Abstract
To meet the demands of developing lead drugs for the profusion of human genes being sequenced as part of the human genome project, we developed a high-throughput assay construction method in yeast. A set of optimized techniques allows us to rapidly transfer large numbers of heterologous cDNAs from nonyeast plasmids into yeast expression vectors. These high- or low-copy yeast expression plasmids are then converted quickly into integration-competent vectors for phenotypic profiling of the heterologous gene products. The process was validated first by testing proteins of diverse function, such as p38, poly(ADP-ribose) polymerase-1, and PI 3-kinase, by making active-site mutations and using existing small molecule inhibitors of these proteins. For less well-characterized genes, a novel random mutagenesis scheme was developed that allows a combination selection/screen for mutations that retain full-length expression and yet reverse a growth phenotype in yeast. A broad range of proteins in different functional classes has been profiled, with an average yield for growth interference phenotypes of ∼30%. The ease of manipulation of the yeast genome affords us the opportunity to approach drug discovery and exploratory biology on a genomic scale and shortens assay development time significantly.
[The sequence data described in this paper have been submitted to the data library under accession no. AF359244.]
The pharmaceutical industry is in the midst of an information overload triggered by the sequencing of the human genome (Lander et al. 2001; Venter et al. 2001). The challenge now is to elucidate the function of the encoded gene products and to determine their possible involvement in disease. Approaches that measure protein binding (two-hybrid analysis) or changes in expression (microarrays) across the entire genome have been initiated to connect sets of genes functionally. A prime example is the use of microarrays to monitor genome-wide differences in transcription between normal and diseased tissue (DeRisi et al. 1996; Schena et al. 1998; Zweiger 1999; Diehn et al. 2000). Already, enormous strides have been made using such expression profiling to associate unique transcriptional patterns with stages of development in certain cancers (Golub et al. 1999; Perou et al. 1999, 2000; Sgroi et al. 1999; Alizadeh et al. 2000; Bittner et al. 2000; Ross et al. 2000). In addition to serving as diagnostic markers for disease, the unique gene patterns identified might also provide drug development targets for therapeutic intervention by pharmaceutical compounds. However, of the hundreds of up- or down-regulated genes observed, only a small percentage might actually play a functional role in the disease being studied.
One way to simplify the problem is to sift through the genes whose expression is altered and identify those genes that might encode activities that are similar to those that have been important historically in drug development. Over the years, researchers have shown that the malfunction of certain classes of proteins, for example, kinases, proteases, phosphodiesterases, phosphatases, and G protein coupled receptors (GPCRs), occurs in a variety of diseases. These findings are not surprising given that these proteins play key roles in signaling pathways that act to coordinate internal cellular functions with the external environment (Cohen 1999; Kowaluk and Jarvis 2000; Pawson and Nash 2000; Stein and Waterfield 2000). Often these proteins belong to large gene families whose members share significant sequence identity, although many members have no established biological function. That is, even if they possess domains suggesting a given biochemical activity, their substrate(s) and role in cellular physiology are unknown.
How then does one begin to build specific biochemical assays for hundreds of genes with no clearly defined substrate? Additionally, many proteins will be intractable for various other reasons, such as that they are unstable in vitro or require membrane localization for activity. Moreover, the sheer number of genes indicates the need for an approach encompassing an increase in scale and parallel processing. In this respect, cell-based assays have an inherent advantage over biochemical assays in that they eliminate the time investment required to gain enough knowledge about each protein to prepare a purified target or to modify the target for activation. More importantly, in cell-based assays, proteins are examined in a cellular context that simulates more closely the normal physiological state (Hertzberg 1993; Silverman et al. 1998).
