Abstract
Campylobacter jejuni is the leading cause of bacterial food-borne diarrhoeal disease throughout the world, and yet is still a poorly understood pathogen. Whole genome microarray comparisons of 11 C. jejuni strains of diverse origin identified genes in up to 30 NCTC 11168 loci ranging from 0.7 to 18.7 kb that are either absent or highly divergent in these isolates. Many of these regions are associated with the biosynthesis of surface structures including flagella, lipo-oligosaccharide, and the newly identified capsule. Other strain-variable genes of known function include those responsible for iron acquisition, DNA restriction/modification, and sialylation. In fact, at least 21% of genes in the sequenced strain appear dispensable as they are absent or highly divergent in one or more of the isolates tested, thus defining 1300 C. jejuni core genes. Such core genes contribute mainly to metabolic, biosynthetic, cellular, and regulatory processes, but many virulence determinants are also conserved. Comparison of the capsule biosynthesis locus revealed conservation of all the genes in this region in strains with the same Penner serotype as strain NCTC 11168. By contrast, between 5 and 17 NCTC 11168 genes in this region are either absent or highly divergent in strains of a different serotype from the sequenced strain, providing further evidence that the capsule accounts for Penner serotype specificity. These studies reveal extensive genetic diversity among C. jejuni strains and pave the way toward identifying correlates of pathogenicity and developing improved epidemiological tools for this problematic pathogen.
Campylobacter jejuni is the principal cause of human bacterial gastroenteritis worldwide (Blaser 1997). Disease outcomes from C. jejuni infection vary from mild, noninflammatory, self-limiting diarrhoea to severe, inflammatory, bloody diarrhoea lasting for several weeks (Wassenaar and Blaser 1999). In addition, C. jejuni is also associated with the development of the peripheral neuropathies Miller Fisher and Guillain-Barré (GBS) syndromes (Nachamkin et al. 1998), which further adds to the economic burden of C. jejuni infection (Buzby et al. 1997). Given the range of disease outcomes and the importance of C. jejuni as a major economic and social problem, it is surprising that the ecology, epidemiology, and, in particular, the pathogenesis of the organism are so poorly understood. This lack of understanding was the driving force for instigating the recently completed C. jejuni genome project (Parkhill et al. 2000).
The sequencing of the C. jejuni NCTC 11168 genome revealed few mechanisms by which the organism can generate genetic diversity. C. jejuni is almost unique among sequenced bacterial pathogens in that the NCTC 11168 genome sequence contains no transposons, phage remnants, or insertion sequence elements and very few repeat sequences. However, several lines of evidence have indicated relative intraspecific genotypic and phenotypic diversity compared to other enteropathogens. For example, two serotyping schemes detect a wide variety of serotypes: the Penner heat-stable antigen typing scheme detects more than 60 serotypes (Penner and Hennessy 1980), whereas the Lior heat-labile antigen typing scheme recognizes >100 serotypes (Lior et al. 1982). Genotypic diversity has been shown by ribotyping, pulsed field gel electrophoresis, and several PCR-based techniques (Fayos et al. 1992; Lam et al. 1995; Hanninen et al. 1999; Kokotovic and On 1999). Unfortunately, these methods are unable to further characterize the genetic basis for this observed variability. Diversity between C. jejuni strains has also been observed at the phenotypic level, particularly for characteristics implicated in pathogenicity, such as adherence to and invasion of epithelial cell lines (Konkel and Joens 1989; Everest et al. 1992; Wooldridge and Ketley 1997), toxin production (Wassenaar 1997), serum resistance (Blaser et al. 1986), exopolysaccharide production (Karlyshev et al. 2000a), sialylation of LOS (Linton et al. 2000b), and the ability to colonize chicks (Korolik et al. 1998; Barrow and Page 2000).
The Penner serotyping system has been used worldwide for grouping C. jejuni strains for more than 20 years (Penner and Hennessy 1980). Important epidemiological correlations have been made on the basis of this typing system, including the overrepresentation of serotype O:19 strains isolated in GBS patients (Yuki and Miyatake 1998) and a cluster of GBS cases linked with serotype O:41 (Prendergast et al. 1998). Problems associated with the Penner serotyping system include difficulty in standardizing preparation of antisera, the expense of specific sera, and a significant proportion of untypable strains. Surprisingly, despite the widespread use of the Penner serotyping system, the nature of the serotypic determinant was until recently unclear. Data now indicate that the heat-stable Penner serotyping antigen is the newly recognized capsular polysaccharide (Karlyshev et al. 2000a).
DNA microarrays have been used to compare interstrain, intraspecific variations in bacteria at the genomic level, referred to as genomotyping. For example, to investigate the differences between Mycobacterium tuberculosis, Mycobacterium bovis, and the various BCG vaccine daughter strains, their genomic compositions were studied by comparative hybridization experiments using a M. tuberculosis H37Rv DNA microarray (Behr et al. 1999). These experiments highlighted many regions of the virulent M. tuberculosis H37Rv reference strain that were lacking in M. bovis and the BCG strains. More recently, use of a whole genome microarray from two sequenced strains revealed diversity among clinical isolates of the gastric pathogen Helicobacter pylori, which is closely related to C. jejuni. This study identified a class of candidate virulence genes by their coinheritance with the well-characterized H. pylori cag pathogenicity island (Salama et al. 2000).
To date there appears to be very little sequence diversity between housekeeping genes from different C. jejuni strains, particularly between human isolates (Dingle et al. 2001; Suerbaum et al. 2001). Genes that encode surface-exposed antigenic proteins are known to vary in sequence, presumably to evade the immune system. The flagellin structural genes flaA and flaB both have hypervariable regions that probably correspond to surface-exposed parts of the proteins. However, genes that encode other nonimmunogenic virulence factors appear to be closely conserved. For example, OmpH1 and Peb1A are both surface exposed on the bacteria, but there does not appear to be the selection for diversity in the ompH1 and peb1A genes that is seen with the flagellin structural genes (for review, see Meinersmann 2000). Furthermore, the NCTC 11168 genome contains very few repeat sequences (<1% of the genome) (Parkhill et al. 2000). Therefore, C. jejuni would appear to be an ideal organism to study by genomotyping.
One of the drawbacks of microarray technology is the initial high cost involved in the synthesis of gene-specific primers to amplify each gene in a genome. The 10-fold sequencing of the C. jejuni NCTC 11168 genome provided the useful by-product of an ordered library of sequence-defined pUC18 clones covering the entire genome (Parkhill et al. 2000). Optimum clones for all putative coding sequences (CDS) were selected, and PCR products were amplified from these clones using a single pair of vector primers. These PCR products were spotted onto glass slides to produce a low-cost C. jejuni whole genome microarray. Although only 34.5% of the PCR products were gene-specific, this type of microarray is suitable for performing genomotyping, where the data generated are qualitative rather than quantitative. The aim of our study was to use this microarray to investigate the extent of genetic diversity among C. jejuni isolates, and more specifically, the relationship between capsule locus variability and Penner serotype.
