Abstract
We developed a high-throughput technique for the generation of cDNA libraries in the yeast Saccharomyces cerevisiae which enables the selection of cloned cDNA inserts containing open reading frames (ORFs). For direct screening of random-primed cDNA libraries, we have constructed a yeast shuttle/expression vector, the so-called ORF vector pYEXTSH3, which allows the enriched growth of protein expression clones. The selection system is based on the HIS3 marker gene fused to the C terminus of the cDNA insert. The cDNAs cloned in-frame result in histidine prototrophic yeast cells growing on minimal medium, whereas clones bearing the vector without insert or out-of-frame inserts should not grow on this medium. A randomly primed cDNA library from human fetal brain tissue was cloned in this novel vector, and using robot technology the selected clones were arrayed in microtiter plates and were analyzed by sequencing and for protein expression. In the constructed cDNA expression library, about 60% of clones bear an insert in the correct reading frame. In comparison to unselected libraries it was possible to increase the clones with inserts in the correct reading frame more than fourfold, from 14% to 60%. With the expression system described here, we could avoid time-consuming and costly techniques for identification of clones expressing protein by using antibody screening on high-density filters and subsequently rearraying the selected clones in a new “daughter” library. The advantage of this ORF vector is that, in a one-step screening procedure, it allows the generation of expression libraries enriched for clones with correct reading frames as sources of recombinant proteins.
In recent years, the human genome sequencing projects have generated enormous amounts of sequence data. With the human genome sequence now available, the challenge of understanding the function of the newly discovered genes has to be addressed. High-throughput technologies have been developed that allow the monitoring of gene activity on the transcriptional level by analysis of complex expression patterns of a specific tissue (Harrison et al. 1995; Schena et al. 1996, 1998; Perret et al. 1998). The next step is the profiling of protein products encoded by expressed cDNA clones to obtain more information on their regulation, biochemical function, and potential interaction partners. This requires the simultaneous expression of protein from a large number of cDNA clones, which has been performed in the Escherichia coli bacterial system (Büssow et al. 1998).
Using robot technology, a human fetal brain cDNA expression library was screened for clonal protein expression in high-throughput on automatically gridded high-density protein arrays. Because the unselected frequency of in-frame clones in libraries with random orientation is statistically only 1/6 of all clones, it would be useful to develop a vector that enables the direct selection of open reading frames (ORFs), improving the yield of clones expressing protein. We have developed such a system in which a C-terminally fused marker gene is expressed only if the cloned insert carries no internal stop sequences, which may result from frameshifts or 5′ and 3′ untranslated regions. Consequently, the use of random-primed cDNAs is required in this system instead of oligo(dT)-tailed cDNAs that carry their own C-terminal termination codon.
Previously, E. coli vectors have been constructed on the basis of the β-galactosidase coding sequence for the generation of in-frame fusion libraries (Gray et al. 1987). However, only DNA fragments in the range of 100–1000 bp could be enriched using this system. Moreover, expression of the marker gene was also observed when the LacZ gene was not in frame with the cDNA because of the polycistronic mRNA in prokaryotes and the reinitiation of translation. Davis and Benzer (1997) constructed a vector that confers kanamycin resistance to the host on translation of an insert in the correct reading frame. They made three size-fractionated cDNA libraries in E. coli, namely 100–200, 200–300, and 300–500 bp. Only the library that contained small cDNA fragments in the range of 100–300 bp could be enriched to 60%–80% for ORF clones.
For the improvement of the selection efficiency, especially of larger inserts, it would be advantageous to use a eukaryotic host, which, in contrast to E. coli, has a codon usage homologous to that of mammalian cells, thus avoiding translation problems and frameshifts. Therefore, we chose the yeast Saccharomyces cerevisiae as a eukaryotic host, which is able to produce soluble proteins in large amounts (Romanos et al. 1992). The ORF vector pYEXTSH3 carries the HIS3 gene, coding imidazol-glycerol-phosphate-dehydrogenase, which enables the selection of ORFs based on histidine prototrophy. Here, we describe the construction of this vector, the development of the selection procedure, and the construction and characterization of a human fetal brain cDNA library in this expression system.
