Abstract
Mammals achieve gene dosage control by (1) random X-chromosome inactivation in females, (2) parental origin-specific imprinting of selected autosomal genes, and (3) random autosomal inactivation. Genes belonging to the third category of epigenetic phenomenon are just now emerging, with only six identified so far. Here we report three additional genes, Nubp2, Igfals, and Jsap1, that show 50%-methylated CpG sites by Southern blot analyses and primarily monoallelic expression in single-cell allele-specific RT-PCR analysis of bone marrow stromal cells and hepatocytes. Furthermore, we show that, in contrast to X inactivation, alleles can switch between active and inactive states during the formation of daughter cells. These three genes are the first in their category to exist as a tight cluster, in the proximal region of mouse chromosome 17, providing a thus far unique example of a region of autosomal random monoallelic expression.
The mouse t complex encompasses a 20- to 30-Mb region of proximal chromosome 17 and contains many loci required for embryonic development and male germ cell function (Bennett 1975; Silver 1993; Fraser and Dudley 1999; Schimenti 2000). Its puzzling features have been attracting mouse geneticists for more than 75 yr, since the discovery of the tailless mouse mutant (Silver 1993; Schimenti 2000). One of the most interesting mutant loci, called Tme, causes embryonic lethality in heterozygous animals when it is inherited from the mother, but not from the father (Winking and Silver 1984). A reasonable candidate for the Tme gene, Igf2r, has been identified and shown to be expressed only from the maternal allele (Barlow et al. 1991). However, other genetic evidence indicates that another gene may be mutated in Tme mice (Forejt and Gregorova 1992; Rogers et al. 1997). A large anti-sense transcript, called Air, is paternally imprinted, but is most likely a transcript involved in imprinting regulation of Igf2r (Lyle et al. 2000). A neighboring gene, Mas1, may (Villar and Pedersen 1994), or may not be imprinted (Schweifer et al. 1997). Thus, it is not yet clear if this region contains a cluster of imprinted genes, like other imprinted regions (Constancia et al. 1998; Tilghman 1999; John and Surani 2000; Sleutels et al. 2000; Reik and Walter 2001). The search for other imprinted genes in the t complex continues, and may help to understand the complicated Tme phenotype.
Recently, genes that are located on autosomes, like imprinted genes, but show random monoallelic expressions, like the X chromosome (Lyon 1961; Avner and Heard 2001), have been discovered. This has added a novel category to monoallelically expressed genes (Chess et al. 1994; Watanabe and Barlow 1996; Ohlsson et al. 1998). The immunoglobulin gene was long the only one showing autosomal random monoallelic expression, but it has now been joined by T-cell receptor genes (Malissen et al. 1992); olfactory receptor genes (Chess et al. 1994); Natural Killer (NK) cell receptor genes (Held et al. 1995); and IL2 (Hollander et al. 1998), IL4 (Bix and Locksley 1998), Pax5 (Nutt and Busslinger 1999; Nutt et al. 1999), IL5 and IL13 (Kelly and Locksley 2000) genes. Initially these genes were discussed in the same category as immunoglobulin and olfactory receptor genes (Chess et al. 1994; Watanabe and Barlow 1996; Ohlsson et al. 1998). However, more recent findings have modified this interpretation (Held and Kunz 1998; Hollander et al. 1998; Riviere et al. 1998). Unlike the “allelic exclusion” of immunoglobulin and olfactory receptor genes, inactivation of alleles for these genes is not stable, and they do not remain inactive in descendent cells. In addition, the random monoallelic expression of autosomal genes found thus far shows rather independent activation/inactivation of each allele (Bix and Locksley 1998; Nutt et al. 1999). Very recent careful reexamination of the IL2 and Pax5 genes has confirmed random monoallelic expression for IL2, but rejected it for Pax5 (Rhoades et al. 2000). The investigators have proposed that apparent monoallelic expression may simply be due to the low level of gene expression, with stochastic amplification of one allele by PCR (Rhoades et al. 2000).
We have sought to identify new candidate imprinted genes through the analysis of genes expressed in placenta, based on the finding that almost all imprinted genes uncovered thus far are expressed in placenta (Reik and Walter 2001). We constructed a cDNA library from nascent placenta of E7.5 mouse embryo and performed large-scale sequencing and mapping of genes from this library (Ko et al. 1998). To our surprise, genes sampled from the placenta showed unusual clustering in the t complex region of the mouse genome (Ko et al. 1998). We report here our unexpected evidence that three genes, Nubp2, Igfals, and Jsap1, show X-like random monoallelic expression at single-cell levels, but in contrast to X inactivation, alleles can switch between active and inactive states during the formation of daughter cells. We discuss a possible mechanism for such gene expression patterns, and their possible involvement in the biology of the t complex.
RESULTS
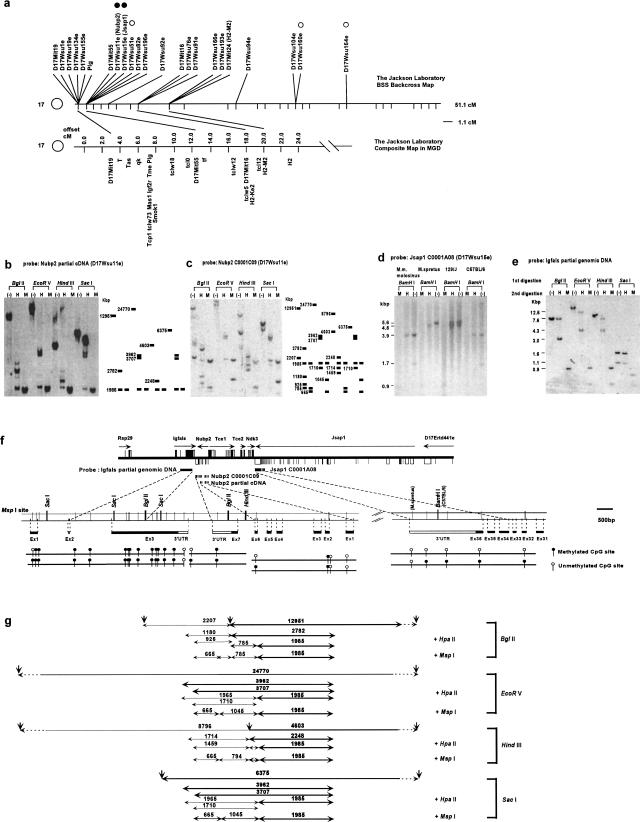
We earlier mapped 155 genes sampled from E7.5 extraembryonic tissue cDNA library on the Interspecific Backcross Mouse Panels, and found that 10 genes are mapped in the t complex region (Ko et al. 1998) (Fig. 1a). To assess whether they are candidates for imprinted genes, we first performed Southern blot analyses of genomic DNAs from a spleen of 129SV/J inbred mouse with methylation-sensitive HpaII and methylation-insensitive MspI restriction enzymes. Of three genes tested, two, D17Wsu11e (Fig. 1b,c) and D17Wsu15e (Fig. 1d), showed partial methylation at CCGG sites. Two other genes (D17Wsu160e and D17Wsu164e), which are located outside of the t complex in chromosome 17, were also examined, but did not show the partial methylation pattern (Fig. 1a). An interesting feature of this partial methylation at CpG sites is that the methylation-sensitive HpaII enzyme seems to cut only about 50% of genomic DNA (see Fig. 1b, BglII digestion, compare HpaII digestion with undigested control DNA; or Fig. 1b, EcoRV digestion, compare HpaII digestion with undigested control DNA). The presence of roughly equal amounts of DNA as methylated or unmethylated indicates that one allele is methylated and the other allele is unmethylated in single cells. However, the possibility remains that both alleles are methylated in 50% of the cells, and both are unmethylated in the remaining cells.
