Abstract
We have isolated and sequenced all 23 members of the 22-kD α zein (z1C) gene family of maize. This is one of the largest plant gene families that has been sequenced from a single genetic background and includes the largest contiguous genomic DNA from maize with 346,292 bp to date. Twenty-two of the z1C members are found in a roughly tandem array on chromosome 4S forming a dense gene cluster 168,489-bp long. The twenty-third copy of the gene family is also located on chromosome 4S at a site ∼20 cM closer to the centromere and appears to be the wild-type allele of the floury-2 (fl2) mutation. On the basis of an analysis of maize cDNA databases, only seven of these genes appear to be expressed including the fl2 allele. The expressed genes in the cluster are interspersed with nonexpressed genes. Interestingly, some of the expressed genes differ in their transcriptional regulation. Gene amplification appears to be in blocks of genes explaining the rapid and compact expansion of the cluster during the evolution of maize.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AF090447, AF031569, and AF090446]
One of the best-characterized sets of storage proteins is derived from the prolamin fraction of maize seed. These proteins, called zeins, are specifically expressed during seed development and act as a reservoir for free amino acids. The relative expression and amino acid composition of seed storage proteins significantly impact the nutritional value of maize as animal feed (Ueda and Messing 1993). The zein-1 fraction, which is isolated with ethanol under nonreducing conditions, contains the α zeins. The α zeins consist of four gene families. The third largest, z1C, comprises mostly 22-kD proteins, whereas the other gene families consist of 19-kD proteins. Therefore, the z1C gene family is frequently referred to as the 22-kD α zein gene family. Expression of the z1C gene family is strongly reduced in opaque-2 (o2) variants (Mertz et al. 1964), because the absence of the O2 gene product in o2 homozygous plants greatly inhibits the transcriptional activation of z1C genes (Schmidt et al. 1992; Ueda et al. 1992; Muth et al. 1996; Wang and Messing 1998). It also has been shown that the regulation of storage protein genes is subject to genomic imprinting (Chaudhuri and Messing 1994) and hypermethylation changes during seed transmission (Lund et al. 1995).
Here we describe the isolation, sequencing, and analysis of all 23 members of the z1C gene family. All sequences have been obtained by constructing genomic libraries of maize inbred BSSS53 including a large-insert library based on bacterial artificial chromosomes (BACs). Twenty-two of the z1C genes are found in a roughly tandem array on the short arm of chromosome 4S. This gene cluster is 168,489 bp and part of a contiguous 346,292-bp chromosomal region sequenced in our laboratory. Additionally, there is one z1C gene copy present in a region proximal to the z1C gene cluster. Protein and RNA analysis for different backgrounds including a null mutation of o2 were used to determine the expression and regulation patterns of gene family members. The results of this research provide insight into chromosome structure, the regulation of multicopy genes, gene density in maize, and the evolution of multigene families in plants.
RESULTS
Construction of Zea mays BSSS53-Specific Genomic Libraries
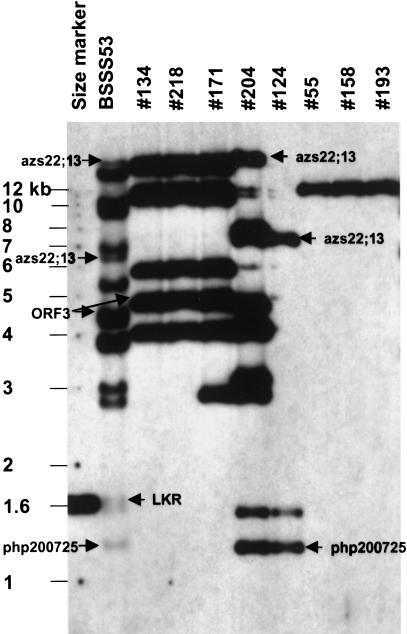
The z1C cluster is located on the short arm of chromosome 4S next to the RFLP marker php200725 at position 23.9 (Chaudhuri and Messing 1995). To capture all of the members of the gene family, two genomic libraries were constructed from partially digested DNA of the inbred maize line BSSS53, one with a cosmid and the other with a BAC vector. Eight BAC clones containing either z1C sequences or the php200725 marker were identified by a PCR assay. DNA from these clones was purified and compared with genomic DNA from BSSS53 maize plants by Southern blot analysis using z1C-specific probes, as well as several other gene-specific probes as described in Figure 1. Five clones, BAC 134, BAC 218, BAC 171, BAC 204, and BAC 124, exhibit common restriction fragment sizes, suggesting that they overlap. In the aggregate, they appear to contain the majority of z1C genes as a cluster within a contiguous chromosomal region. BAC 204 and BAC 124 also contain the php200725 marker, suggesting that this sequence is also contiguous to the z1C gene cluster. Three additional clones, BAC 55, BAC 158, and BAC 193, were found to contain a restriction fragment of the same size hybridizing to a z1C gene probe. The DNA fingerprinting of these clones (not shown) also indicates that these three BAC clones overlap. This analysis suggests that these BAC clones do not contain a cluster of z1C genes and are not contiguous with the z1C cluster found on the other BAC clones.
Figure 1.
Restriction fragment analysis of BAC clones and genomic DNA. Selected 22-kD α zein-positive BAC clones and BSSS53 genomic DNA were digested with HindIII and separated by a 1% agarose gel. After transfer to a nylon membrane, the DNA was sequentially hybridized with the following: (1) a mixture of php200725 and ORF3 (found in the 3′ region of the cluster); (2) azs22;13 (most diverged 22-kD α zein); (3) LKR (lysine-ketoglutarate reductase)-like gene (not present on any BAC clone, as a negative control), and (4) a mixture of six different 22kD α zein cDNAs of BSSS53. Use of different probes allowed us to see which BAC clone(s) contains which gene(s). Moreover, higher hybridization stringency can be applied to eliminate cross-hybridization with 19-kD α zein genes. After each hybridization experiment, the membrane was exposed and a picture taken to record each additional positive band, which results in a composite figure. Because of the different amounts of genomic DNA (∼10 μg) and BAC DNA (∼50 ng each) loaded on the gel, the genomic DNA migrated more rapidly than the BAC samples. Samples from left to right are as follows: a size marker, BSSS53 genomic DNA, BAC 134, BAC 218, BAC 171, BAC 204, BAC 124, BAC 55, BAC 158, and BAC 193. BACs in lanes 8–10 contain the single α zein gene azs22;16, which can also serve as a copy number control for the other clones. Note that the lanes are slightly shifted due to overloading genomic DNA so that smaller restriction fragments can be detected.