RESULTS
Construction of the C. jejuni NCTC 11168 Whole Genome DNA Microarray
A whole genome DNA microarray was constructed containing an element representing each of the 1731 CDSs originally identified from the genome sequence (J. Parkhill, unpubl.). The number of annotated CDSs in the C. jejuni NCTC 11168 genome was subsequently reduced to 1654 (Parkhill et al. 2000). The resulting 77 “nongenes” are therefore each flagged in the gene lists used in the analysis software as a “not annotated gene”. The tiling program, designed to identify the optimum clone for each CDS from the ordered pUC18 plasmid library, identified clones containing a single gene fragment for 597 (34.5%) of the CDSs. For 613 (35.4%) of the CDSs, a further set of clones was identified with an adjacent gene fragment either upstream or downstream of the selected gene. The remaining 521 (30.1%) CDSs are represented by clones containing more than one gene fragment adjacent to the selected gene. The size range of the vector primer-amplified PCR products was 0.5 kb to 2.0 kb. Controls comprised the 5S, 16S, and 23S rRNA genes (with four serial fourfold dilutions of the 23S rRNA gene), the multiple cloning site of the pUC18 vector, and the human β-actin gene. The controls were printed along the top row of each of the 16 subgrids.
General Features of Interstrain Whole Genome Hybridizations
The data generated are available on-line at http://www.sghms.ac.uk/depts/medmicro/bugs/GR-1858. One of the limitations of using a microarray generated using plasmid clones rather than gene-specific primers is the potential difficulty in interpreting the data owing to the presence of overlapping genes on the microarray. If a gene is absent, but is represented by a clone containing adjacent genes that are present, then the red/green hybridization signal intensity ratio may be raised above 0.5 and the gene subsequently scored as present. However, genes scored as absent or highly divergent should be accurate within the limits of array technology.
The results presented identify regions of the NCTC 11168 genome ranging from <0.5 kb to nearly 20 kb to which genomic DNA from one or more of the 11 test strains listed in Table 1 failed to hybridize. Table 1 also shows the percentage of NCTC 11168 genes to which genomic DNA from each strain failed to hybridize. Genes can therefore be identified that are absent or highly divergent in these strains. Such genome content data indicates that of the 11 strains analyzed, strains 81116 and P3 are most distantly related to NCTC 11168, whereas the recent clinical isolate PHLS03 is most closely related to the sequenced strain.
Table 1.
Campylobacter jejuni Strains Used in This Study
| Strain | Relevant characteristics and history | Penner serotype | % genes non-hybridized | Source/Reference |
|---|---|---|---|---|
| 11168 | Sequenced strain, human isolate (Worcester 1977) | O:2 | 0 | NCTC 11168a |
| 81116 | Human isolate, source: water (Chelmsford 1981) | O:6 | 6.95 | NCTC 81116b |
| 81-176 | Human isolate, source: raw milk (Minnesota 1981) | O:23/36 | 3.93 | G. Perez-Perezc |
| X | Human isolate, gastroenteritis, source: poultry (London 1997) | UTd | 1.27 | A. Karlysheve |
| G1 | Human isolate, GBSf | O:1 | 1.21 | N. Gregsong |
| P1 | Penner serotype reference strain | O:1 | 3.69 | NCTC 12500h |
| P2 | Penner serotype reference strain | O:2 | 3.69 | NCTC 12501h |
| P3 | Penner serotype reference strain | O:3 | 6.95 | NCTC 12502h |
| P4 | Penner serotype reference strain | O:4 | 3.87 | NCTC 12561h |
| PHLS01 | Human isolate, phage type 35 | O:2 | 3.51 | PHLS (C0118480)h |
| PHLS02 | Human isolate, phage type 1 | O:2 | 3.39 | PHLS (C0178110)h |
| PHLS03 | Human isolate, phage type 44 | O:2 | 0.54 | PHLS (C0125990)h |
Nature Collection of Type Cultures, Colindale, London
Veterinary Laboratories Agency, Weybridge, UK
Vanderbilt University, Nashville, Tennessee
Untypeable
LSHTM, London UK
Strain from patient who later developed Guillain-Barré syndrome
Guy's Medical School, London
Public Health Laboratory Service, Colindale, London
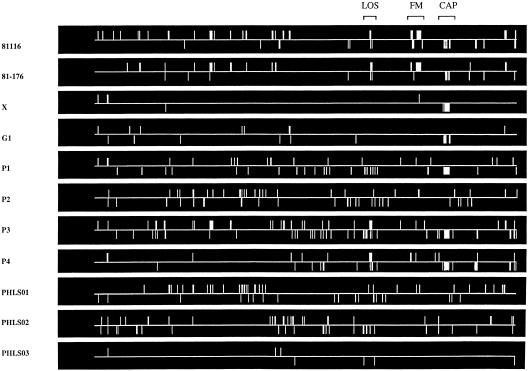
Figure 1 indicates the position of absent or highly divergent genes over the whole genome of each of the 11 isolates, assuming gene order conservation among strains. The complete lists of genes either absent or highly divergent in each of the 11 strains tested are also available on-line at http://www.sghms.ac.uk/depts/medmicro/bugs/GR-1858. Of the 1654 annotated genes, 1300 were common to all strains (core genes), with 354 genes missing from at least one strain (variable genes). As expected, conserved regions included genes for metabolic, biosynthetic, cellular, and regulatory processes, but also for several putative virulence determinants, including genes encoding the cytolethal distending toxin (Pickett et al. 1996), flagellar structural proteins (Wassenaar et al. 1991), phospholipase A (Grant et al. 1997), the PEB antigenic surface proteins (Pei et al. 1991), and proteins potentially involved in host pathogen interactions such as CiaB (Konkel et al. 1999), CadF (Konkel et al. 1997), and CheY (Yao et al. 1997). Variable genes of known function that are not involved in surface structure include the iron acquisition siderophore receptor (Cj0178), which is absent or highly divergent in 81116 and 81-176, and the enterochelin uptake operon ceuBCDE, which is absent or highly divergent in strain PHLS02. The predicted DNA restriction and DNA modification systems in NCTC 11168 appear to be particularly variable, with many of these systems absent or highly divergent in strains 81116, 81-176, X, P1, P2, P3, and P4 (Table 2).
Figure 1.
Comparison of the genomes of 11 different Campylobacter jejuni strains with the NCTC 11168 genome. Genes identified as absent by microarray hybridization analysis are indicated as a vertical line depicting the position of the gene in a linear representation of the 1.64-Mb NCTC 11168 genome from Cj0001to Cj1654. Lines above the horizontal line indicate genes on the forward strand, and lines below indicate genes on the reverse strand. (LOS) Lipo-oligosaccharide biosynthesis locus, (FM) flagellar modification locus, and (CAP) capsule biosynthesis locus.
Table 2.
Divergence in Restriction/Modification Systems in Campylobacter jejuni Strains
| Gene | Functiona | Clone content (%) | Phase variableb | 81116 | 81-176 | X | G1 | P1 | P2 | P3 | P4 | PHLS 01 | PHLS 02 | PHLS 03 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cj0031 | type IIS restriction/ modification enzyme, N-terminal half | Cj0031 (31) | √ | + | + | + | + | + | + | + | + | + | + | + |
| Cj0032 | type IIS restriction/ modification enzyme, C-terminal half | Cj0031 (8) Cj0032 (95) | √ | − | + | − | + | − | + | (+) | − | + | + | (+) |
| Cj0208 | DNA modification methylase | Cj0207 (1) Cj0208 (66) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj0690 | possible restriction/ modification enzyme | Cj0690 (34) | − | + | + | + | + | (+) | + | + | + | + | + | |
| Cj0722 | DNA methylase | Cj0721 (16) Cj0722 (100) Cj0723 (32) | + | + | + | + | + | (+) | + | + | (+) | + | + | |
| Cj1051 | restriction/modification enzyme | Cj1051 (25) | + | − | + | + | + | − | − | + | + | + | (+) | |
| Cj1549 | type I restriction enzyme R protein | Cj1549 (27) | − | + | + | + | + | + | + | − | + | + | + | |
| Cj1551 | type I restriction enzyme S protein | Cj1551 (56) | − | + | + | + | + | + | + | − | + | + | + | |
| Cj1553 | type I restriction enzyme M protein | Cj1553 (39) | + | + | + | + | + | + | + | − | + | + | + |
A + or − indicates that the red/green hybridization signal intensity ratio is >0.5 or <0.5. respectively.