Figure 1.
DNA methylation assay. (a) Genetic map of chromosome 17 on the Jackson Laboratory BSS backcross panel (top) compared with the composite map of t complex region with important landmarks (bottom). Open circles indicates a gene whose methylation status was examined, but did not show partial methylation pattern. Filled circles indicate genes that show partial methylation patterns. (b–d) Southern blot of 129SV/J mouse genomic DNA digested first with BglII, EcoRV, HindIII, and SacI, followed by the second digestion with HpaII (H), MspI (M), and no digestion (–). (b) A 531-bp cDNA fragment containing exons 1–5 of Nubp2 was used as a probe. The schematic diagram with DNA fragment sizes (bp) is shown (right).(c) A 1378-bp cDNA fragment (a full-length C0001C09 clone) containing exons 1–7 of Nubp2 was used as a probe. The schematic diagram with DNA fragment sizes (bp) is shown (right). (d) Southern blot of M. musculus molosinus, Mus spretus, 129SV/J, and C57BL/6J mouse genomic DNAs, digested first with BamHI, followed by the digestion with MspI (M), HpaII (H), and no digestion (–). A 2046-bp cDNA fragment containing exons 36–34 of Jsap1 was used as a probe. (e) A 2141-bp HindIII–EcoRI genomic DNA fragment containing exon 3 of Igfals was used as a probe. (f) A schematic organization of genes (Rsp29, Igfals, Nubp2, Tce1, Tce2, Ndk3, Jsap1, and D17Ertd441e) on the completely sequenced BAC clone (126c8: [Kargul et al. 2000]) and the location of the DNA probes, which were used for the DNA methylation assay (top). Black box indicates coding exon and white box indicates noncoding exons (Kargul et al. 2000). A magnified view of the exon–intron structures for Igfals, Nubp2, and Jsap1, with the restriction enzyme sites of MspI, BglII, SacI, HindIII, and BamHI, is also shown (middle). Positions of polymorphic MspI site in the 3′-UTR of Jsap1 are indicated by the mouse strain name in parentheses. Methylation status (methylated, black circle; unmethylated, white circle) of each MspI site deduced from Southern blot analyses (b–e) are shown (bottom). (g) To correlate Southern blot results (b–e) to the methylation status of the MspI site (f), expected genomic fragments and their sizes (bp) are shown. Bold letters and lines indicate the DNA fragments in (c), and the others indicate the DNA fragments in (e).
D17Wsu11e, later named nucleotide binding protein 2 (Nubp2), encodes a protein bearing an ATP/GTP binding motif (Nakashima et al. 1999). The knockout of the orthologous gene in yeast is lethal, indicating that the gene is essential for a basic cellular function, possibly in cell proliferation (Nakashima et al. 1999). D17Wsu15e was very recently identified as jun N-terminal kinase (JNK)/stress-activated protein kinase-associated protein 1 (Jsap1) by a yeast two-hybrid screen using JNK3 MAPK as bait (Ito et al. 1999). The protein shows very high binding affinity to JNK3, and overexpression of full-length JSAP1 in COS-7 cells leads to a considerable enhancement of JNK3 activation, indicating that Jsap1 functions as a scaffold protein in the JNK3 signaling cascade (Ito et al. 1999).
To characterize further the methylation state of CpG sites in this region, we screened BAC libraries with Nubp2 and Jsap1. One BAC clone (126C8) carrying 175 kb of genomic DNA was identified and completely sequenced (Kargul et al. 2000). Among six novel genes and four known genes deduced from the BAC sequence, a gene encoding the acid-labile subunit of insulin-like growth factor (IGF)-binding proteins (Igfals) (Boisclair et al. 1996) was identified 700 bp downstream of the Nubp2 gene (Fig. 1f). IGFALS binds to and dramatically extends the half-life of the IGF–IGF-binding protein complex, indicating that it plays an important role in the control of cell growth (Boisclair et al. 1996). Southern blot analysis revealed that Igfals also showed 50%-methylated CpG sites (Fig. 1e). Because the complete sequence information of the BAC126C8 was available, all MspI sites were identified and compared with the MspI or HpaII digestion patterns in the Southern blots (e.g., Fig. 1g). This analysis allowed us to clearly identify the methylation status of each MspI site (Fig. 1f).
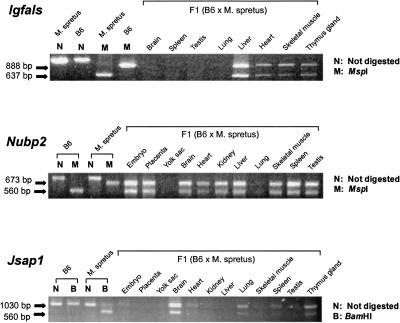
The presence of 50%-methylated CpG sites indicates these three genes, Nubp2, Igfals, and Jsap1, as candidates for imprinted genes with monoallelic expression. To test this notion, we performed allele-distinguishing RT-PCR analyses on RNAs derived from various organs of an F1 hybrid adult mouse obtained by crossing female C57BL/6J (B6) to male Mus spretus (Spr). PCR primers were designed to cross introns, so that any genomic DNA contamination could not influence the results. Both parental alleles of the Igfals gene were expressed in the organs where the gene is expressed (Fig. 2). Similarly, both Nubp2 and Jsap1 were also biallelically expressed in the tissues where these genes are expressed (Fig. 2). These results indicate that the gene is not imprinted, at least in the tissues we have examined. An alternative hypothesis that is consistent with both 50%-methylation status of these genes and biallelic gene expressions is that these genes show random monoallelic expression at the single-cell level, analogous to that seen in X-chromosome inactivation.
Figure 2.
Allele-specific gene expression analysis at organ level. Gene-specific cDNA fragments were amplified by RT-PCR from total RNAs prepared from various organs of adult F1 (C57BL/6J female × Mus spretus male) hybrid mice. To distinguish each parental allele, the PCR products were digested with restriction enzymes (MspI for Igfals, MspI for Nubp2, BamHI for Jsap1) and run on the agarose gels.