There has been widespread recognition that cell-based assays designed in model organisms such as the yeast Saccharomyces cerevisiae provide greater ease of genetic manipulation and can be screened rapidly at a low cost (Kirsch 1993; Silverman et al. 1998; Munder and Hinnen 1999). Although the yeast cell wall has been thought to limit permeability of small molecules during screening, there is now evidence to indicate that compound efflux, as opposed to permeability, is the culprit, and this can be controlled by deletion of the major ABC transporters in yeast (Broach and Thorner 1996; Kolaczkowski et al. 1998; Kaur and Bachhawat 1999). Yeast cell-based assays have been designed in which mammalian proteins with yeast homologs, such as GPCRs, or enzymes such as Topoisomerase II, are made to function in yeast (van Hille and Hill 1998). These types of assays require that the mammalian gene product couple to yeast pathways in such a way as to provide a functional readout that can be used for screening. This approach has been used traditionally to isolate genes from higher organisms by functional complementation of yeast mutations (Becker et al. 1991; Tugendreich et al. 1994; Jonassen et al. 1998). A requirement for this application is that the biochemical activity affected by the yeast mutation has a counterpart in a mammalian pathway (Becker et al. 1991; Jonassen et al. 1998).
On the other hand, the presence of a yeast counterpart is not absolutely necessary for a foreign protein to be active in yeast. For instance, although apoptosis is a process generally associated with multicellular organisms, new mammalian genes that play roles in this pathway have been discovered through the use of genetic approaches in yeast (Shaham et al. 1998; Greenhalf et al. 1999). For example, the growth arrest and cell death induced by the mammalian apoptosis effector Bax is overcome when the known apoptosis inhibitors, Bcl-2 and Bcl-x (L), are coexpressed in yeast (Sato et al. 1994; Greenhalf et al. 1996). The reversal of the growth interference phenotype was used to isolate mammalian genes that were shown later to inhibit induction of apoptosis in mammalian cells (Xu and Reed 1998; Greenhalf et al. 1999). These studies confirm the potential use of yeast growth interference assays to study (and to even identify) new genes involved in key cellular pathways in disease, even if it happens that those pathways are not found in yeast.
We have developed a high-throughput process in yeast that enables us to assay many heterologous genes for which no suitable assay can be applied easily. We describe here the initial phase of our process that establishes a phenotypic readout induced by expression of a heterologous gene. In later phases of assay development, we use genetic strategies to elaborate further on the phenotype or to broaden the response of yeast to a wider spectrum of heterologous cDNAs. The enhancement of the yeast phenotype is based on the synergistic or potentiating effect of such mutations. This type of “synthetic dosage lethality effect” has been described for overexpression of yeast proteins (Kroll et al. 1996). We describe here an example of such an interaction for the mammalian MAP kinase, p38.
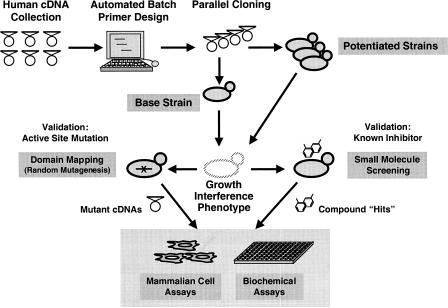
The assay development process reported here is depicted in Figure 1 and encompasses the following suite of methods: (1) a rapid, uniform, and parallel cloning method for the transfer of heterologous cDNAs into yeast expression vectors; (2) a quick method to convert high- or low-copy episomes into linear, integratable vectors; (3) an automated batch processing procedure for analyzing the data from highly sensitive liquid growth assays; (4) a random mutagenesis scheme for validation of the yeast phenotype and for structure/activity analysis of proteins that are characterized poorly or have no known active site; and (5) analysis of the selected mutants and small molecule inhibitors in mammalian secondary assays. The process of profiling cDNAs in a rapid, parallel, and systematic manner eliminates the costly need for a separate assay development strategy for each cDNA and therefore shortens the time between “gene and screen”. These kinds of rapidly constructed assays (which in principle could be derived from any set of heterologous genes, including those from plants or pathogens) allow identification of small molecules as potential leads for drug development and as tools to better understand gene function.
Figure 1.