RESULTS AND DISCUSSION
Construction and Function of pYEXTSH3
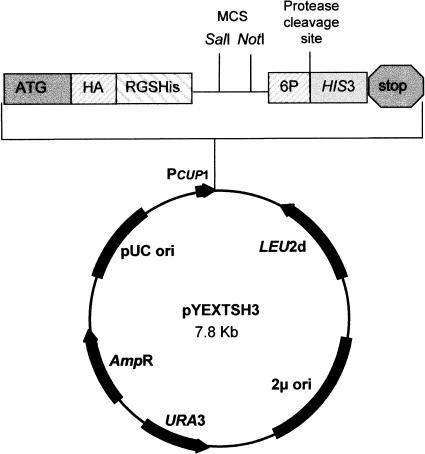
The ORF vector pYEXTSH3 is a derivative of the commercially available expression vector pYEXbx (Clontech), including the Cu2+-inducible CUP1 promoter from the yeast metallothionein gene, which induces expression of the encoded protein (Fig. 1). In addition, the vector contains the E. coli ampicillin resistance (Ampr) gene for selection in E. coli and the yeast selectable markers URA3 and LEU2d. For the improved expression of randomly primed cDNAs, we introduced a translation initiation sequence that was adapted to S. cerevisiae. For the detection and purification of expressed gene products, the plasmid includes a tandem epitope tag positioned N-terminal to the multiple cloning site (MCS), consisting of the hemaglutinine epitope (HA) and the oligo histidine domain (RGS-His6). The modified MCS includes a stop codon in each reading frame and allows the directional cloning (SalI/NotI) of cDNAs. For the cleavage of HIS3p from the fusion protein we additionally inserted the recognition sequence (6P) of the PreScission protease. The yeast HIS3 gene was introduced C-terminal to the MCS (Fig. 2). The translation of the DNA proceeds from the start codon ATG through the tag sequences into the HIS3 gene. Only if an insert is cloned in the correct reading frame and if it does not contain any stop codon will complete fusion protein be produced (Fig. 2B). In this case, expression of the gene product leads to histidine prototrophy in a his3 S. cerevisiae strain. Clones bearing the plasmid pYEXTSH3, but without inserts (Fig. 2A), remain auxotrophic, as no HIS3 gene is expressed because of translation stops in all three reading frames. The yeast cells are also expected to be auxotrophic if they contain a cDNA that is inserted in an incorrect reading frame, leading to internal stop codons and resulting in the termination of translation before reaching the HIS3 sequence (Fig. 2C). In general, mRNA sequences carry many stop codons in reading frames other than the correct one.
Figure 1.
Restriction map of pYEXTSH3. The vector includes the E. coli AmpR gene and the yeast selectable markers URA3 and HIS3. The modified MCS (SalI/NotI) contains a stop codon in each reading frame. The epitope tags HA and RGS-His6 were inserted at the N terminus of the MCS.
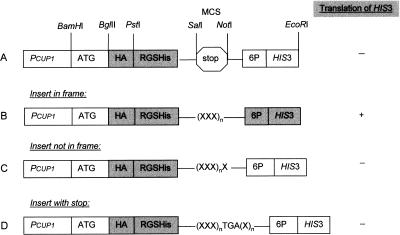
Figure 2.
Design and function of the ORF vector pYEXTSH3. (A) pYEXTSH3 includes a tandem epitope tag, a translation start and stop codon, and the HIS3 marker gene, which is under the control of the CUP1 promoter. In this plasmid, translation terminates before reaching HIS3. (B–D) The cloning of cDNAs insert, represented as XXXn, results in the following outcomes at the C terminus: (B) If the size of the cDNA insert is a multiple of three and both N and C termini are cloned in the correct reading frame, HIS3 is translated. (C) The translation terminates after translation of a few triplets in HIS3, if the insert is a multiple of 3 + 1, as the reading frame is shifted. (D) If the cloned cDNA includes an internal stop, there is no translation of HIS3.