(+) Indicates the possibility that the gene may be absent or highly divergent even though the red/green hybridization signal intensity ratio is > 0.5 (see Methods).
Putative gene function based on annotation (Parkhill et al. 2000).
Genes contain intragenic homopolymeric tracts indicating potential phase variable expression (Parkhill et al. 2000).
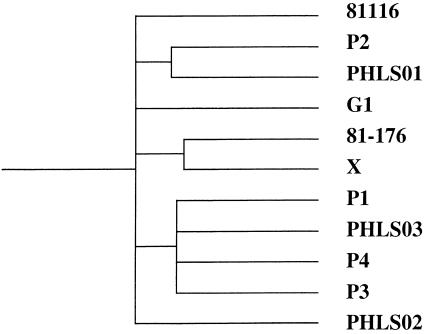
Cluster analysis to compare the genome content of the 11 strains studied was performed (Fig. 2). Such analysis indicates that even though strains P2, PHLS01, PHLS02, and PHLS03 share the same serotype, the genome content of these strains is otherwise dissimilar.
Figure 2.
Cluster analysis performed using GeneSpring 3.2.12 software showing the relationship of all 11 Campylobacter jejuni strains studied to each other based on their genomic composition compared to the sequenced strain NCTC 11168.
Variation in Surface Structures
Many of the C. jejuni NCTC 11168 genes absent or highly divergent in one or more strains are associated with the synthesis and/or modification of surface structures. Data for the three large gene clusters representing capsular polysaccharide biosynthesis (Cj1413–Cj1448), LOS biosynthesis (Cj1131–Cj1152), and the region involved in biosynthesis and posttranslational modification of the flagellin subunits FlaA and FlaB (Cj1305–Cj1345) are represented in Figure 1. Strains 81116 and 81-176 lack much of the NCTC11168 flagellin modification locus, which is conserved in the other nine strains tested. This has been confirmed for strain 81116 by PCR analysis (A.V. Karlyshev, unpubl.). The capsule and LOS hybridization patterns for the 11 strains are detailed in Tables 3 and 4, respectively.
Table 3.
Divergence in the 42.6 kb NCTC 11168 Capsule Biosynthetic Locus in the Campylobacter jejuni Strains Studied
| Gene name | Functiona | Clone content (%) | Phase variableb | 81116 | 81-176 | X | G1 | P1 | P2 | P3 | P4 | PHLS 01 | PHLS 02 | PHLS 03 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cj1413 | kpsS | Cj1413 (67) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj1414 | kpsC | Cj1413 (16) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj1414 (50) | ||||||||||||||
| Cj1415 | cysC | Cj1414 (20) | (+) | + | + | + | + | + | + | + | + | + | + | |
| Cj1415 (100) | ||||||||||||||
| Cj1416 (17) | ||||||||||||||
| Cj1416 | sugar | Cj1416 (82) | + | + | + | + | + | + | + | + | + | + | + | |
| nucleotidyltransferase | ||||||||||||||
| Cj1417 | GMP synthetase | Cj1416 (22) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj1417 (98) | ||||||||||||||
| Cj1418 | phosphoenolpyruvate | Cj1418 (39) | + | + | + | + | + | + | + | + | + | + | + | |
| synthase | ||||||||||||||
| Cj1419 | methyltransferase | Cj1418 (9) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj1419 (77) | ||||||||||||||
| Cj1420 | unknown | Cj1420 (73) | √ | + | + | + | + | + | + | + | (+) | + | + | + |
| Cj1421 | sugar transferase | Cj1421 (33) | √ | (+) | + | + | + | + | + | + | (+) | + | + | + |
| Cj1422 | sugar transferase | Cj1422 (89) | √ | − | + | − | + | + | + | (+) | − | + | + | + |
| Cj1423 | sugar-phosphate | Cj1423 (100) | − | (+) | (+) | + | + | + | (+) | (+) | + | (+) | + | |
| nucleotidyltransferase | Cj1424 (100) | |||||||||||||
| Cj1425 (3) | ||||||||||||||
| Cj1424 | gmhA2 | Cj1423 (22) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj1424 (100) | ||||||||||||||
| Cj1425 (4) | ||||||||||||||
| Cj1425 | galactose kinase | Cj1424 (22) | − | + | (+) | − | − | + | + | + | + | + | + | |
| Cj1425 (100) | ||||||||||||||
| Cj1426 (20) | ||||||||||||||
| Cj1426 | unknown | Cj1425 (2) | √ | − | + | − | − | − | + | − | (+) | + | + | + |
| Cj1426 (100) | ||||||||||||||
| Cj1427 (46) | ||||||||||||||
| Cj1427 | sugar-nucleotide | Cj1427 (75) | − | + | − | + | + | + | (+) | + | + | + | + | |
| epimerase/dehydratase | ||||||||||||||
| Cj1428 | fucose synthetase | Cj1427 (28) | − | (+) | − | + | − | + | − | − | + | + | + | |
| Cj1428 (100) | ||||||||||||||
| Cj1429 (6) | ||||||||||||||
| Cj1429 | unknown | Cj1428 (14) | √ | − | + | − | + | − | + | − | − | + | + | + |
| Cj1429 (100) | ||||||||||||||
| Cj1430 (42) | ||||||||||||||
| Cj1430 | nucleotide-sugar | Cj1429 (29) | + | + | + | + | + | + | + | + | + | + | + | |
| epimerase/dehydratase | Cj1430 (100) | |||||||||||||
| Cj1431 (7) | ||||||||||||||
| Cj1431 | glycosyltransferase | Cj1431 (43) | − | − | − | − | + | + | − | − | + | + | + | |
| Cj1432 | glycosyltransferase | Cj1432 (45) | − | − | − | − | − | + | − | − | + | + | + | |
| Cj1433 | unknown | Cj1432 (2) | √ | − | − | − | + | + | + | − | − | + | + | + |
| Cj1433 (71) | ||||||||||||||
| Cj1434 | galactosyltransferase | Cj1434 (55) | − | + | − | + | + | + | (+) | − | + | + | + | |
| Cj1435 | phosphoserine | Cj1435 (100) | (+) | + | + | + | + | + | (+) | + | + | + | + | |
| phosphatase | Cj1436 (4) | |||||||||||||
| Cj1436 | aminotransferase | Cj1436 (89) | − | + | − | + | + | + | (+) | (+) | + | + | + | |
| Cj1437 | aminotransferase | Cj1437 (84) | − | (+) | − | + | − | + | − | − | + | + | + | |
| Cj1438 (2) | ||||||||||||||
| Cj1438 | galactosyltransferase | Cj1438 (42) | − | (+) | − | + | + | + | + | − | + | + | + | |
| Cj1439 | UDP-galactopyranose | Cj1439 (75) | − | − | − | + | − | + | − | − | + | + | + | |
| mutase | ||||||||||||||
| Cj1440 | galactosyltransferase | Cj1440 (52) | − | (+) | − | + | (+) | + | − | − | + | + | + | |
| Cj1441 (11) | ||||||||||||||
| Cj1441 | kfiD | Cj1440 (3) | − | − | − | − | + | + | − | − | + | + | + | |
| Cj1441 (73) | ||||||||||||||
| Cj1442 | glycosyltransferase | Cj1442 (47) | + | + | + | − | + | (+) | + | + | (+) | + | + | |
| Cj1443 | kpsF | Cj1443 (63) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj1444 | kpsD | Cj1444 (74) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj1445 | kpsE | Cj1445 (65) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj1447 (38) | ||||||||||||||
| Cj1447 | kpsT | Cj1445 (25) | + | + | + | + | + | (+) | + | + | (+) | + | + | |
| Cj1447 (100) | ||||||||||||||
| Cj1448 (13) | ||||||||||||||
| Cj1448 | kpsM | Cj1447 (44) | + | + | (+) | (+) | + | + | + | + | + | + | + | |
| Cj1448 (100) |
A + or − indicates that the red/green hypridization signal intensity ratio is >0.5 or <0.5, respectively.