Steps for parallel profiling of mammalian and other heterologous cDNAs in yeast cell-based assays. Heterologous cDNAs from cDNA collections are transferred in parallel to yeast expression vectors and transformed into yeast strains optimized for drug screening. These strains can be further manipulated genetically (potentiated) to obtain phenotypes for those expressed cDNAs that do not produce a response in the original base screening strain. A growth interference phenotype resulting from the expression of the cDNA from the inducible GAL1 promoter is used as a readout to screen for chemical inhibitors. The phenotype is validated with respect to the biochemical activity of the protein by making active site mutations and testing existing inhibitors. For poorly characterized gene products, the structure–activity relationship is determined using a novel random mutagenesis scheme. Finally, both the mutants and the small molecule inhibitors obtained by using this approach are tested in mammalian secondary assays.
RESULTS
An Inverse PCR Method Facilitates the Rapid, Uniform, and Parallel Cloning of Large Numbers of Heterologous cDNAs into Yeast Expression Vectors
A method was developed to uniformly move groups of cDNAs from repositories already cloned into nonyeast vectors into pARC25B, our specially engineered yeast expression vector. We call this method GRIPP, for Gap Repair with Inverse PCR'd Plasmid (Fig. 2, upper shaded box on the left labeled GRIPP cloning). In this method, the yeast expression vector is amplified by inverse PCR and recombined with a cDNA that has been released from its host vector by restriction digest, as indicated in Figure 2. Colonies are selected by complementation of leu2 auxotrophy. A marked stimulation of colony number is observed (5- to 50-fold) when the transformation includes insert as compared with a control reaction containing the PCR-amplified yeast expression vector alone. The presence of a correct recombination event is verified by whole-cell PCR amplification with primers that flank the insert.
Figure 2.
Transfer of heterologous cDNAs from libraries or nonyeast host plasmids into the pARC vector series by way of recombinational cloning and using the GRIPP method; the vectors allow quick transition in copy number from 2μ and CEN to integration-competent vectors. (Left upper shaded box) A proprietary method for transferring large numbers of heterologous cDNAs from nonyeast host vectors into yeast expression vectors (GRIPP Cloning); (right upper shaded box) cloning steps required to transfer individual mammalian cDNAs from libraries into yeast expression vectors (Recombinational Cloning). A schematic of the yeast expression vector used in all the yeast cell-based assays (pARC vector series) is shown directly below the GRIPP and recombinational cloning boxes. (Lower shaded box) Steps by which the yeast expression vector is integrated quickly into the chromosome for single-copy expression. Two SfiI restriction sites are shown flanking the CEN or 2μ origin of replication (pARC35B or 25B, respectively), and the two boxes containing white arrows indicate sequences homologous to the yeast LYS2 gene; (left) the plasmid has been cut with SfiI and recombines into the chromosome at the LYS2 locus at the sites of homology, deleting the entire LYS2 ORF in the process; (right) the fully integrated plasmid and direction of transcription is shown. The GFP/KanMX cassette used to recombinationally tag the different mammalian cDNAs is also shown.
As shown in Table 1, 16 cDNAs were cloned in parallel using GRIPP. Four colonies were chosen from each transformation and the presence of the desired insert checked by whole-cell PCR amplification. GRIPP is a highly efficient process, because an insert-containing transformant was recovered (in at least one of the four colonies) in 15 out of 16 cases. For the one that was missed, 12 more colonies were chosen, and 4 of these had an insert. Typically, out of four colonies, 75%–100% contain the proper insert. For each cDNA, a single recombinant was selected, the plasmid was “rescued” into Escherichia coli by transformation, and the recombination junctions were sequenced, because of our observations that these regions are prone to mutation. Indeed, 3 of the 16 recombinants had mutations near the ends of the recombined ORFs. We then picked a few more clones for each of these and were able to find isolates with no mutations. Typically, 25 cDNAs can be processed from restriction digestion to sequencing in a 2-wk period of time by a single researcher using no automation. Thus, by using the GRIPP method, large numbers of cDNAs can be moved rapidly from host vectors into yeast expression vectors in a uniform manner.
Table 1.