The S. cerevisiae expression strain GRF18 (his3, leu2) was used for selection following transformation. To check the selection system, three different test inserts of human GAPDH were generated by PCR amplification. These GAPDH amplicons, which all contained the same N terminus but differed in their C-terminal regions, were cloned via SalI/NotI in-frame with the tag sequences and the start codon in pYEXTSH3. In the first case, the GAPDH insert was introduced in the correct reading frame relative to the HIS3 marker (Fig. 2B). The second construct included the stop codon of the GAPDH gene (Fig. 2D), and the third amplicon was modified by inserting an additional base pair at the C-terminal end of GAPDH, leading to a frameshift (Fig. 2C). These plasmids were transformed into yeast and the expression clones were selected directly on minimal medium (SD-his) supplemented with CuSO4 to induce protein expression. The vector without insert was used as a negative control (Fig. 2A), and a modified vector (pYEXT2HIS3), in which the stop codons between SalI and NotI were deleted and which also contains an N-terminally tagged HIS3 gene, was transformed in yeast as a positive control. As expected, the insertion of the cDNA in-frame resulted in histidine-prototrophic yeast cells growing on minimal medium, whereas the clones containing the vector without insert, or with the complete gene including the stop codon, did not grow. However, we observed that 3% of the selected transformants with the out-of-frame GAPDH could grow on minimal medium. This phenomenon might be caused by a translational frameshift and was reduced by increasing selection stringency (from 0.1 to 0.5 mM CuSO4). The survival ratio and the transformation efficiency of the in-frame construct were improved significantly by reducing the copper concentration in the selection medium. These results are shown in Table 1.
Table 1.
Transformation Efficiency of pYEXTSH3 and pYEXT2HIS3
| Vector construct | Concentration of CuSO4 | 0.3 mM | 0.5 mM | ||
|---|---|---|---|---|---|
| w/o CuSO4 | 0.05 mM | 0.1 mM | |||
| pYEXT2HIS3 | 1.2 × 103 | 1.2 × 103 | 2 × 104 | 4.8 × 103 | 1.2 × 103 |
| (A) pYEXTSH3 | 0 | 0 | 60 | 2.7 × 102 | 0 |
| (B) pYEXTGH3 (RF+) | 7 × 102 | 4 × 102 | 5 × 104 | 2.8 × 104 | 2 × 102 |
| (C) pYEXTGnH3 (RF−) | 10 | 3 × 102 | 10 | 0 | |
Transformation efficiency of pYEXTSH3 constructs (A–C) and the control plasmid pYEXT2HIS3 in Saccharomyces cerevisiae GRF18 is shown to be dependent on concentration of CuSO4. The efficiencies are given in cfu/μg DNA.
(RF+), insert in frame with HIS3.
(RF−), insert not in frame with HIS3.
Generation of the Yeast Expression Library
To subclone the cDNAs in this vector, two linkers containing SalI and NotI sites, respectively, were ligated to a randomly primed cDNA library from human fetal brain tissue, resulting in SalI–NotI overhangs. The library was cloned into pYEXTSH3 and transformed in S. cerevisiae strain GRF18. The clones were selected on SD-his agar supplemented with 0.1 mM CuSO4. In an initial experiment to characterize the library, 2000 colonies were picked by a robot into 384-well plates. To determine whether the library was enriched for cloned sequences representing ORFs, 96 clones were selected randomly and analyzed by PCR according to the method described in Lueking et al. (2000), by sequencing and protein expression. The obtained sequences were used for BLASTN searches against the public databases (NCBI). Fifty-eight clones (60%) were found to contain genes that were expressed and contained an ORF fused in-phase with HIS3. A selection of analyzed clones is listed in Table 2. In contrast to unselected libraries, it was possible to increase the percentage of clones in the correct reading frame fourfold, from 14% (Büssow et al. 2000) to 60%.
Table 2.