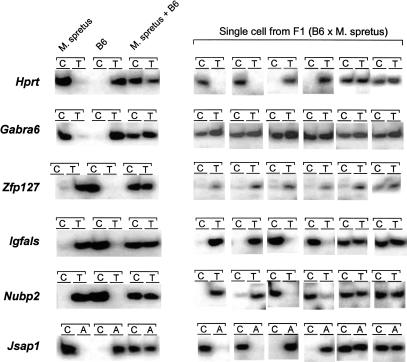
To examine this possibility, gene-specific RT-PCR and subsequent single nucleotide primer extension (SNuPE) assays (Singer-Sam 1995) were performed on single bone marrow stromal cells isolated from F1 hybrid mice (B6 × Spr). As controls, we chose Gabra6 (Takahashi and Ko 1993) as an autosomal biallelically expressed gene, Hprt (Konecki et al. 1982) as an X-chromosome random monoallelically expressed gene, and Zfp127 (Jong et al. 1999) as a paternally expressed imprinted gene. Expression of Gabra6 was biallelic, as expected (although 6% of the cells showed predominant expression of the paternal allele; Fig. 3 shows examples, Table 1 shows a summary). The Zfp127 gene showed predominant expression of paternal allele in 94% of the cells, as expected. In contrast, the Igfals gene showed biallelic expression in only 18% of the cells, but monoallelic expression, maternal (41%) or paternal (41%), in most cells. Nubp2 and Jsap1 showed similar results (Fig. 3, Table 1). These expression patterns were similar to those for the Hprt gene, which showed biallelic expression in 24% of the cells, predominant expression of the maternal allele in 28% of the cells, and predominant expression of the paternal allele in 48% of the cells.
Figure 3.
Allele-specific gene expression analyses by SNuPE assays. Each assay consists of two primer–extension reactions with allele-specific radiolabeled nucleotide (e.g, in case of Hprt, C [α32P-dCTP] represents paternal Mus spretus allele and T [α32P-cTTP] represents maternal C57BL/6J [B6] allele). For each gene, the first six lanes are controls (lanes 1, 2: M. spretus cDNA only; lanes 3, 4: B6 cDNA only; lanes 5, 6: equal mixture of M. spretus and B6 cDNAs). Lanes 7–18 show six representative results of SNuPE assay on single cells isolated from bone marrow stromal cells of adult F1 (C57BL/6 female × M. spretus male) hybrid mice.
Table 1.
Summary of Allele-Specific Gene Expression Analysis in Single Cells
| Expression ratio (Maternal allele/paternal allele) | Number of cells with corresponding expression patterns | |||||
|---|---|---|---|---|---|---|
| Regular gene: Gabra6 | X-linked gene: Hprt | Imprinted gene: Zfp127 | Igfals | Nubp2 | Jsap1 | |
| Paternal > Maternal | ||||||
| <0.0625 | 0 | 0 | 0 | 2 | 1 | 1 |
| 0.0625–0.125 | 0 | 0 | 6 | 2 | 0 | 0 |
| 0.125–0.25 | 0 | 6 | 7 | 0 | 3 | 1 |
| 0.25–0.5 | 1 | 6 | 2 | 3 | 6 | 2 |
| Paternal = Maternal | ||||||
| 0.5–1 | 13 | 2 | 0 | 2 | 7 | 4 |
| 1–2 | 3 | 4 | 1 | 1 | 8 | 0 |
| Paternal < Maternal | ||||||
| 2–4 | 0 | 5 | 0 | 6 | 6 | 1 |
| 4–8 | 0 | 2 | 0 | 1 | 3 | 2 |
| 8–16 | 0 | 0 | 0 | 0 | 1 | 0 |
| >16 | 0 | 0 | 0 | 0 | 0 | 1 |
| Total number of cells | 17 | 25 | 16 | 17 | 35 | 12 |
In additional experiments, we also performed gene-specific RT-PCR and subsequent SNuPE assays on single hepatocytes of F1 hybrid animals. The results are essentially the same, though the fraction of cells that show biallelic expression is consistently higher in hepatocytes than in the bone marrow stromal cells (data now shown). This is perhaps due to the polyploidy of more than 80% of adult liver hepatocytes (Severin et al. 1984).
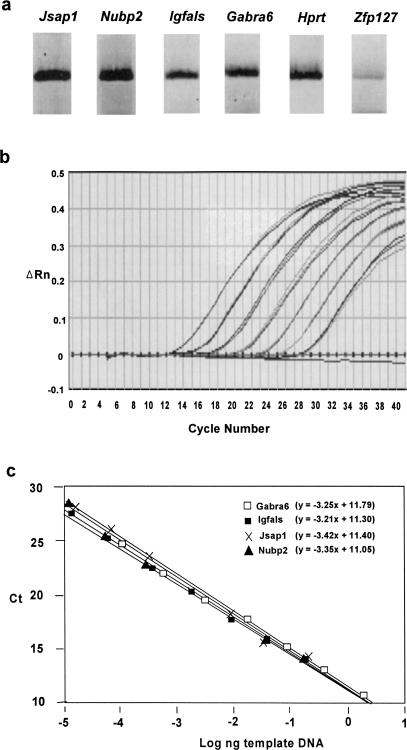
One concern about single-cell RT-PCR analysis was that the apparent monoallelic expression might not truly reflect the expression pattern of the gene, but might rather be an artifact caused by stochastic variations of PCR amplification. It has recently been shown that this is indeed the case for Pax5 (Rhoades et al. 2000), in which the expression of the gene is very low, so that RT-PCR may selectively amplify either allele by chance and provide a false indication of random monoallelic expression (Rhoades et al. 2000). To address this possibility, we first compared the level of RT-PCR products among the examined genes, and found that amplifiable expression levels of genes consistently gave gene-specific monoallelic or biallelic expression. For example, in Figure 4a, the PCR products of Jsap1, Nubp2, and Igfals were comparable with that of control biallelically expressed gene, Gabra6. The Hprt gene also shows a similar level. Zfp127 gives much less RT-PCR product. To further verify this, we measured the relative mRNA abundance of each gene in the bone marrow stromal cells by real-time RT-PCR analysis. First, a standard curve was generated using Jsap1-specific PCR product as a template (Fig. 4b,c). The standard curve was linear, as shown by a correlation coefficient (R2) value of >0.99 over a wide range of template DNA input (Fig. 4c). The narrow standard deviation (0.03–0.25) and small coefficient of variation (<1) of triplicate determinations also indicate that the measurement is highly reproducible. Similar amplification plots and standard curves were obtained for Igfals, Nubp2, and Gabra6 (Fig. 4c). Statistical analysis using a multiple regression model to compare two standard curves did not reveal any significant difference in the sensitivity (y intercept) and amplification efficiency (slope) between Gabra6 and other genes. Therefore, these assays satisfy the critical standard to ensure the proper comparison of mRNA abundance by real-time RT-PCR (Bustin 2000). We then performed real-time RT-PCR analyses of these genes on RNAs from bone-marrow stromal cells. Based on the standard curve, the amounts of mRNA for each gene was calculated as 4.6 × 10−5 ng (Igfals), 8.4 × 10−5 ng (Nubp2), 1.5 × 10−4 ng (Jsap1), and 1.0 × 10−4 ng (Gabra6). Taken together, three genes, Igfals, Nubp2, and Jsap1, show expression levels similar to that of Gabra6, which clearly showed biallelic expression patterns in the single-cell SNuPE assay. Therefore, it is highly unlikely that monoallelic expression patterns of the comparably expressed Igfals, Nubp2, and Jsap1 are due to selective RT-PCR amplification of one or another allele.