(+) indicates the possibility that the gene may be absent or highly divergent even though the red/green hybridization signal intensity ratio is >0.5 (see Methods).
Putative gene function based on annotation (Parkhill et al. 2000).
Genes contain intragenci homopolymeric tracts indicating potential phase variable expression (Parkhill et al. 2000).
NB: Cj1446 was originally designated a CDS, but was unannotated in the final genome annotation (Parkhill et al. 2000).
Table 4.
Divergence in the 20.4 kb NCTC 11168 Lipo-oligosaccharide Biosynthetic Locus in the Campylobacter jejuni Strains Studied
| Gene name | Functiona | Clone content (%) | Phase variableb | 81116 | 81-176 | X | G1 | P1 | P2 | P3 | P4 | PHLS 01 | PHLS 02 | PHLS 03 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cj1131 | galE | Cj1131 (73) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj1132 | unknown | Cj1132 (80) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj1133 (11) | ||||||||||||||
| Cj1133 | waaC | Cj1132 (20) | (+) | + | (+) | (+) | + | (+) | + | + | − | + | + | |
| Cj1133 (100) | ||||||||||||||
| Cj1134 (19) | ||||||||||||||
| Cj1134 | htrB | Cj1134 (67) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj1135 (4) | ||||||||||||||
| Cj1135 | glycosyltransferase | Cj1135 (53) | − | + | + | + | + | + | − | + | + | + | + | |
| Cj1136 | glycosyltransferase | Cj1136 (89) | − | (+) | + | + | + | + | − | − | + | + | + | |
| Cj1137 (3) | ||||||||||||||
| Cj1137 | unknown | Cj1136 (13) | − | (+) | + | + | − | (+) | (+) | − | + | (+) | + | |
| Cj1137 (100) | ||||||||||||||
| Cj1138 (18) | ||||||||||||||
| Cj1138 | galactosyltransferase | Cj1138 (67) | − | − | + | + | (+) | + | (+) | − | + | + | + | |
| Cj1139 | wlaNc | Cj1138 (30) | √ | − | − | + | + | + | + | − | − | + | + | + |
| Cj1139 (96) | ||||||||||||||
| Cj1140 | cst | Cj1140 (93) | − | − | + | + | + | + | (+) | − | + | + | + | |
| Cj1141 (47) | ||||||||||||||
| Cj1141 | neuBld | Cj1140 (26) | − | + | + | + | (+) | + | − | − | + | + | + | |
| Cj1141 (100) | ||||||||||||||
| Cj1142 (6) | ||||||||||||||
| Cj1142 | neuCld | Cj1141 (29) | − | (+) | + | + | + | + | − | − | + | + | + | |
| Cj1142 (98) | ||||||||||||||
| Cj1143 | neuAld | Cj1143 (42) | − | − | + | + | + | + | − | − | + | + | + | |
| Cj1144 | unknown | Cj1143 (15) | √ | − | − | + | + | + | + | − | − | + | + | + |
| Cj1144 (75) | ||||||||||||||
| Cj1145 | unknown | Cj1143 (3) | √ | (+) | + | + | + | + | − | − | − | (+) | + | + |
| Cj1144 (100) | ||||||||||||||
| Cj1145 (100) | ||||||||||||||
| Cj1146 (76) | ||||||||||||||
| Cj1146 | waaV | Cj1146 (89) | + | + | + | + | + | + | (+) | (+) | + | (+) | + | |
| Cj1148 (9) | ||||||||||||||
| Cj1148 | waaF | Cj1148 (80) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj1149 (2) | ||||||||||||||
| Cj1149 | gmhA | Cj1149 (92) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj1150 (27) | ||||||||||||||
| Cj1150 | rfaE | Cj1149 (23) | + | + | + | + | + | + | (+) | (+) | + | + | (+) | |
| Cj1150 (94) | ||||||||||||||
| Cj1151 | rfaD | Cj1150 (35) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj1151 (83) | ||||||||||||||
| Cj1152 | phosphatase | Cj1152 (76) | + | + | + | + | + | + | + | + | + | + | + | |
| Cj1153 (100) |
A + or − indicates that the red/green hypridization signal intensity ratio is >0.5 or <0.5, respectively.
(+) indicates the possibility that the gene may be absent or highly divergent even though the red/green hybridization signal intensity ratio is >0.5 (see Methods).
Putative gene function based on annotation (Parkhill et al. 2000).
Genes contain intragenic homopolymeric tracts indicating potential phase variable expression (Parkhill et al. 2000).
NB: Cj1447 was originally designated a CDS, but was unannotated in the final genome annotation (Parkhill et al. 2000).
The C. jejuni capsular polysaccharide was recently identified as the serodeterminant of the Penner serotyping system (Karlyshev et al. 2000a). The organization of the C. jejuni capsule locus bears similarity to the Escherichia coli paradigm, in which conserved genes associated with transport of the polysaccharide (kps genes; see Table 3) flank a central region that in E. coli is the variable biosynthetic region involved in the generation of diverse polysaccharide structures (Roberts 1996). It was predicted that if this central region does indeed contain the genes involved in C. jejuni capsular polysaccharide biosynthesis, then this region would vary between strains of different Penner serotypes (Karlyshev et al. 2000a). Indeed, the seven non-O:2 serotype strains revealed extensive diversity, with up to 17 NCTC 11168 genes absent or highly divergent from this central region (Table 3). A striking feature of the genome comparisons is the complete conservation of the 42.6-kb capsule biosynthesis locus in four strains with the same Penner serotype (O:2) as the sequenced strain (Table 3). This clearly indicates that this central biosynthetic region is responsible for the variation in capsular polysaccharide detected by the Penner serotyping system. As expected, the kps genes flanking the central capsule region, which encode the general capsule transport mechanism, were conserved in all the strains studied (Table 3).