Physical Map of the z1C Gene Family and php200725 Regions
Sequencing was initially performed by use of cosmids that either contained the linked marker php200725 or z1C genes. The contig (GenBank no. AF090447) from overlapping cosmids III.3C12 and V.9D7 is 65,155 nucleotides long, does not contain any zein genes, but does contain the genetically linked RFLP marker php200725. Cosmid II.2E10 (GenBank no. AF090447), referred to as subcluster B, is 36,590-bp long and contains 5 tandemly arranged zein genes. Another contig, derived from a set of overlapping cosmids (GenBank no. AF031569) that have been sequenced previously (subcluster A), is 78,101-bp long and contains 10 zein genes (Llaca and Messing 1998).
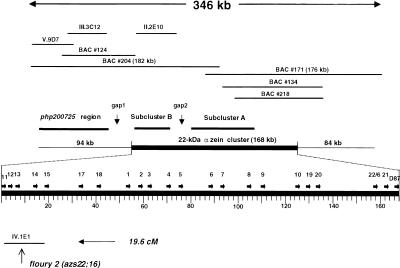
However, the cosmid library did not yield any clones to physically link these clones. Therefore, all BAC clones were sequenced at their ends and compared with the cosmid sequences. Given the sizes of the BAC clones as determined by pulsed-field gel electrophoresis, a contig of 346 kb was constructed that was used as a reference for a physical map of the zein genes relative to the php200725 marker (Fig. 2). Contrary to previous genetic mapping data (Chaudhuri and Messing 1995), all z1C genes are on the same side of php200725. Although the nature of the discrepancy is unclear, the physical analysis is more reliable because it is based on cloned DNA. The sequence ends of the two largest BAC clones, BAC 204 and BAC 171, possessed sequence identity to subcluster A and overlapped by 12,127 nucleotides. To confirm sequences from cosmid clones and obtain sequences in gaps and flanking regions (Fig. 2), BAC 204 and BAC 171 were sequenced and found to collectively possess a sequence of 346,292 bp of contiguous maize DNA sequence. To date, the BAC 204/BAC 171 contig is the longest piece of contiguous maize genomic DNA to be completely sequenced (same GenBank number as cosmid II.2E10, GenBank no. AF090447).
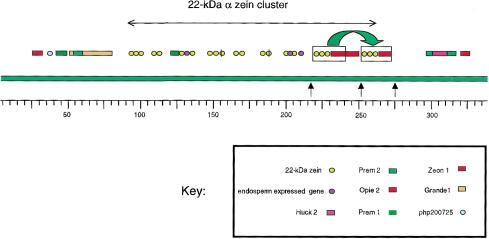
Figure 2.
Physical map of the 346-kb region derived from BAC clones. Two BAC clones, BAC 204 and BAC 171 (see Fig. 1) were used to construct a physical map of the 22-kD α zein cluster region (346-kb contig). Bars above and below the overlapping BAC clones represent the other BAC clones and the cosmid clones that have been sequenced in this study. A leftward arrow on the bottom of the figure points to cosmid clone IV.1E1, containing the fl2 allele that is located 19.6 cm closer to the centromere. However, we do not know the orientation of the Fl2 gene or the zein cluster relative to each other or to the centromere. The physical map is divided into three parts, with the middle portion highlighted by a heavy bar. This 168-kb region is enlarged below to illustrate the tandem array of the 22 clustered 22-kD α zein genes. Gene copies are designated only by the numbers following the (azs22;) prefix. The two other parts of the sequence represent the 94-kb upstream and the 84-kb downstream regions. The upstream region contains the php200725 region sequenced from two overlapping cosmids, V.9D7 and III.3C12, and has been marked by a bar above the sequence. The position of subclusters B and A have been marked by bars as well. Note that sequences in gap1 and gap2 have been derived from BAC clone 204.
The php200725 cosmid contig is positioned from 1 to 65,155 and subcluster B, cosmid II.2E10, from 80,292 to 116,863. There is a 15,138-bp space (gap1) between cosmid sequences from the php200725 region and subcluster B and a 29,511-bp space between subclusters B and A (gap2). Gap1 contains a Zeon-1 retroelement (Hu et al. 1995) and gap2 contains two additional z1C genes and a Prem-1 retroelement (Turcich and Mascarenhas 1994). Beside the 17 zein genes in the cosmid clones and gap2, five additional 22-kD α zein genes were discovered in the sequence of BAC 171. Two of these z1C genes encode proteins that have been identified previously by their position in isoelectric focusing (IEF) gels, in which they were designated as zp22/6 and zp22/D87. On the basis of these data, we can conclude that 22 z1C genes are tandemly arrayed and are physically closely linked to the genetic marker php200725.
The Unlinked z1C Gene Copy Corresponds to the fl2 Locus
In addition to the five BAC clones containing the z1C gene cluster, three independent but overlapping BAC clones contained a single restriction fragment hybridizing to a z1C probe. On the basis of restriction fragment analysis of these DNAs, the same chromosomal region, lacking the sequences flanking the zein gene(s), appears be present on cosmid clone IV.1E1 (data not shown). Because of its smaller size, the cosmid was sequenced to determine how many z1C gene copies are present and what immediate surrounding sequences flank those zein gene copies. The IV.1E1 insert is 30,593 bp in length (GenBank no. AF090446) and contains a single z1C gene (deemed azs22;16) near its center (Fig. 2). Comparison with GenBank database sequences also suggests that azs22;16 is homologous to the genomic clone pCC515 (Coleman et al. 1997). The sequence of pCC515 was derived from maize inbred W64Afl2 and is responsible for the fl2 mutation. To test whether azs22;16 is the normal allele of the fl2 mutation, azs22;16 was mapped by two single nucleotide polymorphisms (SNPs) in a backcross population of (Mo17 X BSSS53) X Mo17 relative to php200725. CDO520 was used as a third marker. The distance of 19.6 cM (21 recombination events of 107) is in agreement with the 20-cM distance between fl2 and php200725 on the maize genetic map (http://www.agron.missouri.edu). Furthermore, one of the codon differences between azs22;16 and pCC515 is a substitution of valine for alanine in position −21; introduction of a 22-kD α zein gene containing this mutation was shown previously to produce a fl2 phenotype in transgenic maize (Coleman et al. 1997).
Expression of the z1C Gene Family
Two cDNA libraries derived from tissues including immature endosperm have been sequenced recently to establish a maize EST database, one from early embryo tissue of IHO90 (Illinois High Oil), and one from early seed tissue of Ohio43 (http://www.zmdb.iastate.edu/). This EST database also contains many zein mRNA sequences including those of z1C genes. Coding regions from the z1C genes were subjected to a BLASTN analysis of the maize EST database. EST matches of 98% or greater fall into seven groups, each representing an expressed gene in the two inbred lines. IHO had four and Ohio43 had five different z1C genes expressed with two of the expressed genes in common (Table 1). Additionally, we compared the z1C genes with completely sequenced cDNA clones from four other inbred lines in GenBank; ESTs represent only single sequence reads of cDNAs from the 5′ or 3′ end. Although we do not have comprehensive cDNA data for all of these inbreds, the seven genes active in either IHO90 or Ohio43 appear to be active in at least some other inbreds as well (Table 1).