GRIPP (Gap Repair by Inverse PCR'd Plasmid) is Highly Efficient for Transferring Large Numbers of Heterologous cDNAs from Non-Yeast Host Vectors into Yeast Expression Vectors
| Gene name | Full or LBD | GRIPP recombination | Sequencing ends of inserts | GFP tagging | ||
|---|---|---|---|---|---|---|
| Percentage with inserts (of four chosen) | Correct sequence of recombinant (of one chosen) | Nature of mutation | GFP recombination successful | GFP immunoblot shows correct protein band | ||
| NR1H4 | Full | 100% | Yes | Yes | No | |
| LXR alpha | Full | 75% | Yes | Yes | No | |
| NR2E3 | Full | 100% | Yes | Yes | Yes | |
| RORC | Full | 100% | No | “+1 f.s. at 3′ end” | Yes | Not done |
| HNF4 alpha | Full | 100% | No | “+1 f.s. at 5′ end” | Yes | Not done |
| NR5A2 (CPF) | Full | 75% | Yes | Yes | Yes | |
| Nurr1 | Full | 0% of 4, 25% of 12 | No | ATG → TTG | Yes | Not done |
| CAR | Full | 100% | Yes | Yes | Yes | |
| STAT6 | Full | 100% | Yes | Yes | Yes | |
| STAT4 | Full | 100% | Yes | Yes | Yes | |
| Estrogen Receptor 1 | LBD | 50% | Yes | Yes | Yes | |
| NAK1 | LBD | 50% | Yes | Yes | Yes | |
| ROR-beta | LBD | 100% | Yes | Yes | Yes | |
| RORalpha1 | LBD | 100% | Yes | Yes | Yes | |
| Estrogen Receptor beta | LBD | 100% | Yes | Yes | No | |
| poly (ADP-ribose) polymerase (PARP-1) | N/A | 75% | Yes | Yes | Yes | |
The parallel transfer of 16 cDNAs into the pARC25B vector series is shown. The process is highly efficient and results in the correct recombinational event 94% of the time when four colonies were analyzed. The process of transfer of an ORF into the vector is 100% if multiple clones are analyzed. However, mutations can occur near the sites of recombination as indicated in the table for HNF4alpha, Nurr1, and RORC.
Strains carrying the integrated cDNAs were transformed with a GFP KanMX PCR product to recombinationally GFP-tag the cDNA. Transformants were checked first for a proper recombination event by whole cell PCR, and then for a protein band of the correct size by immunoblotting using anti-GFP antibodies. We found that 10 of the 13 cDNAs analyzed (i.e., with intact ORFs) were expressed as full-length GFP fusion proteins in yeast. The fusion proteins enabled the analysis of expression and cellular localization, but were not used for screening purposes.
In this study, heterologous cDNAs were also obtained from cDNA libraries. In this case, as detailed in the Methods, ORFs were amplified with PCR using primers containing approximately 45 nt of homology for recombination into gapped vectors according to methods described previously (Fig. 2, upper shaded box on the right labeled Recombinational Cloning; Szostak et al. 1983; Raymond et al. 1999).
A Special Yeast Expression Vector Allows Rapid Conversion of High- or Low-Copy Episomes into Integration-Competent Vectors
To assess rapidly whether a given heterologous cDNA will yield a growth interference phenotype in yeast it is advantageous to express the cDNA from a multicopy plasmid under the control of an inducible GAL1 promoter (Botstein and Fink 1988). However, for subsequent inhibitor screens we found that the assay was more responsive for screening when the GAL1-driven cDNA was expressed from a chromosomally integrated locus (data not shown). To facilitate both the quick episomal screening of yeast for growth interference phenotypes and subsequent chromosomal integration for screening, yeast vectors were engineered such that the 2μ or CEN element could be removed easily from the expression vector by restriction digest using SfiI (Fig. 2, pARC vector series). In those cases in which the cDNA contains one or more SfiI sites, other rare-cutter restriction sites were engineered into subsequent versions of the expression vector. Removing these replication elements linearizes the vector for chromosomal integration by exposing 45-nt segments that match to the ends of the LYS2 ORF in the yeast genome. As shown in Figure 2 (bottom shaded box), the entire plasmid is integrated via γ integration (Sikorski and Hieter 1989), which replaces the LYS2 locus. Successful integrants are selected first by complementation of Leu2 auxotrophy and then the colonies are tested to determine which colonies have become lysine auxotrophs (by replica plating onto α amino adipate plates). In the final step, the Lys−colonies are streaked to isolate single colonies on rich media and checked by whole-cell PCR amplification for proper chromosomal integration with vector and LYS2-specific primers.