C. jejuni LOS consists of a relatively conserved inner core with a terminal outer core region that often varies in structure between strains and can mimic human gangliosides (Moran and Penner 1999). C. jejuni NCTC 11168 genes involved in inner core biosynthesis include waaC, waaV, waaF, rfaE, and rfaD. A positive hybridizing signal was obtained with genomic DNA from 10 of 11 C. jejuni strains for the elements presenting waaC, and for all strains for waaV, waaF, rfaE, and rfaD (Table 4). However, genes involved in C. jejuni NCTC 11168 LOS outer-core biosynthesis are absent or highly divergent in a number of strains. Examples include Cj1139 or wlaN, a β-1,3-galactosyltransferase responsible for the addition of the terminal galactose residue of the NCTC 11168 ganglioside GM1-like LOS (Linton et al. 2000a). This gene is absent in strains 81116, 81-176, P3, and P4 (Table 4). Other putative outer-core sugar transferases such as glycosyltransferases (Cj1135 and Cj1136) and galactosyltransferase (Cj1138) are lacking in strains 81116, 81-176, P3, and P4 (Table 4). Also of note are the genes involved in the sialylation of LOS (neuA1, neuB1, and neuC1), which are absent in strains 81116, P3, and P4 (Table 4; Linton et al. 2000b).
Validation of Microarray Data
To validate microarray data, PCR and sequence analysis of a number of variable genes from the strains studied was performed. A strategy was used to amplify PCR products across putative deletions defined by the microarray data, then to sequence these PCR products to define the precise location of the deletion. Analysis of sequence data generated was performed by BLAST search (Altschul et al. 1997). Two single gene deletions (Cj0008 and Cj1585) and two small gene clusters (Cj0055–Cj0058 and Cj0565–Cj0569) were selected in the four strains 81116, 81-176, X, and G1. Cj0008 is a gene with unknown function, but an ortholog of HP0938 in the H. pylori 26695 genome. Cj1585 is a probable oxidoreductase. The cluster Cj0055–Cj0058 consists of two genes of unknown function and two genes coding for possible periplasmic proteins, all with no match in the H. pylori 26695 genome, and the cluster Cj0565–Cj0569 contains genes with no known function and no match with the H. pylori 26695 genome, suggesting that these clusters may be specific to Campylobacter species. These studies are summarized in Table 5 and clearly validate the microarray hybridization results, which have given consistently reproducible data. Additionally, the microarray data confirm reported data on the neuB genes involved in sialylation in strains 81116, G1, P1, P2, and P3 (Linton et al. 2000b), the wlaN gene involved in LOS biosynthesis, in strains P1, P2, P3, and P4 (Linton et al. 2000a), and deletions in both the flagellin modification locus and the capsule biosynthesis locus in strains 81116, 81-176, X, and G1 (A.V. Karlyshev, unpubl.).
Table 5.
Validation of Microarray Data by PCR and Sequence Analysis
| Strain | Cj0008 | Cj1585 | Cj0055–Cj0058 | Cj0565–Cj0569 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| array | PCR | Sequencing | Array | PCR | Sequencing | Array | PCR | Sequencing | Array | PCR | Sequencing | |
| 81116 | − | − | − | − | −a | −b | − | − | − | − | −a | −c |
| 81-176 | + | + | + | − | −a | −b | + | + | + | − | −a | −c |
| X | − | − | − | + | + | + | + | + | + | + | + | + |
| G1 | − | − | − | + | + | + | + | + | + | + | + | + |
A + or − indicates the presence or absence, respectively, or a gene indicated by microarray data, alongside the subsequent analysis data.
Putative insertion identified by PCR, rather than a deletion.
Sequence analysis suggests the presence of an anaerobic dimethyl sulphoxide reductase ortholog adjacent to Cj1584.
Analysis of sequence data adjacent to Cj0564 shows no matches with any database entry.
DISCUSSION
Using a single pair of vector primers, we have constructed a low-cost whole genome C. jejuni NCTC 11168 DNA microarray representing all 1654 annotated genes that were amplified from the pUC18 clones sequenced as part of the genome project (Parkhill et al. 2000). Eleven C. jejuni strains of diverse origin were analyzed with respect to the genome content of the sequenced strain, revealing extensive genetic diversity within this species. One caveat with the data generated with this microarray is that some genes may be scored as present, when in fact they are absent or highly divergent. This is because of either adjacent gene sequences on some of the PCR products on the microarray or cross-hybridization with another part of the genome. If the gene represented by the PCR product on the array is absent, but an adjacent gene is present, this will potentially increase the red/green hybridization signal intensity ratio. If this ratio is increased above 0.5, using our data analysis filtering method, the gene will be scored as present. Conversely, any gene scored as absent should be accurate within the limits of array hybridization specificity and sensitivity. Therefore, although the microarray has limitations, it is suitable for genomic comparisons where data generated is qualitative. The major advantage of using clones directly from genome projects is an inexpensive and efficient alternative for the construction of DNA microarrays, and this principle could be applied widely where random shot-gun libraries are used for sequencing.
Of the 1654 annotated genes analyzed, 1300 (78.6%) appeared to be common to all strains and can be defined as C. jejuni core genes. As expected, these genes mainly encoded housekeeping functions such as metabolic, biosynthetic, cellular, and regulatory processes, as in the case of the closely related pathogen H. pylori (Salama et al. 2000). By contrast, determinants encoding surface-located structures such as the flagellin synthesis and modification system, LOS, and capsule were absent or highly divergent in one or more strains studied. These determinants probably relate to functions important in survival, transmission, and pathogenesis, indicating that selective pressures have driven profound evolutionary changes to create a diverse Campylobacter species. However, not all putative virulence determinants, including several surface proteins, showed variability. Genes encoding cytolethal distending toxin (Pickett et al. 1996), flagellar structural proteins (Wassenaar et al. 1991), phospholipase A (Grant et al. 1997), the PEB antigenic surface proteins (Pei et al. 1991), and proteins potentially involved in host pathogen interactions such as CiaB (Konkel et al. 1999), CadF (Konkel et al. 1997), and CheY (Yao et al. 1997) were present in all strains, indicating that these proteins are indispensable for C. jejuni to cause disease in humans.
Whole genome analysis shows extensive genome diversity among the 12 C. jejuni strains studied, with at least 354 genes (21.4%) missing from at least one strain. This is likely to be an underestimate of the number of variable genes in C. jejuni, owing to the relatively small number of strains analyzed and the nature of the microarray, which sometimes contained DNA adjacent to the target gene. The percentage of variable genes is remarkably similar to that observed for the closely related gastric pathogen H. pylori, in which 22% of genes were variable in the 15 strains studied (Salama et al. 2000). The hybridization conditions and criteria for designating nonhybridizing genes were, as far as is possible to determine, the same in both studies. Curiously, in contrast to H. pylori, the genome sequence of C. jejuni NCTC 11168 revealed no transposons, phage remnants, or insertion sequence elements and very few repeat sequences (Parkhill et al. 2000). The mechanisms to generate genetic diversity in C. jejuni therefore remain unclear. However, C. jejuni is naturally competent, and this linked to a high recombination rate may contribute to genetic diversity. Recent multilocus sequence typing for C. jejuni has indicated that intraspecies and interspecies horizontal genetic exchange for this species are common (Dingle et al. 2001). The restriction/modification systems in C. jejuni appear divergent, with only G1, PHLS01, PHLS02, and PHLS03 of the 11 strains being similar to NCTC 11168. In H. pylori, numerous strain-specific restriction/modification systems have been found, and DNA methylation patterns have been shown to vary substantially among strains (Ando et al. 2000; Xu et al. 2000). Recently, the sequencing of a second E. coli strain has also shown restriction/modification systems to be variable (Perna et al. 2001). Elevated divergence of these systems may be associated with the positive selection of subpopulations that contribute to genetic diversity.