Figure 4.
Quantitation of gene expression levels. (a) RT-PCR products from cell clones, which were used to perform a SNuPE assay for each gene. (b) Normalized reporter signal (ΔRn) versus cycle number is plotted for the input of Jsap1 template ranging from 3.2 × 10–4, 1.6 × 10–3, 8 × 10–3, 4 × 10–2, 2 × 10–1, 1, and 5 ng. The amplification curves represent the highest to lowest PCR products from left to right. Three replicates for each point are performed with no template control shown at the baseline through 40 cycles. (c) The standard curves of Igfals, Nubp2, Jsap1, and Gabra6. Ct versus log nanograms of the template input is plotted.
A second concern is the presence of cells showing biallelic expression of even the X-linked Hprt gene in some stromal cells. It has been accepted that X-linked genes show clear random monoallelic expression and do not show the biallelic expression in single cells. One possible explanation is the inclusion of more than one cell in a single PCR tube during the preparative pipetting procedure. Alternatively, biallelic expression may arise because epigenetic modification of genes is partially relaxed by the age of this F1 hybrid (1 yr). Indeed, it has been reported that X-chromosome inactivation becomes looser and reactivation occurs with age (Wareham et al. 1987). Nonetheless, Hprt, as well as Nubp2, Igfals, and Jsap1, shows a decisive statistically significant difference from the biallelically expressed Gabra6 gene (p < 0.01), but with no statistically significant difference between these three genes and Hprt (p > 0.2; Table 1; see also Methods section). Taken together, the results strongly indicate that the autosomally located Nubp2, Jsap1, and Igfals genes show random monoallelic expression that is similar to X-linked genes.
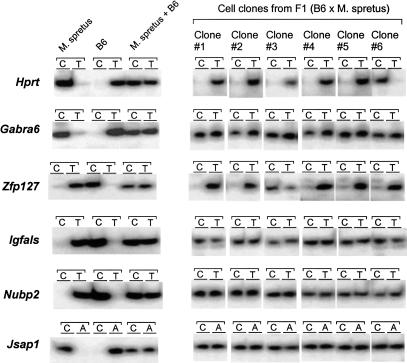
Are the Nubp2, Jsap1, and Igfals genes “like” X-linked genes? One of the characteristic features of X-linked genes is that the inactivation status of alleles is stably maintained throughout many cell divisions. To examine whether the inactivated status of alleles of Nubp2, Igfals, and Jsap1 is stably maintained, we performed allele-specific expression analyses of clonal cell populations. Bone marrow stromal cells from an F1 hybrid mouse were dissociated and cultured at very low cell density. Single cells were cultured to form small colonies of ∼100 cells. Six colonies were further separately cultured until they became confluent in 10-cm dishes, and were then used for SNuPE assays. As shown in Figure 5, the X-linked Hprt gene and imprinted Zfp127 gene showed monoallelic expression, as expected. In contrast, all three genes identified in this study showed biallelic expression. This indicates that epigenetic control of these monoallelically expressed genes is not stable, but changes during the course of multiple cell divisions. Thus, clearly distinct from X-linked gene inactivation, Nubp2, Igfals, and Jsap1 belong to a category of monoallelic gene expression, also exemplified by IL2 (Hollander et al. 1998) and IL4 (Bix and Locksley 1998).
Figure 5.
Allele-specific gene expression analyses of clonal cells by SNuPE assays. Each assay consists of two primer–extension reactions with allele-specific radiolabeled nucleotide (see Fig. 4). For each gene, the left panel shows controls (Mus spretus cDNA only; B6 cDNA only; equal mixture of M. spretus and B6 cDNAs). The right panel shows SNuPE assays on cell clones derived from single bone marrow stromal cells of adult F1 (C57BL/6 female × M. spretus male) hybrid mice. SNuPE assays are performed by using the same number of the clonal cell population for each gene.
DISCUSSION
In this paper, we have shown that three genes, Igfals, Nubp2, and Jsap1, which are clustered in the t complex of the mouse chromosome 17, showed random monoallelic gene expression in single cells. In general, there are four different types of monoallelic gene expression: X inactivation, genomic imprinting, allelic exclusion, and autosomal random monoallelic expression. Three genes reported here appear to fall into the fourth category of monoallelic gene expression. We have provided evidence that this monoallelic gene expression is not due to experimental artifacts such as preferential amplification of one allele by single-cell RT-PCR.
These three new genes provide the first example of clustering of unrelated genes regulated in this way, a common feature of other monoallelically expressed genes such as X-linked and imprinted genes (Sleutels et al. 2000). Previous studies of random monoallelic gene expression did not extend to the methylation status of genome alleles. This work provides the first evidence that correlates autosomal random monoallelic expressions with the 50%-methylation status of the genomic region, as seen in X-linked and imprinted genes. It may be relevant that two reciprocally imprinted genes, Igf2 and H19, show random monoallelic expression in certain cell types (Ohlsson et al. 1999) and sometimes biallelic expression of either or both genes at the single-cell level by RNA-FISH analysis (Jouvenot et al. 1999). Even for X-linked genes, it has been recently reported that escape of some X-linked genes from inactivation varies among individuals (Carrel and Willard 1999). It is conceivable that all monoallelic gene expressions are maintained by similar mechanisms, with very small differences among them.
Among monoallelic gene expressions, only X-chromosome inactivation has a clear biological rationale, because X-chromosome inactivation is necessary to compensate gene dosage with two X chromosomes in females and one X chromosome in males. Evolutionary reasons for the existence of imprinted genes have also been extensively discussed (Reik and Walter 2001), although an answer as clear as that for X chromosome inactivation has not been addressed. Allelic exclusion on immunoglobulin and TCR genes prevents the development of single B and T lymphocytes with both alleles expressed. Similarly, olfactory receptors have a good reason to be monoallelic, because single olfactory cells should express a unique single receptor (Chess et al. 1994). Other known autosomally coded random monoallelic genes, such as IL2 and IL4, have less clear justification except for their immune function. In the case of three genes reported here, the reason for random monoallelic expression is even less clear. Three genes, Igfals, Nubp2, and Jsap1, have no apparent common feature, apart from a common involvement in cell growth controls. Indeed, this could be the case, especially because Igfals is involved in the IGF pathway, which includes two well-established imprinted genes, Igf2 and Igf2r.
