rotor (Beckman). Three different protein concentrations were prepared, corresponding to an absorbance at 280 nm of 0.25–0.75 in 30 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 1 mM TCEP, and measured at three different speeds, which were derived based on the molecular mass of possible oligomerization states of each of the purified protein. At each speed, equilibrium was achieved within 30 h, and a multiple fit alignment was performed with the XL-I software package, using data from three speeds and protein concentrations, to obtain the buoyant molecular mass M(1 - νρ).
Limited Trypsin Proteolysis and Mass Spectrometry.
To identify minimum required regions of the Scm3:Cse4:H4 complex, purified complex was treated with either trypsin or subtilisin at room temperature for 10 min [1∶250 (wt/wt)], and submitted for ion trap mass spectrometry analysis at the Howard Hughes Medical Institute Mass Spectrometry Laboratory at University of California, Berkeley. We identified trypsin-resistant domains of Cse4 (residues 103–184) and H4 (residues 25–103) and the subtilisin-resistant domain of Scm3 (residues 53–115) (Fig. S1B).
Crystallization and Structure Determination of the Scm3:Cse4:H4 Complex and the (Cse4 : H4)2 Complex.
The purified complex of Scm3(41–115):Cse4(103–184):H4(23–103) was initially crystallized in 0.1 M sodium citrate, pH 4.5, 6% PEG 4000, and 0.1 M sodium iodide. The needle-like crystals in space group P22121, a = 32.7, b = 65.5, c = 121.1, α = 90, β = 90, γ = 120, gave recordable diffraction to a minimum Bragg spacing of 2.3 Å at the Advanced Photon Source NE-CAT beamlines (Argonne National Laboratory). Molecular replacement computations with H3:H4 heterodimer coordinates were done with Phaser (35). Model building and refinement were done with Coot (36) and Refmac (37). The final refined model contains Scm3 (residues 44–103), Cse4 (residues 108–180), and H4 (residues 24–97) with R/Rfree = 22.0/25.7. Crystals of the (Cse4 : H4)2 complex in space group R3, a = 169.5, b = 169.5, c = 81.2, α = 90, β = 90, γ = 120 were obtained by screening a complex of Scm3(41–115):Cse4(93–184):H4(1–103). Crystals appeared in 0.1 M Tris-HCl, pH 8.5, 0.2 M NaCl, and 25% PEG 3350, and gave recordable diffraction to a minimum Bragg spacing of 2.6 Å on the NE-CAT beamlines. Molecular replacement with H3:H4 heterodimer coordinates identified four dimers at the asymmetric unit, with no extra electron density for Scm3. The final model of the (Cse4 : H4)2 complex has R/Rfree of 21.8/27.6.
Supplementary Material
Acknowledgments.
We thank the staff at the Advanced Photon Source NE-CAT beamlines for advice and assistance with data collection and interpretation, and David King (HHMI Mass Spectrometry Facility, University of California, Berkeley) for mass spectrometry. U-S.C. is a special fellow of the Leukemia and Lymphoma Society; S.C.H. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: Coordinates and structure factors for the reported crystal structures of the Scm3:Cse4:H4 trimer and the (Cse4 : H4)2 tetramer have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2YFV and 2YFW, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106389108/-/DCSupplemental.
References
- 1.McAinsh AD, Tytell JD, Sorger PK. Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- 2.Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 3.Palmer DK, O’Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 5.Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 6.Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC. The centromere: Hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- 7.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 8.Camahort R, et al. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Stoler S, et al. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci USA. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunleavy EM, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 11.Foltz DR, et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pidoux AL, et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci USA. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoyan T, Gloeckner G, Diekmann S, Carbon J. Multifunctional centromere binding factor 1 is essential for chromosome segregation in the human pathogenic yeast Candida glabrata. Mol Cell Biol. 2001;21:4875–4888. doi: 10.1128/MCB.21.15.4875-4888.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi T, et al. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Bergmann JH, et al. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 20.Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Natsume R, et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 23.Keith KC, et al. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol Cell Biol. 1999;19:6130–6139. doi: 10.1128/mcb.19.9.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Z, et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu H, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011 doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dalal Y, Furuyama T, Vermaak D, Henikoff S. Structure, dynamics, and evolution of centromeric nucleosomes. Proc Natl Acad Sci USA. 2007;104:15974–15981. doi: 10.1073/pnas.0707648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci USA. 2010;107:20317–20322. doi: 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camahort R, et al. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown MT. Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene. 1995;160:111–116. doi: 10.1016/0378-1119(95)00163-z. [DOI] [PubMed] [Google Scholar]
- 31.Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 32.Mellor J, Rathjen J, Jiang W, Barnes CA, Dowell SJ. DNA binding of CPF1 is required for optimal centromere function but not for maintaining methionine prototrophy in yeast. Nucleic Acids Res. 1991;19:2961–2969. doi: 10.1093/nar/19.11.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 35.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.