ORF Library Clones with Sequence Database Matches in the Correct Reading Frame
| Clone | Accession no. | Protein name | Cds/First matched amino acid in database sequence | Predicted protein size (kDa) |
|---|---|---|---|---|
| 001-B01/3 | D63997 | Golgi autoantigene, GCP170 | 270-4862/590 | 63 |
| 001-F01/7 | NM006031.1 | Pericentrin mRNA | 1-10018/8720 | 48 |
| 001-H01/9 | HSU26396 | α-spectrin mRNA | 1-2159/1411 | 28 |
| 001-B02/11 | AF155821 | Protein serine/threonine kinase CPG16 | 160-1467/717 | 38 |
| 001-A01/2 | AB002373 | KIAA0375 | 84-4706/601 | 147 |
| 001-D02/14 | Z26634 | Akryn 2, neuronal mRNA | 1->11775/9237 | 93 |
| 001-F02/16 | AB007890 | KIAA0430 | 1-3767/627 | 115 |
| 001-E03/23 | NM018013 | Protein FLJ10159 | 1-807/1 | 29 |
| 001-G03/25 | NM014787 | KIAA0076 | 87-5183/4023 | 40 |
| 001-D03/22 | NM003128.1 | Spectrin b, non-erythrocytic 1 | 311-7405/1893 | 202 |
| 001-G04/33 | X79536 | HnRNP core protein A1 | 27-989/1 | 35 |
| 002-D08/63 | NM001272.1 | Chromodomain DNA helicase | 211-6331/5290 | 38 |
| 002-F09/38 | HSM802205 | DKFZ p434M2... | 1-3043/2180 | 32 |
| 002-B12/14 | NM007027.1 | Topoisomerase II binding protein | 341-4648/3103 | 57 |
| 002-D12/28 | AB033044 | KIAA1208 | 32-2626/108 | 92 |
Following PCR analysis of 96 clones, we determined the average insert size of expressed cDNAs in this library to be 1.6 kb. The size range of the inserts was 200–3000 bp. The frequency of sequences cloned in the correct reading frame seems not to be dependent on the size of the cloned cDNAs, as was seen to be the case for bacterial expression (Davis and Benzer 1997), in which the proportion of in-frame clones decreased with increasing size of the cloned cDNAs, and only inserts with a maximum size of 500 bp were expressed. However, we observed in our yeast expression system that the efficiency of selection for sequences cloned in ORFs is influenced by the selection conditions. On optimization of the selection stringency, we obtained 60% of clones with in-frame inserts. The remaining 40% had either no insert, an internal stop codon, or were in the incorrect reading frame but, nevertheless, showed a histidine prototrophic phenotype.
On sequence analysis of these 40%, 16% had no insert but resulted from in-frame religation of the vector to itself. Fourteen percent of the clones were shown to have internal stops and, in this case, the HIS3 gene product should not have been produced. Perhaps its expression is the result of a translational frameshift or reinitiation of translation involving interactions between the cloned cDNA sequences and the yeast transcription/translation machinery. Although recognized as playing a key role in the control of a few eukaryotic mRNAs (Hinnebusch 1997), reinitiation has tended not to be regarded as a significant factor in eukaryotic gene expression. Also, the sequence context surrounding the stop codons has a strong influence on the efficiency of reinitiation. AU richness in the penultimate upstream codon and in the downstream region has been shown to be correlated with efficient reinitiation (Grant and Hinnebusch 1994). Internal initiation and leaky ribosomal scanning are alternative pathways used instead of the standard scanning mechanism and can be a basis for translational regulation in response to environmental stress (Rhoads and Lamphear 1995; Gan and Rhoads 1996).
The 10% of clones remaining contained inserts that were not in-frame with respect to the downstream fused HIS3 gene but nevertheless led to a histidine prototrophic phenotype. We suggest that, in these clones, the translation of HIS3 was caused by a ribosomal frameshift. Different types of frameshifting have been described in S. cerevisiae (Farabaugh 1996). They are found most commonly during the translation process, when the fusion of two protein subunits that are not in the same reading frame but belong to the same protein is required. It is not known which characteristics play a role in the frameshifting events in the present experiments. It would be interesting to test different host strains and further selection markers, instead of HIS3, to reduce the number of false-positive clones. The application of a dominant resistance marker could possibly improve the screening efficiency for in-frame clones because more stringent selection conditions could be used.
Antibody Screening on Protein Filters
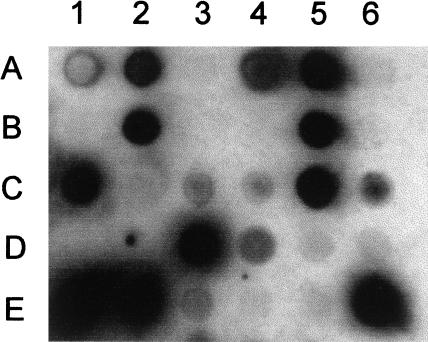
The picked yeast clones were checked additionally for protein expression on high-density protein arrays. The cDNA clones were spotted onto polyvinylidene difluoride (PVDF) membranes that were processed subsequently for protein expression using the monoclonal antibody RGS(His6 (QIAGEN), which recognizes the N-terminal sequence RGS-His6 of recombinant expression products (Fig. 3). Approximately 60% of the clones expressed proteins, which were detected by the antibody screening on protein arrays, confirming the results from the sequence analysis of the selected cDNA. For example, the clone in position A2 in the spotting pattern shown in Figure 3 corresponds to the clone 001-F01/7 (Table 2).
Figure 3.
Protein expression screening of the HFBr library in S. cerevisiae GRF18 on protein filters. The protein filters were screened for cDNA clones expressing recombinant fusion proteins using the RGS-His6 antibody. The clone in position D6 is the negative control containing a vector without an insert, and E6 is the positive control expressing the human GAPDH gene.