Table 1.
mRNAs of 22-kDa α Zein Genes in Different Inbred Lines
| BSSS53 | IHO90* | Ohio43* | Inbred** | cDNA | Accession |
|---|---|---|---|---|---|
| azs22;4 | no | yes | W64A, | cZ22.3, | VO1480, |
| W22 | T;SF4 | X55722 | |||
| azs22;8 | no | yes | W64A | pcM1 | M12141 |
| azs22;10 | yes | yes | n/a | n/a | n/a |
| azs22;14 | no | yes | W64A | cZ22A1 | VO1475 |
| azs22;16 | yes | yes | W64A | cZ22.1 | VO1478 |
| zp22/6 | yes | no | n/a | n/a | n/a |
| zp22/D87 | yes | no | IHP | B49 | VO1477 |
Groups with greater than 98% similarity.
Non-EST projects, cDNA clones.
n/a, Not available.
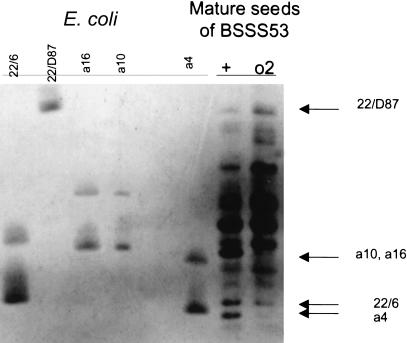
Of the seven expressed genes, five were sampled for protein analysis in the presence and absence of Opaque 2 (O2). O2 encodes a b-zip class of transcription factor that specifically recognizes the promoter of z1C genes (Schmidt et al. 1992). If expression of all z1C genes is controlled by the O2 transcription factor, their proteins should be absent in the homozygous opaque 2 (o2) variant. To identify the five selected z1C gene products in an extract from BSSS53 endosperm tissue, the coding regions of these genes were cloned into the pET5a expression vector, expressed in Escherichia coli, and purified as described in the Methods section. Following ethanol extraction to remove non-zein proteins, the migration patterns of the bacterially expressed proteins were compared with those of proteins isolated from BSSS53 and BSSS53(o2) by use of IEF gel electrophoresis (Fig. 3). BSSS53(o2) is an isogenic line of BSSS53 with an introgressed mutation of the o2 locus, that is, the O2 gene is no longer expressed (R. Song, V. Llaca, and J. Messing in prep.). This IEF analysis suggested that two of the five genes, zp22/6 and zp22/D87, are expressed in the absence of O2. In contrast, azs22;4, asz22;10, and asz22;16 (fl2 allele) appear to require O2, as the corresponding bands are missing in the BSSS53(o2) lanes (Fig. 3).
Figure 3.
Expression of cloned zein genes. Proteins from individual cell cultures and mature seeds of BSSS53 and BSSS53 (o2) were prepared and subjected to IEF gel electrophoresis (as described in Methods). Shown from right to left are samples a4 (azs22;4), a10 (azs22;10), a16 (azs22;16), zp22/D87, and zp22/6. Note that proteins azs22;10 and azs22;16 run in the same position in the gel and that proteins from E. coli always show two bands. The lower band corresponds to the actual endosperm protein and the upper band is possibly a denatured form.
Distance Analysis of the Members of the z1C Gene Family
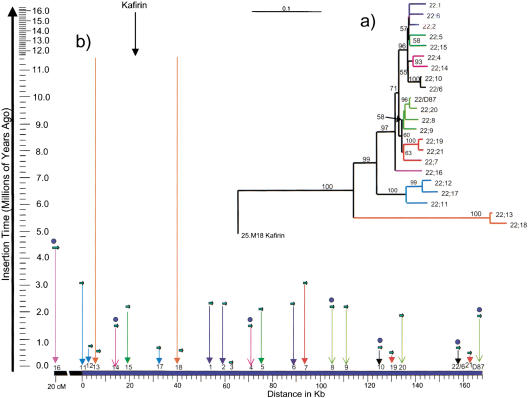
To investigate the amplification of the z1C gene copies in an evolutionary context, the coding sequences of all members of the z1C gene family were compared in a pair-wise fashion using substitution rates for grass nuclear genes (Gaut 1998). On the basis of this analysis, the ancestral z1C gene arose before allotetraploidization of maize (Gaut and Doebley 1997) 11.5 million years ago, but was duplicated within the last 0.5 million years to yield azs22;13 and azs22;18 (Fig. 4). One of the oldest duplications is the fl2 allele (∼4.3 million years ago) that persisted as a single copy ∼20 cM closer to the centromere. The other members of this gene cluster fall into smaller clades. However, the divergence of these genes does not correlate with their amplification. For instance, azs22;10, asz22;19, and azs22;20 diverged at different times, but became amplified as a group together with a large opie2 retrotransposon (SanMiguel et al. 1996). Interestingly, another opie2 element was inserted between the duplication (Fig. 5). Another example is the azs22;14 and azs22;15 pair, which is a duplicate of azs22;4 and azs22;5. Interestingly, in both examples, the 5′ copy is an active gene in both sets (Fig. 4). One set of genes arose ∼2 million years ago, another 0.5 million years ago, indicating that the expansion of the gene family is a recent event in the evolution of the maize genome.
Figure 4.
Distance analysis of the 22-kD α zein gene sequences from BSSS53. (a) A phylogenetic tree (see Methods section) has been inserted in the right corner. Gene names are abbreviated as 22;n instead of azs22;n. The coding sequences of all 22-kD α zein genes except for azs22;3 are represented in the tree. The azs22;7 sequence has been derived by subtracting an insertion sequence. Bootstrap numbers are shown above or at the internodes. The tree scale bar represents 10% divergence. (b) The estimated divergence time of each α zein gene (millions of years) vs. point of insertion (kb) is shown. Small green arrows represent the different copies of the zein genes; active copies have a blue dot above them. Phylogenetically related zein copies have identical arrows (color and head style) corresponding to the colored tree branches. The positions of green arrows for azs22;13 and azs22;18 represent their recent divergence, whereas the orange arrows represent their origin. Genes from the same clade have been color coded to illustrate that amplification did not occur in a simple physical order.
Figure 5.
DNA sequence elements in the 346-kb contig. The entire 346-kb sequence is represented as a green bar with size scale below. Above the bar, the 22-kD α zein genes are shown in relation to other sequence features. Color-coded bars visualize the position of the various predicted genes, retroelements, and the 22-kD α zein genes. A key for these is provided. The truncation of azs22;3 and the insertion in azs22;7 are marked by the vertical lines through the small circles. The 3′ terminal duplicated sequence is marked with boxes. The portion depicting the 168-kb zein gene cluster is marked by a bidirectional arrow above. Within this region, the two subclusters are marked by bidirectional arrows as well. In addition, upward dotted arrows indicate from left to right the positions of the direct repeats, DR1, DR2, and DR3 that flank the recent amplified genes.