Compared to most genomes from enteric bacteria, the NCTC 11168 DNA sequence has a characteristically constant 30.6% G + C content over the genome. However, it is noticeable that where there are differences, these occur in loci that appear to vary between strains. For example, the NCTC 11168 G + C content in the capsule biosynthesis region is 27.6% over 42.6 kb, in the LOS biosynthesis region it is 27.8% over 20.4 kb, and the restriction modification genes Cj1549 and Cj1553 have a G + C content of 34.3% and 37.8%, respectively. The significantly variable G + C content of these regions indicates that they may have been acquired by horizontal genetic exchange and introduced relatively recently during the evolution of the C. jejuni genome. Additionally, phase-variable, or contingency genes, that were originally identified directly from sequencing the random shotgun library of NCTC 11168 (Parkhill et al. 2000), appear to be absent in several strains (Tables 2–4). These include four of six such genes from the capsule locus, both phase-variable genes in the LOS locus, and a phase-variable restriction/modification system, indicating that these genes are dispensable but are probably required to generate diversity on the C. jejuni cell surface.
Analysis of the flagellin modification locus shows similar hybridization efficiencies among all strains except for the laboratory passaged strains 81116 and 81-176 (Fig. 1). The loss of this region, which has been confirmed for 81116 using PCR analysis (A.V. Karlyshev, unpubl.), may be caused by homologous recombination through the near identical genes Cj1318 and Cj1336 that flank the deletion in NCTC 11168 (Parkhill et al. 2000). Such a recombinant event leading to this deletion may be expected in strains that have been extensively passaged in the laboratory over a 20-year period.
The Penner serotyping system has been used for differentiating C. jejuni strains for more than 20 years (Penner and Hennessy 1980). It was presumed that the Penner serotyping antigen arose from the C. jejuni surface-encoded O antigen. Recently, we demonstrated that the capsule was responsible for the Penner serotyping antigen (Karlyshev et al. 2000a). The DNA microarray data presented here provides further evidence that the capsule accounts for Penner serospecificity in that all four strains of O:2 serotype showed complete conservation of the capsule biosynthesis locus compared to the sequenced strain NCTC 11168 (also serotype O:2; Table 3). The seven non-O:2 serotype strains lacked between 5 and 17 genes located in the putative capsule biosynthetic locus of NCTC 11168. By contrast, scrutiny of the LOS biosynthesis region showed that two non-O:2 serotypes (strains X and G1) had complete conservation of the LOS biosynthesis locus compared to the sequenced strain and that the O:2 serotype strains P2 and PHLS01 lack a single gene compared to NCTC 11168 (Table 4). Ten percent of C. jejuni strains are untypable using the Penner serotyping method (e.g., strain X in this study). Further genetic analysis of the capsule region among many more C. jejuni serotypes should enable the genetic basis of Penner serospecificity to be determined, allowing the development of improved genotyping methods to distinguish between C. jejuni strains. The microarray data support the hypothesis that the region flanked by the kps genes (see Table 3) is the capsule biosynthetic region, having a similar overall arrangement to group II/III capsules in E. coli (Roberts 1996).
C. jejuni strain diversity combined with variable host responses results in a complex spectrum of disease outcomes, ranging from asymptomatic colonization to severe inflammatory diarrhoea. In addition, chronic sequelae are also associated with C. jejuni infection, including irritable bowel syndrome and neuropathological complications such as Guillain-Barré and Miller Fisher syndromes. Variable genes identified through whole genome analysis may encode factors contributing to different disease outcomes and to the survival of the organism in different ecological niches. Comparison of a larger collection of strains with known clinical histories should allow associative traits to be determined for subpopulations of strains that are important in specific disease outcomes. A precise understanding of genetic differences between C. jejuni strains should also allow the identification of improved epidemiological markers and the development of rational approaches to reduce this problematic pathogen from the food chain.
METHODS
Bacterial Strains and Growth Conditions
The C. jejuni strains used in this study and relevant background information are shown in Table 1. Strains 81116 and 81-176 are laboratory strains that have been extensively studied. Strain X was isolated from a patient with noncomplicating gastroenteritis, whereas strain G1 was isolated from a patient with gastroenteritis who later developed GBS. Strains P1, P2, P3, and P4 are the first four Penner serotype reference strains, and the PHLS strains were selected as recent clinical isolates, all with serotype O:2. Bacteria were grown on blood agar plates, consisting of Columbia Agar Base (Oxoid) supplemented with 7% (v/v) defibrinated horse blood (TCS Microbiology), in a microaerobic atmosphere at 37°C.
Construction of a C. jejuni NCTC 11168 DNA Microarray
A tiling program was designed to select a minimum set of clones representing all 1731 initially identified CDSs from the 8–10-fold redundant pUC18 library from the C. jejuni NCTC 11168 genome project (Parkhill et al. 2000). Plasmid DNA from each of the selected clones was transferred to 19 96-well plates. PCR amplicons were prepared in 100-μL reactions in 96-well plates using standard vector primers (M13 forward and M13 reverse). All PCR amplifications and liquid handling were performed using a RoboAmp 4200 (MWG-Biotech). The amplicons were evaluated on agarose gels to ensure they were of the expected size and concentration. A small sample of PCR products was sequenced to confirm that the expected gene had been amplified. Then 50 μL of each successful PCR reaction was ethanol-precipitated and resuspended in 10 μL of 50% (v/v) DMSO in H2O. The microarray was prepared by the robotic printing of these vector primer-generated PCR products representing each gene in the C. jejuni NCTC 11168 genome onto CMT-GAPS-coated glass slides (Corning), using a Microgrid I microgridding robot (Bio Robotics). We used a 4 × 4 split pin configuration with a center-to-center spot spacing of 400 μm, producing spots with a diameter of ∼150 μm. After printing, free amino groups on the coated glass slides were acetylated with 5.5 g of succinic anhydride in 350 mL of borate-buffered 1-methyl-2-pyrrolidinone at pH 8. Slides were stored in microscope slide boxes in a cool, dry environment and used within 1 mo.
Labeling of Genomic DNA
Genomic DNA was isolated from 48-h plate-grown C. jejuni strains using the Wizard Genomic DNA Purification Kit (Promega). Whole genomic DNA was used as a template for direct incorporation of fluorescent analogs (Cy3 and Cy5 dCTP) by a randomly primed polymerization reaction. Briefly, 3 μg of template DNA and 9 μg of random hexamers (GIBCO BRL) in a reaction volume of 41.5 μL were denatured at 95°C for 5 min and snap cooled on ice. Then 5 μL of 10× Klenow buffer (USB Corporation), 1.5 μL of Cy-dye-labeled dCTP (Amersham Pharmacia Biotech), 1 μL of dNTP solution (5 mM each dATP, dGTP, and dTTP, plus 2 mM dCTP), and 10 U of Klenow (the large fragment of DNA polymerase I, exonuclease-free; USB Corporation) were added. Test strain DNA was labeled with Cy5-dCTP and the control NCTC 11168 strain DNA with Cy3-dCTP. The labeling reactions were incubated at 37°C for 90 min.