What is then the raison d'être for autosomal random monoallelic expression in general? First, it is possible that genes important for critical developmental decisions may need to be regulated by monoallelic expression (Holliday 1990). When shutting off a particular gene is critical for cell fate, a single active gene will be easier to close down than two active genes. Second, one can also argue that autosomal random monoallelic expression is a bystander effect spreading from nearby imprinted genes. Although the exact physical distance between this gene cluster and Igf2r is not known, genetic mapping data indicate that they are at most within a few megabases (Ko et al. 1998; R. Nagaraja and D. Schlessinger, pers. comm.). Third, monoallelic gene expression may be a natural phenomenon for genes with a low level of expression. A gene driven by a limited amount of a critical transcription factor shows stochastic expression (Ko 1992; McAdams and Arkin 1997), which can lead to monoallelic gene expression (Cook et al. 1998). In a similar way, all monoallelic gene expression may originate from intrinsically stochastic features of gene expression regulation, as recently proposed (Ohlsson et al. 2001).
It has been postulated that monoallelically expressed genes will be more sensitive to reduced dosage, because gene dosage is already a half of the normal amount, which will be further reduced to one-fourth of that by loss-of-function mutation in one allele (Watanabe and Barlow 1996; Cook et al. 1998; Nutt and Busslinger 1999). In fact, some mutants defective in one allele may show a null phenotype, called haploinsufficiency (e.g., cleidocranial dysplasia [Lee et al. 1997; Mundlos et al. 1997]). Therefore, it will be interesting to see whether the knockout of these three genes will show haploinsufficiency phenotypes. If this is the case, random monoallelic expression of these genes may indeed contribute to the complicated genetic behavior of the Tme locus or t complex as a whole.
Finally, we would like to point out that the “switching” or “relaxing” of active allele status could provide organisms with a safeguard against loss-of-function mutations in genes with dosage-controlled expression; at least 50% of descendent cells would always have an active allele even if one allele were damaged.
METHODS
DNA Methylation Assay
For Igfals and Nubp2, 40 μg genomic DNA from 129SV/J mouse spleen was digested with BglII, EcoRV, HindIII, and SacI. One-third of the products were digested with HpaII and another one-third were digested with MspI. After gel electrophoresis on 1% agarose gels, the DNAs were blotted onto nylon membranes. For Jsap1, genomic DNA from a spleen of M. musculus molosinus, M. spretus, 129SV/J, and C57BL/6J was digested with BamHI with HpaII or MspI. A DNA probe for Igfals was prepared by PCR amplification of genomic DNAs. A probe for Nubp2 was prepared by PCR amplification of cDNA clone (C0001C09) with a primer pair (5′-CTGTGCGGTCCCAGCATACC-3′, 5′-GGCTGGACGAAAGCCTATAG-3′). A full-insert of cDNA clones, C0001C09 as a probe for Nubp2 and C0001A08 as a probe for Jsap1, were used (Ko et al. 1998). Hybridization, washing, and detection were performed by the standard method.
Allele-Specific Gene Expression Analysis at the Organ Level
PCR was performed in a 25-μL reaction mixture containing a 0.5-μM gene-specific primer pair spanning the intron/exon boundary (5′-TGCCAGCCGGTGTACACTTACA-3′, 5′-CCC TATTTAGGCCACATCGCTTC-3′ for Igfals, 5′-GGTCATAG AGAACATGAGCGGCTT-3′, 5′-TGTAGGGGAATCAAGAGTC ACC-3′ for Nubp2, and 5′-GTGACAAGGCCGCCAGTAGTT TC-3′, 5′-GGACAATCTTGGGTTGACGGGTATTT-3′ for Jsap1) and cDNA from F1 (C57BL/6J × M. spretus) mice organs. The PCR conditions were as follows: preheating step at 95°C for 3 min, followed by 35 cycles of denaturing step at 95°C for 30 sec, annealing step at 57°C (Igfals), 65°C (Nubp2), and 62°C (Jsap1) for 45 sec, and extension step at 72°C for 45 sec with 3 min final extension. The 4 μL PCR product was digested with MspI, MspI, and BamHI for Igfals, Nubp2, and Jsap1, respectively, at 37°C.
Allele-Specific Gene Expression Analysis on Single Cells
A primary culture of bone marrow stromal cells was prepared from an adult female F1 (C57BL/6J × M. spretus) hybrid by the standard method. The cells at 80% confluence were trypsinized and washed three times with phosphate buffered saline (PBS). After dissociating cells into single cells by trypsin, the cells were individually aspirated under the microscope by mouth pipette with 1.0-mm-diameter capillary pipettes drawn out to 60–70 μm diameter. Single cells were stored in a 0.2-mL reaction tube (MicroAmp Perkin Elmer) containing 0.05% NP-40 and immediately chilled on ice. Samples were stored at −80°C before use.
The RT-PCR was performed on the single cell by the EzrTth RNA PCR kit (Perkin-Elmer) in a 50-μL reaction mixture containing 0.05% NP-40 (Calbiochem), 2.5 mM Mn (OAc)2, 0.2 units of rTth DNA polymerase, 300 μM each dNTPs, 250 mM Bicine (pH 8.2), 575 mM KOAc, and 0.5 μM primer pairs (5′-TGGAACTCGATCTTACCGCCAA-3′, 5′-GTT ATTCCTGAGGTTGAGGTAGCGAA-3′ for Igfals; 5′-CGTC CAGCCCTAGTCACTTC-3′, 5′-TGTAGGGGAATCAAGAGTC ACC-3′ for Nubp2; 5′-GGCAACCCTAGGCACTTAGTACG TGT-3′, 5′-CAGAAGCCCCAAGGGTAGAATTTC-3′ for Jsap1; 5′-CAAAGCCTAAGATGAGCGCAAGT-3′, 5′-CAAATCA AAAGTCTGGGGACGCA-3′ for Hprt; 5′-TGTGGTCTGGTCTG CTAAAGCTC-3′, 5′-ACACAGAGAAACATCACAATCCTAGC AA-3′ for Zfp127; and 5′-CTGCAATACTGTTGCTATTTCC-3′, 5′-AAGTGTAGATATGATGGTAGCC-3' for Gabra6). The reaction mixtures were incubated at 60°C for 30 min for reverse transcription, followed by the PCR reaction (preheating at 94°C for 1 min, 40 cycles of heating at 94°C for 15 sec, annealing and extension at 60°C (Igfals), 68°C (Nubp2), 60°C (Jsap1), 60°C (Hprt), 60°C (Zfp127), and 55°C (Gabra6) for 30 sec, with the final extension for 7 min. For Zfp127, an additional 20 cycles of PCR was performed. The PCR products were run on 3% agarose gel and purified by cutting out the DNA band. An aliquot of purified DNAs were run on 3% agarose gel and stained with SYBRGreen (FMC). The amount of DNAs was estimated by comparing the band intensities to those of DNA size markers.