Typically, ∼10%–45% of the Leu prototroph colonies have simultaneously become Lys−, and these Lys− colonies are usually deleted for the LYS2 locus. The method is on par with the efficiency of other strategies but allows for uniform integration of the cDNAs, saves time, and does not require extending the size of the expression vector or any type of re-PCR of the construct that could introduce mutations.
Recombination is Used to Epitope Tag the cDNA to Determine Expression Level and the Size of the Protein Product
To assure that the gene product of the expressed heterologous cDNA is full-length and properly localized in the cell, cDNAs were epitope tagged with green fluorescent protein (GFP).The GFP tagging and assay development processes were performed in parallel: Generally, the native (untagged) proteins were used in assays for compound screening. Those colonies expressing GFP fusion proteins frequently could be visualized on plates (Cronin and Hampton 1999), but in some cases a fluorescent signal was not observed, possibly because of low expression level and/or toxicity of the heterologous protein. Thus, the expression of cDNAs tagged with GFP was verified by immunoblot analysis using anti-GFP antibodies (see data in Table 1). The GFP tagging was performed by recombination using a protocol described previously (Longtine et al. 1998). As indicated in Table 1, 10 of the 13 cDNAs with intact ORFs produced GFP fusion proteins in yeast of the predicted size as observed by immunoblotting for GFP. We have tagged nearly 70 cDNAs using this process with a >90% success rate (data not shown), showing that it is usually possible to express a given mammalian or viral cDNA in yeast. In addition, the expected cellular localization of many of the fusion constructs was confirmed by fluorescence microscopy (see Fig. 6B as an example). We have also used other reporters including β-galactosidase and yeast auxotrophic markers (e.g., URA3) to generate the fusion proteins, but the data shown in this paper focus on studies using the GFP moiety.
Figure 6.
Selected mutants that were screened for expression of full-length CPF (glowing), or for truncated CPF (nonglowing) are confirmed by immunoblot analysis using anti-GFP antibodies. (A) Immunoblot probed with anti-GFP antibody shows the expression of several selected CPF constructs containing mutations that restore growth to yeast. Two lanes that were loaded with cellular extracts from white colonies carrying stop codons (isolates 49 and 51, labeled “Stop”) showed no protein bands as predicted. (B) The cellular localization of CPF was determined using a fluorescent phase contrast microscope at 100× magnification. (Top) Phase contrast; the large organelle in the cell body is the vacuole; (bottom) CPF-GFP localized properly in the nucleus.
High-Throughput Liquid Growth Assays Are a Sensitive Measure to Test Whether the Expression of Heterologous cDNAs Causes Growth Defects in Yeast
High-throughput liquid growth assays in 96-well plates were used to profile cDNAs phenotypically in parallel as described in the Methods. Cells carried the cDNAs either in high copy or from an integrated locus and were expressed from the inducible GAL1 promoter. The cells were grown first in glucose (uninduced) and then in the presence of increasing concentrations of galactose (induced), and their growth was compared with cells containing high-copy or integrated vector alone as appropriate. End-point growth inhibition values were calculated as described in the Methods.
Table 2 shows the effect on growth interference of a diverse set of heterologous cDNAs expressed in yeast. This set includes nuclear receptors, protein kinases, small G proteins, small G protein activators (GAPs), small G protein exchange factors (GEFs), and many other cDNAs that represent a broad cross-section of different protein functional classes. The table shows the effect of the various cDNAs on yeast cell growth when expressed from the GAL1 promoter either from a high-copy plasmid (2μ) or from a chromosomally integrated copy (Int).
Table 2.