DISCUSSION
Compactness of the z1C Gene Cluster
We cloned and sequenced all 23 members of the z1C gene family that encodes the 22-kD α zein storage proteins in maize. Twenty-two of the genes are found in a tandem array on chromosome 4S, whereas the twenty-third gene is located at a more proximal location on the same chromosome arm. Although other gene clusters have been described in plants, the 22-kD α zein gene cluster is unique because of its size, compactness, and stability. For instance, the major disease-resistance gene complex in lettuce has been estimated to have 24 copies, but it is spread over 3.5 Mbp (Meyers et al. 1998). It has been suggested that the size of this complex in lettuce is related to the genome size, which is slightly smaller than maize. Rice with a genome size only one-sixth of that of maize has a disease-resistance gene cluster of the Xa21 family that has eight genes within 230 kb (Ronald et al. 1992; Song et al. 1997). However, this is still a rather large distance compared with our maize example with 22 genes found within 168 kb. An example of disease-resistance genes in maize is the rp1 locus, located within 1 mb of chromosome 10 (Sudupak et al. 1993; Collins et al. 1999). These genes undergo unequal crossing over very frequently and change in copy number even within one generation. Crossing over between different copies of the gene family also creates new chimeric genes. There is no evidence of such crossing over for the zein cluster of the z1C genes. Sequence comparison with GenBank allowed us to identify a number of orthologous genomic sequences from other inbred lines (Table 2). For example, the size of intergenic spaces of two gene copies is known for W64A and of five gene copies for W22. In both cases, the size of intergenic spaces of the orthologous sequences in BSSS53 is the same. Orthologous sequences are conserved 98% or greater, but paralogous sequences share as little as 78% sequence identity, suggesting that the z1C gene cluster is highly conserved among different inbred lines. Moreover, compared with the cluster of disease-resistance genes, it appears that the z1C gene cluster is more stable.
Table 2.
Orthologous 22-kDa α Zein Genomic Sequences*
| Clone name | Inbred | Accession | BSSS53 | Homology |
|---|---|---|---|---|
| pML1 | A619 | X14334 | azs22;2 | 98% |
| ZSF4C1 | W22 | X55722 | azs22;4 | 98% |
| ZSF4C2 | W22 | X55723 | azs22;5 | 98% |
| ZSF4C3 | W22 | X55726 | azs22;6 | 98% |
| ZSF4C4 | W22 | X55724 | azs22;7 | 98% |
| ZSF4C5 | W22 | X55725 | azs22;8 | 98% |
| Z7 | W22 | KO2066 | azs22;11 | 99% |
| gZ22.8H3 | W64A | X61085 | azs22;12 | 98% |
| gZ22.8H3 | W64A | X61085 | azs22;13 | 98% |
| zA1 | W64A | VO1474 | azs22;20 | 99% |
Sequences with > 98% similarity including pseudogenes.
Possible Mechanism of Gene Amplification
What mechanisms could one then envision concerning how these zein genes have amplified in such a compact fashion within a relatively short time during evolution? In this respect, it is important to consider that amplification and phylogeny of zein gene copies do not correlate, suggesting that they occurred independently (Fig. 4). Because it is more difficult to resolve this with older amplification events, we focus on the sequences around the latest amplification event at the 3′ end. Interestingly, a 2-kb nongenic sequence can be found in three strategic positions (Fig. 5). The first one (DR1) is upstream of the promoter of azs22;10, the second (DR2) is upstream of zp22/6, and the third (DR3) is downstream of zp22/D87. If unequal crossing over occurs between two parental lines containing only DR1 and DR2, either a new copy (DR3) will be generated or one repeat will be lost. In such a scenario, all sequences between the repeats become duplicated or deleted. This could explain how zein genes of different clades are amplified at the same time. It could also account for the expansion of the zein gene cluster in infrequent but synchronized steps. These repeats are reminiscent of LTRs, which can also undergo unequal crossing over, which, in most cases, leads to a deletion resulting in a solo LTR. Recombination between short repeats would also explain the simultaneous absence of zp22/6 and zp22/D87 in many inbreds, in which amplification could have been reversed by a deletion. On the other hand, unequal crossing over within zein genes cannot be excluded either. In contrast, it is likely that the internal deletion of zp22/D87 has arisen from such an event after duplication of the azs22;20 gene.
Gene Density of z1C Gene Cluster Chromosomal Region
The sequenced region has a variable gene density. To analyze gene distribution within the two locations on chromosome 4S, sequences from the 346-kb region containing the zein gene cluster and the 31-kb region containing the Fl2 gene were subjected to BLASTX analysis. The 346-kb region was divided into gene islands to illustrate the variability of gene density (Table 3). The overall gene density is ∼1/10 kb, which is similar to the fl2 locus. If maize had 50,000 genes, the average gene density would amount to 1/50 kb. These relatively gene-rich regions differ drastically from the other large region in maize that has been characterized at the DNA sequence level (280 kb), the Adh1 locus on chromosome 1 (SanMiguel et al. 1996). Beside Adh1, only one other gene (u22) has been identified within the sequenced region containing Adh1. The remaining space is occupied by nested retrotransposons. Insertion of these elements has been estimated to have occurred between 0.5 and 5 million years ago. This is within the same time period in which the z1C gene family has expanded (Fig. 4).
Table 3.
Gene Density at the Zein Gene Cluster and the fl2 Locus
| Interval (kb) | # of genes* | Gene density (Kb/gene) |
|---|---|---|
| 0–25 | 4 | 6.25 |
| 25–93 | 2 | 34 |
| 93–263 | 25 | 6.8 |
| 263–333 | 1 | 70 |
| 333–346 | 3 | 4.3 |
| Total | 35 | 9.9 |
| 0–31 | 3 | 10.15 |
Genes of transposable elements have been excluded; final table entry is for the fl2 locus.