SNuPE assays were performed according to the well-established method (Singer-Sam 1995). For each reaction, equal amounts of template DNAs, as described earlier, were combined with the following 10-μL reaction mixture: 2 μCi of α32P-labeled d-TTP or d-CTP (d-ATP or d-CTP in the case of Jsap1), 1× SNuPE buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 15 mM MgCl2), 0.65 units of Taq DNA polymerase, and 1 μM SNuPE primer: 5′-CAGCGGGCCTTCTGGCTGGAC-3′ for Igfals; 5′-GAGCAGCCTGTTGACCTGGGG-3′ for Nubp2; 5′-TGCCTCCTGCACAGGGCTCCA-3′ for Jsap1; 5′-GCAT GAACCTTCTATGAA-3′ for Hprt5 (Greenwood and Burke 1996), 5′-TGCCTCAGTAGCCTTTGAAAG-3′ for Zfp127, and 5′-ACCAAGTGACCTTGCTAGAAC-3′ for Gabra6 (Takahashi and Ko 1993). SNuPE assays were performed with a single cycle of denaturation at 95°C for 1 min, annealing at 50°C (for Nubp2 at 60°C) for 2 min, and primer extension at 72°C for 1 min. After 15% denaturing polyacrylamide gel electrophoresis at 250 V for 20 min, the radioactive gels were exposed to a Phosphorscreen (Molecular Dynamics) for 20 min. The screen was read by PhosphorImager (Molecular Dynamics) and signal intensities determined by the ImageQuant software (Molecular Dynamics). Each band on a scanned gel image was circumscribed with an equivalent size box. Signal intensity was transferred to Microsoft Excel and converted to allele-specific gene expression levels according to the established calculation method (Singer-Sam 1995). When the expression ratio of B6/Spr is higher than 2, it is considered as B6 allele-predominant monoallelic expression. When the expression ratio of B6/Spr is lower than 0.5, it is considered as Spr allele-predominant monoallelic expression. When the expression ratio of B6/Spr is between 0.5 and 2, it is considered as biallelic gene expression. Statistical analysis was performed on the SAS program package by applying Fisher's (3 × 2) exact test (2-tail, 5% significance level) to the expression ratio of B6/Spr of each gene versus the expression ratio of B6/Spr of Hprt or versus that of B6/Spr of Gabra6.
Allele-Specific Gene Expression Analyses on Clonal Cells
The 80% confluent bone marrow stromal cells were diluted 105-fold and plated into a 35-mm culture dish. After 12 h, single cells were marked on the surface of dishes. After 7 d of culture, colonies containing more than 50 cells were isolated by a cloning cylinder (Bel-Art Products) and transferred to an 85-mm culture dish individually. After an additional 14 d of culture, cells were trypsinized, washed three times with PBS, and lysed in 5 mL of Trizol Reagent (GIBCO BRL) to isolate RNAs. To remove potential contamination of genomic DNA, we digested total RNAs with 1 unit of DNaseI (Boehringer Mannheim) for 15 min at 37°C, followed by phenol/chloroform extraction, and ethanol precipitation. RT-PCR and SNuPE assays were performed as described earlier.
Real-Time RT-PCR
PCR primers and fluorescent probes were designed by PrimerExpress software (PE Applied Biosystems). To prepare a control DNA for each gene, we performed PCR in a 25-μL reaction mixture containing a 0.5-μM gene-specific primer pair: 5′-GACAACAGCATCTCCAGCATCGAAG-3′, 5′-GTTCTGTA GGGCAAAGTCACGAAGC-3′ for Igfals; 5′-TAGCCTGCGT CCTGATTCCTTGAGAT-3′, 5′-TGAAATGTAGGGGAATCAA GAGTCACCT-3′ for Nubp2; 5′-TGCTCCCACACACTTGCT TAGAACTG-3′, 5′-TACCYGGGAACCTGGCATGAAAGCT-3′ for Jsap1; and 5′-CAAAGATACAATGGAAGTGAGCAGTAC3′, 5′-TAGGAAAGTGTAGATATGATGGTAGCC-3′ for Gabra6. The PCR conditions were as follows: preheating step at 95°C for 3 min, followed by 35 cycles of denaturing step at 95°C for 30 sec, annealing step at 60°C (Igfals, Nubp2, Jsap1) and 58°C (Gabra6) for 45 sec, and extension step at 72°C for 45 sec with a 3-min final extension. The PCR products were run on 3% agarose gel and purified by cutting out the DNA band. An aliquot of purified DNAs was run on 3% agarose gel and stained with SYBRGreen (FMC). The amount of DNAs was estimated by comparing the band intensities to those of DNA size markers.
To estimate the relative abundance of mRNAs, RT and PCR were conducted in one step by using TaqMan EZ RT-PCR kit in a GeneAmp 5700 Sequence Detection System (PE Applied Biosystems) according to the manufacturer's protocol. Briefly, a 50-μL reaction mixture containing 2.5 mM Mn(OAc)2; 5.0 units of rTth DNA polymerase; 300 μM each of dATP, dCTP, and dGTP; 600 μM dUTP; 900 nM nested primer pairs (5′-TGGAACTCGATCTTACCGCCAA-3′, 5′-GTTATTCC TGAGGTTGAGGTAGCGAA-3′ for Igfals; 5′-CGTCCAGCCCT AGTCACTTC-3′, 5′-TGTAGGGGAATCAAGAGTCACC-3′ for Nubp2; 5′-GGCAACCCTAGGCACTTAGTACGTGT-3′, 5′-CAG AAGCCCCAAGGGTAGAATTTC-3′ for Jsap1; and 5′-CTG CAATACTGTTGCTATTTCC-3′, 5′-AAGTGTAGATATGATGGTAGCC-3′ for Gabra6); and 250 nM fluorescent probes (5′-CC AGCTGGAATATCTGCTTCTGTCCAACA-3′ for Igfals; 5′-TTTCTCATCGAGACTGACCCACACATGCT-3′ for Nubp2; 5′-CCATTAACCTGAGGCTAAGCCTGCATCCT-3′ for Jsap1; and 5′-AAAGAAGATGCTTGCTAACACCAGGAGGTTC-3′ for Gabra6) was prepared. The reaction mixtures were incubated at 60°C for 30 min for reverse transcription, followed by the PCR reaction: preheating at 94°C for 1 min, 40 cycles of heating at 95°C for 15 sec, annealing and extension at 60°C (Igfals, Nubp2, Jsap1) and 55°C (Gabra6) for 1 min. ΔRn (normalized reporter signal) that represents the increases in fluorescence due to the cleavage of the reporter dye during PCR, was determined and plotted against cycle numbers (Heid et al. 1996) (Fig. 4b). Ct values (the PCR cycle number required for fluorescence intensity to exceed an arbitrary threshold in the exponential phase of the amplification) were determined for a series of standards. A standard curve was generated by plotting Ct versus the log of the amount of standard PCR product added to the reaction (5 × 10−5–0.2 ng) (Fig. 4c) and used to compare the relative amount of Igfals, Nubp2, Jsap1, and Gabra6 mRNA of the F1 (B6/Spr) hybrid mouse bone marrow stromal cells.