Protein Expression
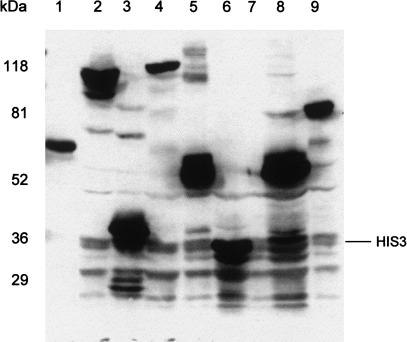
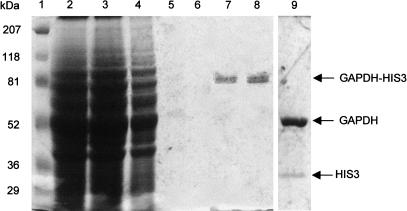
The gene products of some representative clones, which had been sequenced previously, were characterized by protein expression analysis. Whole-cell proteins separated by SDS-PAGE were transferred to PVDF membranes, followed by detection of expression products by Western immunoblotting with the antibody specific for the N-terminal RGS-His6 epitope of the expressed fusion proteins. As shown in Figure 4, the HIS3 gene product was detected at 36 kD (lane 6), whereas no protein was detectable from crude cell lysate of the negative control pYEXTSH3, which does not contain a cDNA insert (lane 7). The gene products of the RGS-His6-tagged fusion genes, GAPDHnHIS3 (construct with frameshift) and GAPDH-HIS3, are shown in lanes 8 and 9, respectively. The translation of the first construct terminates after 10 amino acids because of the stop codon that was introduced by the reading-frame shift, whereas the latter construct resulted in the expression of a full-length fusion protein with the expected molecular weight of 86 kD. The expression products of five cDNA clones are shown in lanes 1–5, in which all clones express a detectable fusion protein.
Figure 4.
Expression of different inserts from HFBr cDNA clones in the ORF-selecting vector pYEXTSH3. Proteins separated by SDS-PAGE (100 μg each lane) were detected from crude cell extracts by immunoblotting with an anti-RGS-His6 antibody. (Lanes 1–5) HIS3 fusion proteins from selected cDNA expression clones 001-B2, 001-H1, 001-F1, 001-F2, and 001-E3; (lanes 6–9) HIS3 from pYEXT2His3, pYEXTSH3 without insert (negative control), pYEXTGnH3 (GAPDH not in-frame), and pYEXTGH3 (GAPDH in-frame, positive control). The positions of the prestained Bio-Rad broad range marker are indicated on the left.
In Figure 5 the purification (lanes 2–8) and cleavage (lane 9) of the GAPDH-HIS3 protein is shown as an example. The RGS-His6 tagged fusion protein was affinity purified under native conditions using Ni-NTA agarose, and the C-terminally fused HIS3 protein was cleaved after the 6P site by protease digestion. After 16 h incubation at 4°C, the fusion protein was cleaved completely without any degradation products (lane 9).
Figure 5.
Purification and cleavage of the GAPDH-HIS3 fusion protein. The fusion protein was expressed in S. cerevisiae GRF18 and purified using Ni-NTA agarose. Proteins were visualized by Coomassie staining. (Lane 1) Prestained Bio-Rad broad range marker; (lane 2) cell lysate; (lane 3) flow-through; (lanes 4–6: wash1-3; (lanes 7,8) eluate 1 and 2; (lane 9) cleavage of the C-terminally fused HIS3 protein by PreScission Protease.
In summary, the screening system described here is a useful one-step screening procedure for the generation of expression libraries enriched with clones that produce authentic polypeptides. Using the eukaryotic host S. cerevisiae, we were able to produce soluble proteins in large amounts that can be used to generate protein arrays and for the analysis of antibody specificity and other protein–protein interaction or ligand–receptor systems (Cahill 2001). A further field of application for our system is to establish protein product catalogs from different tissues and development stages. As these proteins are expressed from arrayed cDNA clones their identity can be checked easily by commonly used DNA techniques or by mass spectrometry (Büssow et al. 2000; Cahill et al. 2000). The cDNA clones can also be used to generate high-density DNA arrays (Clark et al. 1999).