Upstream of the α zein gene cluster is the linked php200725 marker. EST analysis confirms that php200725 is expressed. Near php200725, there are two full-length LTR-retrotransposons belonging to the Prem1 and Prem2 families (Turcich and Mascarenhas 1994). On either side of php200725, an element belonging to the Zeon1 family was identified (Hu et al. 1995). The sequence surrounding php200725 contains six predicted coding sequences and several miniature-inverted-repeat-transposable-elements (MITEs), which unlike retrotransposons, are known to invade genic regions (Wessler et al. 1995). Most of the predicted genes occur within the first 25 kb. The following 68 kb have a relatively low gene density, mainly because of the full-length LTR retrotransposons. There are three additional genes of unknown function interspersed with the zein genes (Fig. 5) that are also expressed in maize endosperm (data not shown). This would amount to a gene density of 6.8 kb/gene over a rather long distance (170 kb). There are relatively few retrotransposons within this region. The most recent transposition into the zein cluster is found at the 3′ end, where we found insertions of the Opie2 retrotransposon (SanMiguel et al. 1996). Downstream of the α zein gene cluster, within the next 70 kb, we find only one predicted gene, a cytochrome P450-like gene. This low gene density is mainly the result of the presence of large retrotransposons and is followed again by a gene-rich region (Table 3). The gene density at the fl2 position, 20 cM away from php200725, also seems to be relatively high. Beside the single z1C gene in the center of a 30-kb region, two additional genes are predicted. A single 8-kb LTR, a copia-type retrotransposon, is located in the 3′ region and two predicted gene sequences with no known function are located in the first 10 kb. The predicted genes are flanked by multiple MITEs and are separated from the 22-kD α zein gene by a fractured Prem1 retroelement.
Basis of Changes in Gene Expression of the Members of the Gene Family
Expressed copies of zein genes are interspersed with inactive copies at variable distances. This is consistent with many other examples of gene clusters. For instance, in the human major histocompatability complex (MHC), 3 of about 20 class I genes are expressed (Trowsdale 1993). In addition, many of the inactive zein genes have accumulated mutations within the coding region, with most of them converting the glutamine codons CAG and CAA to stop codons. It has been shown that premature stop codons decrease mRNA stability (Van Hoof and Green 1996) and that a single in-frame stop codon reduces the mRNA concentration significantly (Liu and Rubenstein 1993). Moreover, of 16 genes lacking significant mRNA levels, only 5 have more than one in-frame stop codon. Eleven genes might have been inactivated only recently. Therefore, many copies of the zein gene cluster might serve as a gene reservoir, in which normal expression of individual members could be restored by recombination in different inbred lines of maize.
However, the most striking change in gene expression is found in the transcriptional control of these genes. Our experiments do not exclude the possibility that zp22/6 and zp22/D87 are still activated by O2. In contrast, the promoter regions of both genes have the cis-acting elements of the O2 transcription factor that have been shown in transient expression systems to be sufficient for the transcription of reporter genes (Muth et al. 1996). However, a lack of the O2 gene product prevents the expression of azs22;10, the progenitor of zp22/6, whereas expression of zp22/6 remains active. It is clear that 19-kD α zein genes are activated by an alternate transcription protein complex because they are also expressed in the absence of O2. Unlike zp22/6, they lack the cis-acting elements for O2, but share the prolamin box, P-box with the GTGTAAAG motif, at about the same distance from the transcriptional start site. Although this element is present in all of the α zein genes, another sequence-specific interaction must account for a transcriptional factor not yet characterized at the gene level. We therefore proposed a recruiter model for the expression of zein genes (Wang and Messing 1998). This is based on the biochemical data concerning the interaction between the prolamin box-binding factor PBF-1 and O2 and the fact that their binding sites are just 20 bp apart. In this scenario, the tissue-specific transcription factor PBF-1 is expressed after mitotic divisions cease in the starchy endosperm. This represents the onset of storage protein and starch synthesis. However, transcriptional activation is modulated by additional trans-acting factors, which are specific for a subset of promoters (e.g., 22- and 19-kD zeins). Modulation depends on the affinity of these additional trans-acting factors (e.g., O2 and O7) to PBF-1 and their promoter-binding sites.
Orthologous and Paralogous Sequences in the Regulation of Gene Expression
It is interesting to note that maize arose as an allotetraploid (Gaut and Doebley 1997). This provides us with examples in which genes from the two subgenomes have led to changes in promoter specificity. Orthologous genes like R1 and B1 are helix-loop-helix type transcription factors that have arisen from the same ancestral gene (Gaut and Doebley 1997), but are now expressed in different tissues at different times during plant development (Ludwig and Wessler 1990; Goff et al. 1992). However, there have also been paralogous gene duplications of transcription factors like R1, and P1, a myb-like transcription factor, that have changed their expression (Walker et al. 1995; Zhang et al. 2000). Therefore, it is possible that an ortholog or paralog of O2 has evolved that might act on only slightly different promoter sequences. Genetic analysis would be consistent with this explanation, as nonallelic opaque mutations have been isolated. For instance, combinations of o2 and o7 give additive effects on α zein gene expression (Di Fonzo et al. 1979).
Another variable parameter is the target sites of these nonallelic trans-acting factors. For instance, we found that inbred lines like W22, B73, Mo17, CO159, CM37, TX303, and T232 not only lack zp22/6 but also zp22/D87, indicating that the amplification of the 3′ region may represent a haplotype of today's germplasm. A654 and A188, like BSSS53, belong to the other haplotype (R. Song, V. Llaca, and J. Messing, in prep.). Because zp22/6 and zp22/D87 are expressed in the absence of O2, the difference of the O2 effect would be stronger in haplotypes missing these two genes. Therefore, phenotypes affected by zein gene expression might differ with respect to genetic background. All of these examples suggest that plant genomes can adapt very rapidly by duplicating different types of genes either by polyploidization (orthologs) or gene amplification (paralogs) and then fine tuning their expression through a combination of trans- and cis-acting factors (Messing 2001).
Genomic Imprinting as a Possible Stability Factor for the Gene Cluster
The compactness of the gene cluster poses the question of how these genes escaped epigenetic gene silencing that has been observed for multiple tandem copies of either endogenous or exogenous genes (Matzke et al. 1994; Kermicle et al. 1995; Kumpatla et al. 1997; Vaucheret et al. 1998). Epigenetic modifications are thought to be responsible for gene silencing because of the associated hypermethylation of DNA sequences. Paramutated and imprinted genes are also hypermethylated, which represents the inactive state of a gene (Meyer et al. 1993; Ronchi et al. 1995; Walker 1998; Alleman and Doctor 2000). Hypermethylation of zein genes has been reported previously (Lund et al. 1995). However, during female gametogenesis, hypermethylated alleles can be demethylated, reversing the gene-silencing effect of genes that are expressed in the endosperm (Messing and Grossniklaus 1999). This would be consistent with the reciprocal crosses of the hypermethylated alleles of zein genes that change their methylation state depending on the direction of reciprocal crosses (Lund et al. 1995). Therefore, if any of the active zein genes in the cluster are imprinted, the imprint would be removed during female gametogenesis and only the male-transmitted gene would not be expressed during maize endosperm development. Then, one would predict that the single gene azs22;16 in the fl2 position should not become epigenetically modified. Interestingly, genetic analysis of the fl2 mutation has exhibited gene dosage but not a parent-of-origin inheritance. On the other hand, as methylated genes in the cluster are only demethylated after meiosis, the epigenetic modification is still present during meiosis and might suppress unequal crossing over between zein genes. It is believed that epigenetically modified sequences also prevent recombination and transposition (Peschke et al. 1987; Bennetzen et al. 1994; Timmermans et al.). Such suppression of recombination would also be consistent with the conservation of the zein gene number and distances among inbred lines. Occasionally, epigenetic modifications may also be reversed by stress, for example, activation of a transposable element (Peschke et al. 1987) and depend on environmental factors as in paramutation (Mikula 1995), which would account for infrequent unequal crossing over in the zein gene cluster. Therefore, one could envision two classes of genes in plants that are subject to genomic imprinting. One might require imprinting for development like MEA, FIS, and FIE (Ohad et al. 1999; Luo et al. 2000; Vielle-Calzada et al. 2000), whereas the other might require imprinting for allelic structural features. Clearly, having a complete sequence set of a single multigene family and the physical position of all their members in the genome will be of great value as a reference for further comparative genome analysis and gene expression studies.