DNA Microarray Hybridizations
Whole genome DNA comparisons were performed, comparing both laboratory strains and clinical isolates listed in Table 1 against the sequenced strain NCTC 11168. Microarray slides were incubated in a prehybridization buffer (3.5× SSC buffer, 0.1% SDS, 10 mg/mL BSA) at 60°C for 20 min. After prehybridisation, slides were washed in distilled water for 1 min, followed by a 1-min wash in isopropanol. Labeled DNA from the test and control strains were coprecipitated in the presence of 10 μg of tRNA and resuspended in a total volume of 16 μL with final concentration of 4× SSC buffer and 0.3% SDS. The hybridization mixture was denatured at 95°C for 2 min, cooled slowly to room temperature, applied to the microarray, and covered with a 22-mm2 coverslip. The slide was placed in a waterproof hybridization chamber (CMT Hybridization Chambers, Corning) for hybridization in a 60°C waterbath overnight. After hybridization, slides were washed in 1× SSC buffer with 0.06% SDS for 2 min, followed by two washes in 0.06× SSC buffer, each for 2 min.
Data Acquisition and Analysis
Slides were scanned with a ScanArray 3000 (GSI Lumonics) following the manufacturer's guidelines. Fluorescent spot intensities were quantified using ImaGene 3.0 (BioDiscovery Inc.) software. For each spot, background fluorescence was subtracted from the average spot fluorescence to produce a channel-specific value. The data were further analyzed using GeneSpring 3.2.12 (Silicon Genetics) software. The geometric mean of the normalized red/green ratio was calculated for each test strain using data from three array experiments. Spots were excluded for low signal (<100 units in the Cy3 channel), and a cutoff for the normalized red/green ratio of <0.5 was used to create gene deletion lists using the GeneSpring 3.2.12 software.
If a gene was absent, but was represented by a clone containing adjacent genes that were present, then the red/green hybridisation signal intensity ratio could be raised above 0.5 and the gene subsequently scored as present. If a gene shared enough homology with other genes in the genome to allow cross-hybridization events, the same situation could occur. It was important therefore to consider both (a) the composition of each clone representing each gene on the array, and (b) the possibility of cross-hybridization of each clone with the rest of the genome. The possibility of cross-hybridization was analyzed by BLASTN analysis of the sequence of each clone used on the array against the whole genome sequence. This showed that there is very little significant cross-hybridization between C. jejuni NCTC 11168 genes. These data are available on-line at http://www.sghms.ac.uk/depts/medmicro/bugs/GR-1858. A file showing the composition of each clone used to represent each gene on the array is also available on-line. To highlight these considerations in the results presented in Tables 2, 3, and 4, an arbitrary scoring system was used to highlight genes scored as present that may in fact be absent. A gene scored as present was marked (+) rather than + if that gene was represented on the microarray by (a) an adjacent gene clone or multiple gene clone with a red/green ratio between 0.5 and 0.6; (b) a single gene clone or an adjacent gene clone with significant predicted cross-hybridization with a red/green ratio between 0.5 and 0.75; or (c) a multiple gene clone with significant predicted cross-hybridization with a red/green ratio between 0.5 and 0.9.
Validation of Microarray Data by PCR and Sequencing
PCR primers were designed to approach both the 5′ and 3′ edges of predicted deletions so that the amplified region would span the missing locus for those strains in which it is deleted. Primer design was aided by the use of Artemis 3.0 software (Rutherford et al. 2000) to visualize the edges of the predicted deletions. Both standard PCR using primer pairs and PCR using a single primer (Karlyshev et al. 2000b) were performed. The resulting amplicons were sequenced using the dideoxynucleotide chain termination method with a PRISM BigDye sequencing kit (PE Applied BioSystems) with primers designed to be internal to the original PCR primer(s).
Acknowledgments
We gratefully acknowledge Lynne Batty, Hannah Eno, and Jane Turner for technical assistance and Rob Davies for developing the tiling program for selecting optimum clones from the C. jejuni NCTC 11168 plasmid library. C. jejuni 81-176 chromosomal DNA was a generous gift from Tohru Takata and Guillermo Perez-Perez (Vanderbilt University, Nashville, TN). Strain 81116 was supplied by Diane Newell (Veterinary Laboratories Agency, Weybridge, UK), strain G1 by Norman Gregson (LSHTM, London), and strains PHLS01, PHLS02, and PHLS03 by Richard Thwaites and Jennifer Frost (Public Health Laboratoy Service, London, UK). This work was supported by the Wellcome Trust and the BBSRC, UK.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL brendan.wren@lshtm.ac.uk; FAX 44-20-7636-8739.
Article published on-line before print: Genome Res., 10.1101/gr.185801.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.185801.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T, Xu Q, Torres M, Kusugami K, Israel DA, Blaser MJ. Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol Microbiol. 2000;37:1052–1065. doi: 10.1046/j.1365-2958.2000.02049.x. [DOI] [PubMed] [Google Scholar]
- Barrow PA, Page K. Inhibition of colonisation of the alimentary tract in young chickens with Campylobacter jejuni by pre-colonisation with strains of C. jejuni. FEMS Microbiol Lett. 2000;182:87–91. doi: 10.1111/j.1574-6968.2000.tb08879.x. [DOI] [PubMed] [Google Scholar]
- Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, Small PM. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- Blaser MJ. Epidemiologic and clinical features of Campylobacter jejuni infections. J Infect Dis. 1997;176 (Suppl. 2):S103–S105. doi: 10.1086/513780. [DOI] [PubMed] [Google Scholar]
- Blaser MJ, Perez GP, Smith PF, Patton C, Tenover FC, Lastovica AJ, Wang WI. Extraintestinal Campylobacter jejuni and Campylobacter coli infections: Host factors and strain characteristics. J Infect Dis. 1986;153:552–559. doi: 10.1093/infdis/153.3.552. [DOI] [PubMed] [Google Scholar]
- Buzby JC, Allos BM, Roberts T. The economic burden of Campylobacter-associated Guillain-Barré syndrome. J Infect Dis. 1997;176 (Suppl. 2):S192–S197. doi: 10.1086/513785. [DOI] [PubMed] [Google Scholar]
- Dingle KE, Colles FM, Wareing DR, Ure R, Fox AJ, Bolton FE, Bootsma HJ, Willems RJ, Urwin R, Maiden MC. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol. 2001;39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everest PH, Goossens H, Butzler JP, Lloyd D, Knutton S, Ketley JM, Williams PH. Differentiated Caco-2 cells as a model for enteric invasion by Campylobacter jejuni and C. coli. J Med Micro. 1992;37:319–325. doi: 10.1099/00222615-37-5-319. [DOI] [PubMed] [Google Scholar]
- Fayos A, Owen RJ, Desai M, Hernandez J. Ribosomal RNA gene restriction fragment diversity amongst Lior biotypes and Penner serotypes of Campylobacter jejuni and Campylobacter coli. FEMS Microbiol Lett. 1992;74:87–93. doi: 10.1016/0378-1097(92)90741-6. [DOI] [PubMed] [Google Scholar]