METHODS
Strains, Media, and Transformation
E. coli strains XL1-Blue (Stratagene) and DH10B (Life Technology) were used as host strains for cloning and amplification of plasmid DNA. The transformation of recombinant plasmids was performed by electroporation using the Gene Pulser (Bio-Rad). E. coli transformants were selected on LB (0.5% yeast extract, 1% NaCl, 15% bactotryptone) medium supplemented with 100 μg/mL ampicillin. The S. cerevisiae strain GRF18 (α, leu2-3, leu2-2112, his3-15, can1, mal) was used as the eukaryotic expression strain. The ORF vector constructs and the cDNA expression library were transformed in S. cerevisiae by the lithium acetate method (Gietz et al. 1992), and the resulting transformants were selected on SD-his induction medium (2% dextrose, 0.67% Yeast Nitrogen Base [YNB], 40 mg/L leucine, 0.1 mM CuSO4). The S. cerevisiae clones were cultivated in microtiter plates in freezing medium (2% dextrose, 5% glycerol, 0.67% YNB, 40 mg/L histidine, 0.5 M betaine). For protein expression, the clones were transferred in SD-leu medium (2% dextrose, 0.67% YNB, 40 mg/L histidine) and the expression was induced by addition of 0.5 mM CuSO4).
Construction of the Vector
The YEX system uses the Cu2+-inducible CUP1 promoter to control expression of genes inserted in the MCS. The expression vector pYEXbx (Clontech) was modified by introduction of a NotI restriction site and a translation initiation sequence (CAAAATGTCT) downstream of the promoter region, which allows the translation of cDNAs without their own start codon. For detection and purification of the gene product, we introduced a tandem epitope tag positioned N-terminal of the MCS consisting of the hemaglutinine epitope (HA) and the oligo histidine domain (RGS-His6). A DNA fragment that carries the coding sequence of the HIS3 gene (SWISS-PROT P06633) and the PreScission protease recognition (6P) sequence (CTGGAAGTTCTGTTCCAGGGGCCC) at the N terminus was generated by PCR amplification from the plasmid YdpH and cloned NotI/EcoRI in the expression vector. The resulting plasmid pYEXTSH3 can be used for cloning of cDNAs with SalI/NotI overhangs and for the expression of randomly primed cDNAs. The restriction map of pYEXTH3 is shown in Figure 1.
Cloning of the cDNA Expression Library
The cDNA inserts of a random-primed cDNA library from human fetal brain tissues in the vector pSPORT (provided by B. Korn; Resource Centre of the German Genome Project [RZPD], Heidelberg) were cleaved with SalI/EagI and electrophoresed through 1% agarose gel. Fragments in the size range 200 bp–2 kb were eluted from the gel (QIAGEN, QIAquick Gelextraction Kit) and ligated into the SalI and NotI sites of the vector. Following transformation in E. coli DH10B by electroporation, the library was plated onto square agar plates (230 mm × 230 mm, Genetics) and grown for 20 h at 37°C. Approximately 150,000 clones were scraped from the agar, resuspended in P1 buffer (50 mM Tris-HCl at pH 8.0, 10 mM EDTA, 100μg RNase A), and plasmid DNA was isolated. The purified DNA was transformed in S. cerevisiae GRF18.
Generation of the Yeast Expression Library
Following transformation the yeast cells were plated on SD-his induction medium at a density of 1500 clones per agar plate (230 mm × 230 mm) and were incubated 3–4 d at 28°C. Colonies obtained were picked into 384-well microtiter plates and filled with freezing medium by a robot. The colony-picking program was adapted to yeast cells. The robot-picking specifications are as follows: It has a resolution of 5 μm, a velocity of 3 m/sec, and it is able to pick about 3600 clones/h. The cells were grown in the microtiter wells for 3–4 d at 28°C, and afterward the clones were replicated by a 384-pin replicator. All copies were stored frozen at −80°C.
Sequence Analysis
For sequencing, the S. cerevisiae clones were transferred into 96-well plates filled with 65 μL freezing medium and were incubated 4 d at 28°C. The inserts of the cDNA clones were amplified by PCR directly from liquid cultures as described previously (Lueking et al. 2000). For PCR amplification, the primers pYEXbx5 (CATATAGAAGTCATCGA) and pYEXbx3 (TTTGCAG CTACCACATT) were used.