METHODS
Genomic Libraries
An overlapping cosmid library for Zea mays BSSS53 was constructed using the SuperCos system as described elsewhere (Llaca and Messing 1998). A BAC library of Zea mays BSSS53 was constructed with the pBeloBAC II vector (Wang et al. 1997). High-molecular-weight (HMW) DNA was prepared as described previously (Guidet et al. 1990) using 2-week-old maize seedling stems grown under greenhouse conditions. The HMW DNA was partially digested with HindIII, and then subjected to size selection and fractionation with pulsed field gel electrophoresis (PFGE) as described elsewhere (Osoegawa et al. 1998). One additional fractionation was carried out to increase the average insertion size. The desired DNA fraction was electroeluted by the method of Strong et al. (1997). Vector preparation, ligation, and transformation have been described previously (Osoegawa et al. 1998).
The BAC library contained ∼7 × 104 independent recombinants, with an average insert size of 100 kb (∼3 genome equivalents). The library was divided into ∼350 sublibraries, each with ∼200 clones, and amplified. DNA from each sublibrary underwent a PCR-based screening using a gene-specific primer set. Primer sets were designed from different 22-kD α zein genes and php200725. After identification of a PCR product within one of the sublibraries, the positive sublibrary was further divided into subpools that contained 10 to 40 recombinants per subpool. PCR analysis was then performed to identify a positive subpool. The positive subpool was plated, single colonies were isolated, and the PCR assay was used to identify single BAC clones.
Shotgun DNA Sublibraries and Sequencing
Determinations of cosmid and BAC sequences were carried out by the shotgun DNA sequencing method (Messing et al. 1981). However, instead of M13, the pUC119 vector was used to generate the shotgun DNA libraries containing large (4–6 kb) and medium (2–4 kb) fragments (Vieira and Messing 1982). To prepare cosmid and BAC DNA for shotgun library construction, standard alkaline lysis was performed to extract DNA from an overnight culture of cells. Cosmid DNA was purified using a Qiaprep anion exchange column (QIAGEN), whereas BAC DNA was purified by double cesium-chloride equilibrium centrifugation. Pseudo-random cosmid sublibraries were generated as described by Llaca and Messing (1998), whereas BACs were randomly sheared using a Hydroshear system, as specified by the manufacturer (GeneMachines). After production of the shotgun library, one plate of 96 clones was picked and sequenced in one direction (see below). The E. coli chromosomal content of this library was determined by simple sequence analysis, and an assessment of quality and randomness was performed. Less than 5% and 1% E. coli DNA was present in the cosmid and BAC sublibraries, respectively.
Minipreps of subclones were performed using QIAGEN Ultra-well Kits. Sequencing reactions were performed with a combination of multipipetting devices and MJ Research 96-well thermocyclers. Fluorescent automated DNA sequencing was performed using BigDye primers in an ABI 377 or 3700 Sequencer (Applied Biosystems-Perkin Elmer). Base calling, quality assessment, and assembling were carried out in an Origin 2000 Unix computer with software developed by the University of Washington Genome Center. Vector sequences were removed using the program Cross-match, and base calling and quality assessment were performed using phred (Ewing and Green 1998). Sequences were assembled by use of phrap and edited with CONSED (Gordon et al. 1998). Assembly was made at 7–9× coverage. The reliability of the sequences was verified by assessing the locations of ends of shotgun clones and comparing them with the expected insert size, as well as matching them to electronic and actual restriction maps. Gaps and low-quality areas were finished using custom-specific primers. The finished sequence was deposited into GenBank.
Isoelectric Focusing of Individual 22-kD Zeins from BSSS53
Coding regions of 22-kD α zein genes were inserted into the E. coli expression vector pET5a (Promega). The coding sequences corresponding to the portion of the 22-kD α zein proteins without the signal peptide (−21) were amplified by PCR with the following primer pair: amino terminus, 5′-ACACCATATGTTCATTATTCCACAATGCTCA-3′ (underlined sequence is a NdeI site); carboxyl terminus, 5′-TTAAGGATCCTATATAATCTAAAAGATGGCA-3′ (underlined sequence is a BamHI site). The PCR products were treated with NdeI and BamHI and cloned into corresponding sites of the expression vector. The resulting fusion proteins contained an extra methionine at the amino terminus when compared with natural mature 22-kD α zeins, but this extra amino acid did not change the pI value of the protein. The recombinant clones were transformed into the BL21(DE3) plys S strain and the expression of fusion proteins was induced by IPTG according to the manufacturer's instructions. The bacteria were collected by centrifugation, resuspended in 300 μlL TE buffer, and subjected to several cycles of freezing (liquid nitrogen) and thawing (37°C water bath). The solution was adjusted to 70% ethanol and kept at 4°C overnight. The supernatant was recovered by centrifugation and further desalted by a Centricon-10 column (Amico) with 70% ethanol. The protein concentration was determined by the Bio-Rad protein assay kit (Bio-Rad) and ∼10 μg of ethanol extracted protein for each sample was analyzed on an IEF (pH 5–8) gel as described before (Chaudhuri and Messing 1995). Protein bands were visualized by Commassie blue staining.
DNA Sequence Analysis Programs
Sequence comparisons were performed locally using the Lasergene programs from DNAstar, on Macintosh G4 computers. DNA sequences were submitted in FASTA format to the National Center for Biotechnology Information for BLASTN or BLASTX analysis (Altschul et al. 1997). Sequence data was aligned in the Genetic Database Environment (GDE) 2.2 program (Smith et al. 1994) using CLUSTALV with the following settings; K-tuple size 2, window size 6, gap penalty 10, floating penalty 10. Sequences were then adjusted interactively. Small insertions occurring only in one or two sequences were excluded from the phylogenetic analysis.