Acknowledgments
We thank Dr. David Schlessinger for discussion and critical reading of the manuscript, Dr. Denise Barlow for suggestion of the use of 50% methylation assays, and Dr. Toru Takebayashi for advice on statistical analysis. This work was initially supported by NICHD grant (R01HD32248) to M.S.H.K. and was later supported by the NIA/NIH intramural research program.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL: kom@grc.nia.nih.gov; FAX (410) 558-8331.
Article published on-line before print: Genome Res., 10.1101/gr. 194301.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.194301.
REFERENCES
- Avner P, Heard E. X-chromosome inactivation: Counting, choice and initiation. Nat Rev Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- Bennett D. The T-locus of the mouse. Cell. 1975;6:441–454. [Google Scholar]
- Bix M, Locksley RM. Independent and epigenetic regulation of the interleukin-4 alleles in CD4+ T cells. Science. 1998;281:1352–1354. doi: 10.1126/science.281.5381.1352. [DOI] [PubMed] [Google Scholar]
- Boisclair YR, Seto D, Hsieh S, Hurst KR, Ooi GT. Organization and chromosomal localization of the gene encoding the mouse acid labile subunit of the insulin-like growth factor binding complex. Proc Natl Acad Sci. 1996;93:10028–10033. doi: 10.1073/pnas.93.19.10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard HF. Heterogeneous gene expression from the inactive X chromosome: An X-linked gene that escapes X inactivation in some human cell lines but is inactivated in others. Proc Natl Acad Sci. 1999;96:7364–7369. doi: 10.1073/pnas.96.13.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Constancia M, Pickard B, Kelsey G, Reik W. Imprinting mechanisms. Genome Res. 1998;8:881–900. doi: 10.1101/gr.8.9.881. [DOI] [PubMed] [Google Scholar]
- Cook DL, Gerber AN, Tapscott SJ. Modeling stochastic gene expression: Implications for haploinsufficiency. Proc Natl Acad Sci. 1998;95:15641–15646. doi: 10.1073/pnas.95.26.15641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forejt J, Gregorova S. Genetic analysis of genomic imprinting: An Imprintor-1 gene controls inactivation of the paternal copy of the mouse Tme locus. Cell. 1992;70:443–450. doi: 10.1016/0092-8674(92)90168-c. [DOI] [PubMed] [Google Scholar]
- Fraser LR, Dudley K. New insights into the t-complex and control of sperm function. Bioessays. 1999;21:304–312. doi: 10.1002/(SICI)1521-1878(199904)21:4<304::AID-BIES6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Greenwood AD, Burke DT. Single nucleotide primer extension: Quantitative range, variability, and multiplex analysis. Genome Res. 1996;6:336–348. doi: 10.1101/gr.6.4.336. [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Held W, Kunz B. An allele-specific, stochastic gene expression process controls the expression of multiple Ly49 family genes and generates a diverse, MHC-specific NK cell receptor repertoire. Eur J Immunol. 1998;28:2407–2416. doi: 10.1002/(SICI)1521-4141(199808)28:08<2407::AID-IMMU2407>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Held W, Roland J, Raulet DH. Allelic exclusion of Ly49-family genes encoding class I MHC-specific receptors on NK cells. Nature. 1995;376:355–358. doi: 10.1038/376355a0. [DOI] [PubMed] [Google Scholar]
- Hollander GA, Zuklys S, Morel C, Mizoguchi E, Mobisson K, Simpson S, Terhorst C, Wishart W, Golan DE, Bhan AK, et al. Monoallelic expression of the interleukin-2 locus. Science. 1998;279:2118–2121. doi: 10.1126/science.279.5359.2118. [DOI] [PubMed] [Google Scholar]
- Holliday, R. 1990. Genomic imprinting and allelic exclusion. Dev. Suppl.: 125–129. [PubMed]
- Ito M, Yoshioka K, Akechi M, Yamashita S, Takamatsu N, Sugiyama K, Hibi M, Nakabeppu Y, Shiba T, Yamamoto KI. JSAP1, a novel jun N-terminal protein kinase (JNK)-binding protein that functions as a Scaffold factor in the JNK signaling pathway. Mol Cell Biol. 1999;19:7539–7548. doi: 10.1128/mcb.19.11.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John RM, Surani MA. Genomic imprinting, mammalian evolution, and the mystery of egg-laying mammals. Cell. 2000;101:585–588. doi: 10.1016/s0092-8674(00)80870-3. [DOI] [PubMed] [Google Scholar]
- Jong MT, Carey AH, Caldwell KA, Lau MH, Handel MA, Driscoll DJ, Stewart CL, Rinchik EM, Nicholls RD. Imprinting of a RING zinc-finger encoding gene in the mouse chromosome region homologous to the Prader-Willi syndrome genetic region. Hum Mol Genet. 1999;8:795–803. doi: 10.1093/hmg/8.5.795. [DOI] [PubMed] [Google Scholar]
- Jouvenot Y, Poirier F, Jami J, Paldi A. Biallelic transcription of Igf2 and H19 in individual cells suggests a post-transcriptional contribution to genomic imprinting. Curr Biol. 1999;9:1199–1202. doi: 10.1016/S0960-9822(00)80026-3. [DOI] [PubMed] [Google Scholar]
- Kargul GJ, Nagaraja R, Shimada T, Grahovac MJ, Lim MK, Nakashima H, Waeltz P, Ma P, Chen E, Schlessinger D, et al. Eleven densely clustered genes, six of them novel, in 176 kb of mouse t-complex DNA. Genome Res. 2000;10:916–923. doi: 10.1101/gr.10.7.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly BL, Locksley RM. Coordinate regulation of the IL-4, IL-13, and IL-5 cytokine cluster in Th2 clones revealed by allelic expression patterns. J Immunol. 2000;165:2982–2986. doi: 10.4049/jimmunol.165.6.2982. [DOI] [PubMed] [Google Scholar]
- Ko MSH. Induction mechanism of a single gene molecule: Stochastic or deterministic? Bioessays. 1992;14:341–346. doi: 10.1002/bies.950140510. [DOI] [PubMed] [Google Scholar]
- Ko MSH, Threat TA, Wang X, Horton JH, Cui Y, Pryor E, Paris J, Wells-Smith J, Kitchen JR, Rowe LB, et al. Genome-wide mapping of unselected transcripts from extraembryonic tissue of 7.5-day mouse embryos reveals enrichment in the t-complex and under-representation on the X chromosome. Hum Mol Genet. 1998;7:1967–1978. doi: 10.1093/hmg/7.12.1967. [DOI] [PubMed] [Google Scholar]