An Average of 30% of the Heterologous Genes Tested in This Set Cause Growth Interference in Yeast When Expressed from a Chromosomal Locus
| cDNA | Species | Protein family | % Interference | Valid. | |
|---|---|---|---|---|---|
| 2μ | Int | ||||
| RCC1 | Human | Guanine Nucleotide Exchange Factor | ND | 95% | R |
| APS | Murine | Insulin Receptor Substrate | 42% | 3% | |
| IRS-1 | Human | Insulin Receptor Substrate | 70% | ND | |
| NR1H4 | Human | Nuclear receptor | 30% | −2% | |
| LXR alpha | Human | Nuclear receptor | 30% | −1% | |
| NR2E3 | Human | Nuclear receptor | 30% | −4% | |
| RORC | Human | Nuclear receptor | 33% | ND | |
| NR5A2 (CPF) | Human | Nuclear receptor | 91% | 98% | R |
| CAR | Human | Nuclear receptor | 61% | −2% | |
| Estrogen Receptor 1 | Human | Nuclear receptor (LBD Only) | 54% | −3% | |
| NAK1 | Human | Nuclear receptor (LBD Only) | 2% | −1% | |
| ROR-beta | Human | Nuclear receptor (LBD Only) | 3% | 3% | |
| RORalpha1 | Human | Nuclear receptor (LBD Only) | 33% | −2% | |
| Estrogen Receptor beta | Human | Nuclear receptor (LBD Only) | 43% | 1% | |
| PARP-1 | Human | Poly (ADP-ribose) polymerase | 92% | 97% | A, C |
| Rhino2A | Viral | Protease (Viral) | 94% | 80% | A |
| HIV Protease-LacZ | Viral | Protease (Viral) | ND | 97% | A, C |
| RGS2 | Human | RGS (GAP for G alpha) | 70% | 3% | |
| RGS4 | Human | RGS (GAP for G alpha) | 49% | 14% | |
| RGS5 | Human | RGS (GAP for G alpha) | 42% | ND | |
| PTEN | Human | Lipid and Tyrosine Phosphatase | −2% | 1% | |
| PI3_Kinase | Bovine | Lipid Kinase | 81% | 90% | A, C |
| PKC_lambda | Murine | Ser/Thr Kinase | 28% | 2% | |
| PKB_alpha | Human | Ser/Thr Kinase | 42% | −2% | |
| ARK1 | Human | Ser/Thr Kinase (Aurora-related) | ND | 98% | R |
| p38 | Human | Ser/Thr Kinase (MAP Kinase) | 70% | 70% | A, C |
| HGK | Human | Ser/Thr Kinase (MAP4K4) | ND | 42% | |
| PDK1 | Murine | Ser/Thr Kinase (Phosphoinositide-dependent) | 52% | 5% | |
| BUB1 | Human | Ser/Thr Kinase (Spindle Checkpoint) | ND | 80% | A* |
| Kinase in DiGeorge Syndrome Critical Region | Human | Ser/Thr Kinase (tssk-2 homolog) | ND | 76% | R |
| INSR | Human | Tyrosine Kinase (Receptor) | 58% | −2% | |
| CAP/Ponsin-1 | Murine | SH3-containing/Vinculin binding protein | 44% | 2% | |
| RhoA | Human | Small GTPase | ND | 58% | R |
| RhoC | Human | Small GTPase | ND | 80% | R |
| Snapin | Murine | SNARE-associated Protein | 0% | 1% | |
| VAMP2 | Murine | Synaptobrevin | −2% | 7% | |
| SNAP23 | Murine | Synaptosomal Associated Protein (t-SNARE) | 56% | −3% | |
| Syntaxin4 | Murine | Syntaxin/t-SNARE | 37% | 2% | |
| Munc18c | Murine | Syntaxin Binding Protein | 25% | −3% | |
| STAT4 | Human | Transcription factor | 69% | 5% | |
| STAT6 | Human | Transcription factor | 30% | −1% | |
High-throughput liquid growth assays were used to analyze the cDNAs presented in this table. The species and the protein family of the various cDNAs are listed, and whether expression was from a high copy plasmid (2μ) or from a chromosomally integrated copy (Int). Cells expressing either high copy or integrated versions of the cDNA were grown in glucose (uninduced) and in the presence of increasing concentrations of galactose (induced) and the percentage growth interference was calculated as compared to vector with no heterologous cDNA. Those strains that had a chromosomally integrated copy of the cDNA and showed a noticeable growth interference, which we defined as 55% or greater, are bolded. Of the 38 integrated cDNAs, there were 12 that produced a 55% or greater interference phenotype when expressed from a chromosomal locus, or 32% of the total. ND indicates those cases in which growth interference was not determined. We did further experiments to show that the growth interference was due to the inherent activity of the gene product. This was accomplished by showing that an active site mutation (A) or a known small molecule inhibitor (C) relieved the growth interference phenotype (column labeled Valid.). In other cases, we used random mutagenesis (R) to show that missense mutations that eliminated the growth interference phenotype mapped to recognizable functional domains.