Phylogenetic Analysis
A nexus format file was generated in GDE and analyzed on a PowerMac G4 with PAUP* (Phylogenetic Analysis Using Parsimony and other methods) version 4b4 (Swofford 1999). A total of 3256 nucleotides were analyzed using the distance (minimum evolution) criterion. To root the tree, we selected the 22-kD α kafirin gene of clone 25.M18 (GenBank no. AF114171) that was linked to the orthologous sequence of the php200725 marker in Sorghum bicolor. Sorghum is believed to have a common ancestor with one of the progenitors of the allotetraploid maize genome that diverged some 16.5 million years ago (Gaut and Doebley 1997). Kafirins are the storage protein genes in sorghum, which are related to the zein genes in maize, both in terms of sequence and size (DeRose et al. 1989). All nucleotide positions were treated as independent, unordered, multistate characters of equal weight, and alignment gaps were distributed proportionally to unambiguous changes. Trees were generated by a heuristic search with random stepwise addition and 100 replicates using the Tamura-Nei Model and a γ correction of 1.6922. Trees were optimized using tree bisection-reconnection (TBR) branch swapping with MULTREES in effect. The robustness and stability of the tree was estimated using nonparametric bootstrapping (Felsenstein 1985) with 1000 replicates and 100 repetitions.
Acknowledgments
Part of the DNA sequencing was conducted by Steve Young and Steve Kavchok, whose tireless efforts to conclude the sequencing are gratefully acknowledged. We thank Huihua Fu for technical assistance during the construction of the BAC library. We also thank Kathy Ward for her technical assistance. This work has been supported by DOE Grant no. DE-FG05–95ER20194 to J.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL messing@mbcl.rutgers.edu; FAX (732) 445-0072.
Article published on-line before print: Genome Res., 10.1101/gr.197301.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.197301.
REFERENCES
- Alleman M, Doctor J. Genomic imprinting in plants: Observations and evolutionary implications. Plant Mol Biol. 2000;43:147–161. doi: 10.1023/a:1006419025155. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Schrick K, Springer P, Brown WE, SanMiguel P. Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome. 1994;37:565–576. doi: 10.1139/g94-081. [DOI] [PubMed] [Google Scholar]
- Chaudhuri S, Messing J. Allele-specific imprinting of dzr1, a post-transcriptional regulator of zein accumulation. Proc Natl Acad Sci. 1994;91:4867–4871. doi: 10.1073/pnas.91.11.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— RFLP mapping of the maize dzr1 locus that regulates methionine-rich 10-kDa zein accumulation. Mol Gen Genet. 1995;246:707–715. doi: 10.1007/BF00290716. [DOI] [PubMed] [Google Scholar]
- Coleman CE, Clore AM, Higgins R, Lopes MA, Larkins BA. Expression of a mutant alpha-zein creates the floury 2 phenotype in transgenic maize. Proc Natl Acad Sci. 1997;94:7094–7097. doi: 10.1073/pnas.94.13.7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N, Drake J, Ayliffe M, Sun Q, Ellis J, Hulbert S, Pryor T. Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell. 1999;11:1365–1376. doi: 10.1105/tpc.11.7.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose RT, Ma D-P, Kwon I-S, Hasnain SE, Klassy RC, Hall T. Characterization of the kafirin gene family from sorghum reveals extensive homology with zein from maize. Plant Mol Biol. 1989;12:245–256. doi: 10.1007/BF00043202. [DOI] [PubMed] [Google Scholar]
- Di Fonzo N, Fornasari E, Salamini F, Reggiani R, Soave C. Interaction of the mutants floury-2, opaque-7 with opaque-2 in the synthesis of endosperm proteins. J Hered. 1979;71:397–402. [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gaut BS. Molecular clocks and nucleotide substitution rates in higher plants. Evol Biol. 1998;30:93–120. [Google Scholar]
- Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Cone KC, Chandler VL. Functional analysis of the transcriptional activator encoded by the maize B gene: Evidence for a direct functional interaction between two classes of regulatory proteins. Genes & Dev. 1992;6:864–875. doi: 10.1101/gad.6.5.864. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed - a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Guidet F, Rogowsky P, Langridge P. A rapid method of preparing megabase plant DNA. Nucleic Acids Res. 1990;18:4955. doi: 10.1093/nar/18.16.4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Das OP, Messing J. Zeon-1, a member of a new maize retrotransposon family. Mol Gen Genet. 1995;248:471–480. doi: 10.1007/BF02191647. [DOI] [PubMed] [Google Scholar]
- Kermicle JL, Eggleston WB, Alleman M. Organization of paramutagenicity in R-stippled maize. Genetics. 1995;141:361–372. doi: 10.1093/genetics/141.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpatla SP, Teng W, Buchholz WG, Hall TC. Epigenetic transcriptional silencing and 5-azacytosine-mediated reactivation of a complex transgene in rice. Plant Physiol. 1997;115:361–373. doi: 10.1104/pp.115.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CN, Rubenstein I. Transcriptional characterization of an α-zein gene cluster in maize. Plant Mol Biol. 1993;22:323–336. doi: 10.1007/BF00014939. [DOI] [PubMed] [Google Scholar]
- Llaca V, Messing J. Amplicons of maize zein genes are conserved within genic but expanded and constricted in intergenic regions. Plant J. 1998;15:211–220. doi: 10.1046/j.1365-313x.1998.00200.x. [DOI] [PubMed] [Google Scholar]
- Ludwig SR, Wessler SR. Maize R gene family: Tissue-specific helix-loop-helix proteins. Cell. 1990;62:849–851. doi: 10.1016/0092-8674(90)90259-h. [DOI] [PubMed] [Google Scholar]
- Lund G, Ciceri P, Viotti A. Maternal-specific demethylation and expression of specific alleles of zein genes in the endosperm of Zea mays L. Plant J. 1995;8:571–581. doi: 10.1046/j.1365-313x.1995.8040571.x. [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci. 2000;97:10637–10642. doi: 10.1073/pnas.170292997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke AJM, Neuhuber F, Park Y-D, Ambros PF, Matzke MA. Homology-dependent gene silencing in transgenic plants: Epigenetic silencing of loci contain multiple copies of methylated transgenes. Mol Gen Genet. 1994;244:219–229. doi: 10.1007/BF00285449. [DOI] [PubMed] [Google Scholar]
- Mertz ET, Bates LS, Nelson OE. Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science. 1964;145:279–280. doi: 10.1126/science.145.3629.279. [DOI] [PubMed] [Google Scholar]
- Messing J. Do plants have more genes than humans? Trends Plant Sci. 2001;6:195–196. doi: 10.1016/s1360-1385(01)01925-2. [DOI] [PubMed] [Google Scholar]
- Messing J, Grossniklaus U. Genomic imprinting in plants. In: Ohlsson R, editor. Results and problems in cell differentiation: Genomic imprinting. Heidelberg, Germany: Springer Verlag; 1999. pp. 23–40. [DOI] [PubMed] [Google Scholar]