- Konecki DS, Brennand J, Fuscoe JC, Caskey CT, Chinault AC. Hypoxanthine-guanine phosphoribosyltransferase genes of mouse and Chinese hamster: construction and sequence analysis of cDNA recombinants. Nucleic Acids Res. 1982;10:6763–6775. doi: 10.1093/nar/10.21.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Thirunavukkarasu K, Zhou L, Pastore L, Baldini A, Hecht J, Geoffroy V, Ducy P, Karsenty G. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat Genet. 1997;16:307–310. doi: 10.1038/ng0797-307. [DOI] [PubMed] [Google Scholar]
- Lyle R, Watanabe D, te Vruchte D, Lerchner W, Smrzka OW, Wutz A, Schageman J, Hahner L, Davies C, Barlow DP. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat Genet. 2000;25:19–21. doi: 10.1038/75546. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X chromosome of the mouse (Mus musculus) Nature. 1961;190:327–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Malissen M, Trucy J, Jouvin-Marche E, Cazenave PA, Scollay R, Malissen B. Regulation of TCR α and β gene allelic exclusion during T-cell development. Immunol Today. 1992;13:315–322. doi: 10.1016/0167-5699(92)90044-8. [DOI] [PubMed] [Google Scholar]
- McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc Natl Acad Sci. 1997;94:814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- Nakashima H, Grahovac MJ, Mazzarella R, Fujiwara H, Kitchen JR, Threat TA, Ko MSH. Two novel mouse genes—Nubp2, mapped to the t-complex on chromosome 17, and nubp1, mapped to chromosome 16—establish a new gene family of nucleotide-binding proteins in eukaryotes. Genomics. 1999;60:152–160. doi: 10.1006/geno.1999.5898. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Busslinger M. Monoallelic expression of Pax5: A paradigm for the haploinsufficiency of mammalian Pax genes? Biol Chem. 1999;380:601–611. doi: 10.1515/BC.1999.077. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Vambrie S, Steinlein P, Kozmik Z, Rolink A, Weith A, Busslinger M. Independent regulation of the two Pax5 alleles during B-cell development. Nat Genet. 1999;21:390–395. doi: 10.1038/7720. [DOI] [PubMed] [Google Scholar]
- Ohlsson R, Tycko B, Sapienza C. Monoallelic expression: 'There can only be one'. Trends Genet. 1998;14:435–438. doi: 10.1016/s0168-9525(98)01583-2. [DOI] [PubMed] [Google Scholar]
- Ohlsson R, Flam F, Fisher R, Miller S, Cui H, Pfeifer S, Adam GI. Random monoallelic expression of the imprinted IGF2 and H19 genes in the absence of discriminative parental marks. Dev Genes Evol. 1999;209:113–119. doi: 10.1007/s004270050233. [DOI] [PubMed] [Google Scholar]
- Ohlsson R, Paldi A, Graves JA. Did genomic imprinting and X chromosome inactivation arise from stochastic expression? Trends Genet. 2001;17:136–141. doi: 10.1016/s0168-9525(00)02211-3. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: Parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Rhoades KL, Singh N, Simon I, Glidden B, Cedar H, Chess A. Allele-specific expression patterns of interleukin-2 and Pax-5 revealed by a sensitive single-cell RT-PCR analysis. Curr Biol. 2000;10:789–792. doi: 10.1016/s0960-9822(00)00565-0. [DOI] [PubMed] [Google Scholar]
- Riviere I, Sunshine MJ, Littman DR. Regulation of IL-4 expression by activation of individual alleles. Immunity. 1998;9:217–228. doi: 10.1016/s1074-7613(00)80604-4. [DOI] [PubMed] [Google Scholar]
- Rogers I, Okano K, Varmuza S. Paternal transmission of the mouse Thp mutation is lethal in some genetic backgrounds. Dev Genet. 1997;20:23–28. doi: 10.1002/(SICI)1520-6408(1997)20:1<23::AID-DVG3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Schimenti J. Segregation distortion of mouse t haplotypes the molecular basis emerges. Trends Genet. 2000;16:240–243. doi: 10.1016/s0168-9525(00)02020-5. [DOI] [PubMed] [Google Scholar]
- Schweifer N, Valk PJ, Delwel R, Cox R, Francis F, Meier-Ewert S, Lehrach H, Barlow DP. Characterization of the C3 YAC contig from proximal mouse chromosome 17 and analysis of allelic expression of genes flanking the imprinted Igf2r gene. Genomics. 1997;43:285–297. doi: 10.1006/geno.1997.4816. [DOI] [PubMed] [Google Scholar]
- Severin E, Meier EM, Willers R. Flow cytometric analysis of mouse hepatocyte ploidy. I. Preparative and mathematical protocol. Cell Tissue Res. 1984;238:643–647. doi: 10.1007/BF00219883. [DOI] [PubMed] [Google Scholar]
- Silver LM. The peculiar journey of a selfish chromosome: Mouse t haplotypes and meiotic drive. Trends Genet. 1993;9:250–254. doi: 10.1016/0168-9525(93)90090-5. [DOI] [PubMed] [Google Scholar]
- Singer-Sam J. PCR primer: A laboratory manual. NY: Cold Spring Harbor Laboratory Press; 1995. Use of the SNuPE assay to quantitate allele specific sequences differing by a single nucleotide; pp. 339–343. [Google Scholar]
- Sleutels F, Barlow DP, Lyle R. The uniqueness of the imprinting mechanism. Curr Opin Genet Dev. 2000;10:229–233. doi: 10.1016/s0959-437x(00)00062-9. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Ko MSH. The short 3'-end region of complementary DNAs as PCR-based polymorphic markers for an expression map of the mouse genome. Genomics. 1993;16:161–168. doi: 10.1006/geno.1993.1153. [DOI] [PubMed] [Google Scholar]
- Tilghman SM. The sins of the fathers and mothers: Genomic imprinting in mammalian development. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- Villar AJ, Pedersen RA. Parental imprinting of the Mas protooncogene in mouse. Nat Genet. 1994;8:373–379. doi: 10.1038/ng1294-373. [DOI] [PubMed] [Google Scholar]
- Wareham KA, Lyon MF, Glenister PH, Williams ED. Age related reactivation of an X-linked gene. Nature. 1987;327:725–727. doi: 10.1038/327725a0. [DOI] [PubMed] [Google Scholar]
- Watanabe D, Barlow DP. Random and imprinted monoallelic expression. Genes Cells. 1996;1:795–802. doi: 10.1046/j.1365-2443.1996.d01-276.x. [DOI] [PubMed] [Google Scholar]
- Winking H, Silver LM. Characterization of a recombinant mouse T haplotype that expresses a dominant lethal maternal effect. Genetics. 1984;108:1013–1020. doi: 10.1093/genetics/108.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]