Generation of High-Density Filters
For generation of high-density filters the yeast clones were grown in the 384-well microtiter plates and were gridded onto PVDF membrane (Millipore) for protein analysis as described (Meier-Ewert et al. 1994; Büssow et al. 1998) with the following modifications. Before spotting, the filters were soaked in medium (0.67% YNB, 2% dextrose, 40 mg/L histidine, 0.5% calcium propionate). The clones were gridded in duplicate in a 3 × 3 spotting pattern at a density of 9216 clones per 222 mm × 222 mm PVDF membrane with ink guide dots. The prepared filters were placed onto square SD-leu agar plates containing 0.5% calcium propionate (antifungal agent). Following colony growth for 2 d at 28°C, protein filters were transferred to agar plates supplemented with 0.5 mM CuSO4 to induce protein expression and were then incubated for an additional 2 d at 28°C. Then protein filters were placed on Whatman 3MM paper soaked in 4 M NaOH and left at room temperature for 1 h. The membranes were transferred onto Whatman 3MM paper soaked in denaturant solution 0.5 M NaOH, 1.5 M NaCl (10 min); neutralizing solution 1 M Tris-HCl (pH 7.5), 1.5 NaCl (10 min); 2× SSC (15 min). Yeast debris was wiped off with paper towels in TBST-T buffer (20 mM Tris-HCl at pH 7.5, 0.5 M NaCl, 0.05% Tween 20, 0.5% Triton X-100) and the membranes were washed two times briefly with TBS before using for antibody screening.
Antibody Screening on Protein Filters
The processed filters were blocked 1 h in blocking buffer (2% [w/v] BSA/TBST) and were incubated for 1–2 h in primary antibody solution diluted (1 : 2000) in blocking buffer according to the manufacturer's instructions. After washing 2 × 10 min in TBST, filters were incubated in horseradish peroxidase-conjugated secondary antibody diluted in 5% nonfat dry milk powder in TBST for 1 h. After washing four times for 10 min in TBST buffer, the filters were incubated in chemiluminescence substrate (NEN, Renaissance) for 1 min at room temperature. The excess chemiluminescence reagent was removed and the membranes either were exposed to autoradiography film, or a video system and a BioImager (428 nm) were used to generate pictures of filters, respectively.
Protein Expression
Selected clones of S. cerevisiae GRF18 were grown in 20–100 mL SD-leu medium at OD600 of 1.0, and protein expression was induced by addition of 0.5 mM CuSO4 for 3–4 h. The cells were harvested by centrifugation (10 min, 2500g, 4°C) and washed once with PBS (10 mM Na2PO4, 300 mM NaCl at pH 8.0). Washed pellets were resuspended in lysis buffer (10 mM Na2PO4, 300 mM NaCl at pH 8.0, 1% (v/v) Triton X-100) with protease inhibitor PMSF (1 mM). One volume sterile, acid-washed glass beads (0.5 mm) were added and cells were lysed by seven cycles of 1 min full-speed vortexing followed by 1 min cooling on ice. Beads were removed and the extract was cleared by centrifugation (15 min, 10,000g). Whole-cell proteins in the supernatant were separated directly by SDS-PAGE or purified by affinity chromatography (Ni-NTA agarose). Gels were blotted onto PVDF membranes, blocked with 2% BSA/TBST, and incubated for 1–2 h with the primary antibody diluted in blocking buffer according to the manufacturer's instructions. The secondary antibody was horseradish peroxidase-conjugated anti-mouse antibody for use with the NEN chemiluminescence protocol.
Protease Cleavage
The HIS3 fusion proteins were cleaved by using 2 units of PreScission Protease (Amersham Pharmacia Biotech) for each 100 μg of fusion protein in the eluate. The sample was adjusted to 1× cleavage buffer by addition of an appropriate volume of 10× cleavage buffer according to the manufacturer's instructions and was incubated for 4 h at 5°C.
Acknowledgments
We thank Bernhard Korn (RZPD, Heidelberg) for the human fetal brain cDNA, Christine Lang (Technical University, Berlin) for the YdpH plasmid carrying the HIS3 gene and the S. cerevisiae strain GRF18, Martin Horn for assistance of adaptation of the picking procedure to yeast cells, and Heike Göhler and Erich E. Wanker for valuable discussions. This work was funded by Bundesministerium für Bildung und Forschung (BMBF Grants 0311018, 0311323, and 0311870 ) and the Max-Planck-Gesellschaft.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cahill@molgen.mpg.de; FAX 49-30-8413-1128.
Article published on-line before print: Genome Res., 10.1101/gr.181501.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.181501.