- Grant KA, Belandia IU, Dekker N, Richardson PT, Park SF. Molecular characterization of pldA, the structural gene for a phospholipase A from Campylobacter coli, and its contribution to cell-associated hemolysis. Infect Immun. 1997;65:1172–1180. doi: 10.1128/iai.65.4.1172-1180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanninen ML, Hakkinen M, Rautelin H. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1999;65:2272–2275. doi: 10.1128/aem.65.5.2272-2275.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlyshev AV, Linton D, Gregson NA, Lastovica AJ, Wren BW. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol Microbiol. 2000a;35:529–541. doi: 10.1046/j.1365-2958.2000.01717.x. [DOI] [PubMed] [Google Scholar]
- Karlyshev AV, Pallen MJ, Wren BW. Single-primer PCR procedure for rapid identification of transposon insertion sites. BioTechniques. 2000b;28:1078–1082. doi: 10.2144/00286bm05. [DOI] [PubMed] [Google Scholar]
- Kokotovic B, On SL. High-resolution genomic fingerprinting of Campylobacter jejuni and Campylobacter coli by analysis of amplified fragment length polymorphisms. FEMS Microbiol Lett. 1999;173:77–84. doi: 10.1111/j.1574-6968.1999.tb13487.x. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Joens LA. Adhesion to and invasion of HEp-2 cells by Campylobacter spp. Infect Immun. 1989;57:2984–2990. doi: 10.1128/iai.57.10.2984-2990.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel ME, Garvis SG, Tipton SL, Anderson DE, Jr, Cieplak W., Jr Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) from Campylobacter jejuni. Mol Microbiol. 1997;24:953–963. doi: 10.1046/j.1365-2958.1997.4031771.x. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Kim BJ, Rivera-Amill V, Garvis SG. Bacterial secreted proteins are required for the internalization of Campylobacter jejuni into cultured mammalian cells. Mol Microbiol. 1999;32:691–701. doi: 10.1046/j.1365-2958.1999.01376.x. [DOI] [PubMed] [Google Scholar]
- Korolik V, Alderton MR, Smith SC, Chang J, Coloe PJ. Isolation and molecular analysis of colonising and non-colonising strains of Campylobacter jejuni and Campylobacter coli following experimental infection of young chickens. Vet Micro. 1998;60:239–249. doi: 10.1016/s0378-1135(98)00145-x. [DOI] [PubMed] [Google Scholar]
- Lam KM, Yamamoto R, DaMassa AJ. DNA diversity among isolates of Campylobacter jejuni detected by PCR-based RAPD fingerprinting. Vet Micro. 1995;45:269–274. doi: 10.1016/0378-1135(94)00133-h. [DOI] [PubMed] [Google Scholar]
- Linton D, Gilbert M, Hitchen PG, Dell A, Morris HR, Wakarchuk WW, Gregson NA, Wren BW. Phase variation of a β-1,3 galactosyltransferase involved in generation of the ganglioside GM1-like lipo-oligosaccharide of Campylobacter jejuni. Mol Microbiol. 2000a;37:501–514. doi: 10.1046/j.1365-2958.2000.02020.x. [DOI] [PubMed] [Google Scholar]
- Linton D, Karlyshev AV, Hitchen PG, Morris HR, Dell A, Gregson NA, Wren BW. Multiple N-acetyl neuraminic acid synthetase (neuB) genes in Campylobacter jejuni: Identification and characterization of the gene involved in sialylation of lipo-oligosaccharide. Mol Microbiol. 2000b;35:1120–1134. doi: 10.1046/j.1365-2958.2000.01780.x. [DOI] [PubMed] [Google Scholar]
- Lior H, Woodward DL, Edgar JA, Laroche LJ, Gill P. Serotyping of Campylobacter jejuni by slide agglutination based on heat-labile antigenic factors. J Clin Microbiol. 1982;15:761–768. doi: 10.1128/jcm.15.5.761-768.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinersmann RJ. Population genetics and genealogy of Campylobacter jejuni. In: Nachamkin I, Blaser M J, editors. Campylobacter. Washington, D.C.: ASM Press; 2000. pp. 351–368. [Google Scholar]
- Moran AP, Penner JL. Serotyping of Campylobacter jejuni based on heat-stable antigens: Relevance, molecular basis and implications in pathogenesis. J Appl Microbiol. 1999;86:361–377. doi: 10.1046/j.1365-2672.1999.00713.x. [DOI] [PubMed] [Google Scholar]
- Nachamkin I, Allos BM, Ho T. Campylobacter species and Guillain-Barre syndrome. Clin Microbiol Rev. 1998;11:555–567. doi: 10.1128/cmr.11.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Pei ZH, Ellison RTd, Blaser MJ. Identification, purification, and characterization of major antigenic proteins of Campylobacter jejuni. J Biol Chem. 1991;266:16363–16369. [PubMed] [Google Scholar]
- Penner JL, Hennessy JN. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J Clin Microbiol. 1980;12:732–737. doi: 10.1128/jcm.12.6.732-737.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna NT, Plunkett G, III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Pickett CL, Pesci EC, Cottle DL, Russell G, Erdem AN, Zeytin H. Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB gene. Infect Immun. 1996;64:2070–2078. doi: 10.1128/iai.64.6.2070-2078.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast MM, Lastovica AJ, Moran AP. Lipopolysaccharides from Campylobacter jejuni O:41 strains associated with Guillain-Barré syndrome exhibit mimicry of GM1 ganglioside. Infect Immun. 1998;66:3649–3655. doi: 10.1128/iai.66.8.3649-3655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts IS. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: Sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Salama N, Guillemin K, McDaniel TK, Sherlock G, Tompkins L, Falkow S. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc Natl Acad Sci. 2000;97:14668–14673. doi: 10.1073/pnas.97.26.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerbaum S, Lohrengel M, Sonnevend A, Ruberg F, Kist M. Allelic diversity and recombination in Campylobacter jejuni. J Bacteriol. 2001;183:2553–2559. doi: 10.1128/JB.183.8.2553-2559.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar TM. Toxin production by Campylobacter species. Clin Microbiol Rev. 1997;10:466–476. doi: 10.1128/cmr.10.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar TM, Blaser MJ. Pathophysiology of Campylobacter jejuni infections of humans. Microbes Infect. 1999;1:1023–1033. doi: 10.1016/s1286-4579(99)80520-6. [DOI] [PubMed] [Google Scholar]
- Wassenaar TM, Bleumink-Pluym NM, van der Zeijst BA. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 1991;10:2055–2061. doi: 10.1002/j.1460-2075.1991.tb07736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooldridge KG, Ketley JM. Campylobacter–host cell interactions. Trends Microbiol. 1997;5:96–102. doi: 10.1016/S0966-842X(97)01004-4. [DOI] [PubMed] [Google Scholar]
- Xu Q, Morgan RD, Roberts RJ, Blaser MJ. Identification of type II restriction and modification systems in Helicobacter pylori reveals their substantial diversity among strains. Proc Natl Acad Sci. 2000;97:9671–9676. doi: 10.1073/pnas.97.17.9671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao RJ, Burr DH, Guerry P. CheY-mediated modulation of Campylobacter jejuni virulence. Mol Microbiol. 1997;23:1021–1031. doi: 10.1046/j.1365-2958.1997.2861650.x. [DOI] [PubMed] [Google Scholar]
- Yuki N, Miyatake T. Guillain-Barré syndrome and Fisher's syndrome following Campylobacter jejuni infection. Ann NY Acad Sci. 1998;845:330–340. doi: 10.1111/j.1749-6632.1998.tb09685.x. [DOI] [PubMed] [Google Scholar]