For Bub1, the interference phenotype was reduced, but not eliminated, by the active site mutation (J. Couto, unpubl.).
Twelve of the 38 chromosomally integrated cDNAs showed interference of 55% or greater when expressed from a chromosomal locus (see bolded values in Table 2). This translates to about a 30% yield of strains with phenotypes that are robust enough to screen for small molecule inhibitors using a library of compounds. Particular functional classes of proteins behave differently with respect to the likelihood of producing interference phenotypes in yeast; for example, protein kinases had a success rate of 50% as opposed to 9% for nuclear receptors (see Table 2).
Those strains carrying cDNAs causing ∼55% or greater growth interference at full induction (2% galactose) when integrated into the chromosome were developed further for screening purposes (Perkins et al. 2001). This was achieved by varying the inoculum and time of growth to determine the optimal assay parameters for screening against existing or novel small molecule inhibitors of the growth interference phenotype. A signal-to-noise ratio was calculated for each assay that compared the growth of yeast expressing a heterologous cDNA with that of yeast containing empty vector (see Methods). The calculation of this value for growth in liquid media is a highly sensitive metric for the detection of growth changes that might be dismissed as insignificant in a conventional agar plate assay.
Of the 12 cDNAs developed for screening, 6 were amenable to validation by making active site mutations or using existing inhibitors (designated as A or C in the last column of Table 2 for Active site mutation or Compound, respectively). For the six other genes, a novel combination selection/screen was used to correlate the growth interference phenotype with the structure/activity of the protein (indicated in the last column of Table 2 as R for Random mutagenesis, and detailed in a later section of the Results). For those cDNAs in which an active site mutation was engineered or yeast expressing the cDNA was treated with a known inhibitor, growth was restored to nearly wild-type levels, with the exception of BUB1, which showed partial growth restoration (see A* in Table 2 for BUB1). In cases in which the cDNAs were mutagenized randomly, the missense mutations that relieved the growth interference phenotype were in recognizable functional domains.
The Interference Phenotype Caused by Expression of the MAP Kinase, p38, is Reversed by an Active-Site Mutation
As proof-of-principle examples to show that the normal biochemical activity of a protein can result in a growth interference phenotype in yeast, we developed assays for proteins of different functional classes whose active sites were well-defined and for which known inhibitors existed. As shown in Figure 3A, yeast with p38 carried on a 2μ plasmid showed little growth when yeast were plated on galactose (to induce p38 expression). As a control, a mutation in the active site (K53R) was introduced, and this mutation was able to restore growth, as shown in Figure 3A (Hanks et al. 1988). The bottom two plates of Figure 3A show controls that confirm the growth interference is plasmid-linked and that the particular clone maintains the ability to grow on galactose in the absence of p38. Thus, the kinase activity is necessary for growth interference in yeast.
Figure 3.