- Messing J, Crea R, Seeburg PH. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981;9:309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Heidmann I, Niedenhoff I. Differences in DNA methylation are associated with a paramutation phenomenon in transgenic petunia. Plant J. 1993;4:89–100. doi: 10.1046/j.1365-313x.1993.04010089.x. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Chin DB, Shen KA, Sivaramakishnan S, Lavelle DO, Zhang Z, Michelmore RW. The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell. 1998;10:1817–1832. doi: 10.1105/tpc.10.11.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikula BC. Environmental programming of heritable epigenetic changes in paramutant r-gene expression using temperature and light at a specific stage of early development in maize seedlings. Genetics. 1995;140:1379–1387. doi: 10.1093/genetics/140.4.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth JR, Müller M, Lohmer S, Salamini F, Thompson RD. The role of multiple binding sites in the activation of zein gene expression by Opaque-2. Mol Gen Genet. 1996;252:723–732. doi: 10.1007/BF02173979. [DOI] [PubMed] [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell. 1999;11:407–416. doi: 10.1105/tpc.11.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoegawa K, Woon PY, Zhao B, Frengen E, Tateno M, Catanese JJ, de Jong PJ. An improved approach for construction of bacterial artificial chromosome libraries. Genomics. 1998;52:1–8. doi: 10.1006/geno.1998.5423. [DOI] [PubMed] [Google Scholar]
- Peschke VM, Phillips RL, Gengenbach BG. Discovery of a transposable element activity among progeny of tissue culture-derived maize plants. Science. 1987;238:804–807. doi: 10.1126/science.238.4828.804. [DOI] [PubMed] [Google Scholar]
- Ronald PC, Albano B, Tabien R, Abenes L, Wu KS, McCouch S, Tanksley SD. Genetic and physical analysis of the rice bacterial blight disease resistance locus, Xa21. Mol Gen Genet. 1992;236:113–120. doi: 10.1007/BF00279649. [DOI] [PubMed] [Google Scholar]
- Ronchi A, Petroni K, Tonelli C. The reduced expression of endogenous duplications (REED) in the maize R gene family is mediated by DNA methylation. EMBO J. 1995;14:5318–5328. doi: 10.1002/j.1460-2075.1995.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zkharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Schmidt RJ, Ketudat M, Aukerman MJ, Hoschek G. Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell. 1992;4:689–700. doi: 10.1105/tpc.4.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SW, Overbeck R, Woese CR, Gilbert W, Gillevet PM. The genetic data environment: an expandable GUI for multiple sequence analysis. Comput Appl Biosci. 1994;10:671–675. doi: 10.1093/bioinformatics/10.6.671. [DOI] [PubMed] [Google Scholar]
- Song W-Y, Pi L-Y, Wang G-L, Gardener J, Holsten T, Ronald PC. Evolution of the rice Xa21 disease resistance gene family. Plant Cell. 1997;9:1279–1287. doi: 10.1105/tpc.9.8.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong ST, Ohta Y, Litman GW, Amemiya CT. Marked improvement of PAC and BAC cloning is achieved using electroelution of pulsed-field gel-separated partial digests of genomic DNA. Nucleic Acids Res. 1997;25:3959–3961. doi: 10.1093/nar/25.19.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudupak MA, Bennetzen JL, Hulbert SH. Unequal exchange and meiotic instablity of disease-resistance genes in the Rp1 region of maize. Genetics. 1993;133:119–125. doi: 10.1093/genetics/133.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogentic analysis using parsimony (and other methods), version 4.0. Sunderland, MA: Sinauer Associates; 1999. [Google Scholar]
- Timmermans MCP, Das OP, Messing J. Characterization of a meiotic crossover in maize identified by a restriction fragment length polymorphism-based method. Genetics. 1996;143:1771–1783. doi: 10.1093/genetics/143.4.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J. Genomic structure and function in the MHC. Trends Genet. 1993;9:117–122. doi: 10.1016/0168-9525(93)90205-v. [DOI] [PubMed] [Google Scholar]
- Turcich MP, Mascarenhas JP. PREM-1, a putative maize retroelement has LTR (long terminal repeat) sequences that are preferentially transcribed in the pollen. Sex Plant Reprod. 1994;7:2–11. [Google Scholar]
- Ueda T, Messing J. Manipulation of amino acid balance in maize seeds. In: Setlow JK, editor. Genetic engineering. Vol. 15. NY: Plenum Press; 1993. pp. 109–130. [DOI] [PubMed] [Google Scholar]
- Ueda T, Waverczak W, Ward K, Sher N, Ketudat M, Schmidt RJ, Messing J. Mutations of the 22– and 27-kD zein promoters affect transactivation by the Opaque-2 protein. The Plant Cell. 1992;4:701–709. doi: 10.1105/tpc.4.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof A, Green PJ. Premature nonsense codons decrease the stability of phytohemagglutinin mRNA in a position-dependent manner. Plant J. 1996;10:415–424. doi: 10.1046/j.1365-313x.1996.10030415.x. [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Mourrain P, Palauqui JC, Vernhetts S. Transgene-induced gene silencing in plants. Plant J. 1998;16:651–659. doi: 10.1046/j.1365-313x.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Baskar R, Grossniklaus U. Delayed activation of the paternal genome during seed development. Nature. 2000;404:91–94. doi: 10.1038/35003595. [DOI] [PubMed] [Google Scholar]
- Walker EL. Paramutation of the r1 locus of maize is associated with increased cytosine methylation. Genetics. 1998;148:1973–1981. doi: 10.1093/genetics/148.4.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EL, Robbins TP, Bureau TE, Kermicle J, Dellaporta SJ. Transposon-mediated chromosomal rearrangements and gene duplications in the formation of the R-r complex. EMBO J. 1995;14:2350–2363. doi: 10.1002/j.1460-2075.1995.tb07230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Boysen C, Shizuya H, Simon MI, Hood L. Complete nucleotide sequence of two generations of a bacterial artificial chromosome cloning vector. BioTechniques. 1997;23:992–993. doi: 10.2144/97236bm04. [DOI] [PubMed] [Google Scholar]
- Wang Z, Messing J. Modulation of gene expression by DNA-protein and protein-protein interactions in the promoter region of the zein multigene family. Gene. 1998;223:333–345. doi: 10.1016/s0378-1119(98)00245-5. [DOI] [PubMed] [Google Scholar]
- Wessler SR, Bureau TE, White SE. LTR-retrotransposons and MITEs: Important players in the evolution of plant genomes. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- Zhang P, Chopra S, Peterson T. A segmental gene duplication generated differentially expressed myb-homologous genes in maize. Plant Cell. 2000;12:2311–2322. doi: 10.1105/tpc.12.12.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]