REFERENCES
- Büssow K, Cahill D, Nietfeld W, Bancroft D, Scherzinger E, Lehrach H, Walter G. A method for global protein expression and antibody screening on high-density filters of an arrayed cDNA library. Nucleic Acids Res. 1998;26:5007–5008. doi: 10.1093/nar/26.21.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büssow K, Nordhoff E, Lübbert C, Lehrach H, Walter G. A human cDNA library for high-throughput protein expression screening. Genomics. 2000;65:1–8. doi: 10.1006/geno.2000.6141. [DOI] [PubMed] [Google Scholar]
- Cahill DJ. Protein and antibody arrays and their medical applications. J Immunol Meth. 2001;250:81–91. doi: 10.1016/s0022-1759(01)00325-8. [DOI] [PubMed] [Google Scholar]
- Cahill DJ, Nordhoff E, O'Brien J, Klose J, Eickhoff H, Lehrach H. Bridging genomics and proteomics. In: Pennington S, Dunn M, editors. Proteomics. Oxford: BIOS Scientific Publishers; 2000. pp. 1–20. [Google Scholar]
- Clark MD, Panopoulou GD, Cahill DJ, Buessow K, Lehrach H. Construction and analysis of arrayed cDNA libraries. Meth Enzymol. 1999;303:205–33. doi: 10.1016/s0076-6879(99)03015-3. [DOI] [PubMed] [Google Scholar]
- Davis CA, Benzer S. Generation of cDNA expression libraries enriched for in-frame sequences. Proc Natl Acad Sci. 1997;94:2128–2132. doi: 10.1073/pnas.94.6.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh PJ. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan W, Rhoads RE. Internal initiation of translation directed by the 5′-untranslated region of the mRNA for eIF4G, a factor involved in the picornavirus-induced switch from cap-dependent to internal initiation. J Biol Chem. 1996;271:623–626. doi: 10.1074/jbc.271.2.623. [DOI] [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CM, Hinnebusch AG. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol Cell Biol. 1994;14:606–618. doi: 10.1128/mcb.14.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MR, Mazzara GP, Reddy P, Rosbash M. Searching for clones with open reading frames. Meth Enzymol. 1987;154:129–156. doi: 10.1016/0076-6879(87)54074-5. [DOI] [PubMed] [Google Scholar]
- Harrison SM, Dunwoodie SL, Arkell RM, Lehrach H, Beddington RSP. Isolation of novel tissue-specific genes from cDNA libraries representing the individual tissue constituents of the gastrulating mouse embryo. Development. 1995;121:2479–2489. doi: 10.1242/dev.121.8.2479. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of yeast GCN4. A window on factors that control initiator-trna binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- Lueking A, Holz C, Gotthold C, Lehrach H, Cahill DJ. A system for dual protein expression in Pichia pastoris and Escherichia coli. Protein Expr Purif. 2000;20:372–378. doi: 10.1006/prep.2000.1317. [DOI] [PubMed] [Google Scholar]
- Meier-Ewert S, Rothe J, Mott R, Lehrach H. Establishing catalogues of expressed sequences by oligonucleotide fingerprinting of cDNA libraries. In: Hochgeschwender U, Gardiner K, editors. Identification of transcribed sequences. New York: Plenum; 1994. pp. 253–260. [Google Scholar]
- Perret E, Ferran EA, Marinx O, Liauzun P, Dumont X, Fournier J, Kaghad M, Ferrara P, Caput D. Improved differential screening approach to analyse transcriptional variations in organized cDNA libraries. Gene. 1998;208:103–115. doi: 10.1016/s0378-1119(97)00658-6. [DOI] [PubMed] [Google Scholar]
- Rhoads RE, Lamphear BJ. Cap-independent translation of heat shock messenger RNAs. Curr Top Microbiol Immunol. 1995;203:131–153. doi: 10.1007/978-3-642-79663-0_7. [DOI] [PubMed] [Google Scholar]
- Romanos MA, Scorer CA, Clare JJ. Foreign gene expression in yeast: A review. Yeast. 1992;8:423–488. doi: 10.1002/yea.320080602. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW. Parallel human genome analysis: Microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Heller RA, Theriault TP, Konrad K, Lachenmeier E, Davis RW. Microarrays: Biotechnology's discovery platform for functional genomics. Trends Biotechnol. 1998;16:301–306. doi: 10.1016/s0167-7799(98)01219-0. [DOI] [PubMed] [Google Scholar]