The growth interference phenotype caused by p38 expression can be validated by active site mutations or using established p38 inhibitors. (A) A mutation known to abolish p38 kinase activity (K53R) was engineered into the active site of p38, and the allele was tested for its ability to cause growth interference in yeast. (Upper right plate) Wild-type p38 expressed from a 2μ plasmid does not allow growth when yeast are grown on SC-Leu 2% galactose. However, growth occurs when a p38 cDNA carrying an active site mutation was tested under the same conditions (see Key to Strains in text). Subsequent reversion of the mutation using PCR resulted in no measurable growth on SC-Leu 2% galactose plates, thus confirming that no unintended mutation occurred while introducing the active site mutation. (Bottom left plate [control]) Lack of selective pressure to maintain the plasmid (SC-Trp 2% galactose) allows all strains to grow because of plasmid loss; (bottom right plate [control]) in the presence of selection for the 2μ plasmid, but under growth conditions in which the cDNA is not induced (SC-Leu 2% glucose), no growth inhibition is observed. (B) Cells expressing p38 were exposed to varying concentrations of the inhibitors PD169316 or SB203580 and the percentage of growth restoration was analyzed. The EC50 for PD169316 is 0.16 μM ± 0.01 and for SB203580 is 4.6 μM ± 0.44.
We have obtained similar results for poly(ADP-ribose) polymerase (PARP-1; Perkins et al. 2001), PI 3-kinase (E. Perkins and A. Nguyen, unpubl.), HIV Protease (as a β-galactosidase fusion; (E. Yeh, unpubl.), and Rhino2A (Klump et al. 1996; J. Couto, unpubl.). Expression of these proteins caused growth interference in yeast that was relieved by active-site mutations and/or known small molecule inhibitors (Table 2, A and/or C).
In the case of PARP-1 and PI 3-kinase, coexpression of the regulatory mammalian enzymes poly(ADP-ribose) glycohydrolase (Lin et al. 1997) or PTEN (Di Cristofano and Pandolfi 2000), respectively, which are enzymes that remove the protein modification products of PARP-1 or dephosphorylate the product of PI 3-kinase, also restores growth to yeast expressing PARP-1 (Perkins et al. 2001) or PI 3-kinase (A. Nguyen, unpubl.). These experiments provide further evidence that these proteins produce their phenotypes in yeast by virtue of their biochemical activities.
Known Inhibitors of p38 Restore Growth to Yeast
To further confirm that the growth inhibition observed in the yeast assays is the result of the biochemical activity of the expressed protein, existing inhibitors of these proteins were tested for their ability to restore growth in the yeast assay. As described above, p38 caused growth interference when it was expressed from a high-copy plasmid. However, the observed growth interference was reduced greatly when p38 was expressed as an integrated copy, likely because of lower expression. To overcome this problem, we took advantage of the observation in previous studies that the toxicity caused by overexpression of the yeast HOG1 gene (the yeast counterpart of the mammalian p38 gene), was enhanced when the yeast carried mutations in the two phosphatase genes, PTP2 and PTP3 (Wurgler-Murphy et al. 1997). As expected, deleting PTP2 and PTP3 in yeast expressing an integrated copy of the human p38 gene raised the growth interference phenotype to a level that made testing the known inhibitors possible.
In Figure 3B, yeast carrying either a chromosomal copy of human p38 or no human cDNA were exposed to varying concentrations of two known p38 inhibitors. Growth was restored to yeast expressing p38 when they were exposed to increasing concentrations of the two inhibitors PD169316 and SB 203580 (Cuenda et al. 1995); the EC50 value for PD169316 was 0.16 μM ± 0.01, whereas that for SB203580 was 4.6 μM ± 0.44 (S. Thode, unpubl.; see Methods). Similar results were obtained using the known inhibitors for a number of other cDNAs that caused growth interference: PARP-1 by 6(5H) phenanthridinone (Perkins et al. 2001), PI 3-kinase by LY294002 (D. Sun, unpubl.), and HIV Protease (as a β-galactosidase fusion protein) by Saquinavir (D. Sun, unpubl.).
These data confirm that small molecule inhibitors against heterologous proteins function in the yeast cell-based assays. They also show that the growth interference phenotype in yeast is caused by the normal biochemical activity of these proteins, as the phenotype is abolished when their activity is inhibited, either by active-site mutations or by small molecule inhibitors.
A Novel Random Mutagenesis Scheme Allows the Selection/Screen for Mutations that Retain Full-Length Expression and Restore Growth to Yeast
Apart from the well-defined proof-of-principle targets discussed in the section above, growth interference was observed when several less well-characterized cDNAs were expressed in yeast (Table 2). A novel method was developed to map the important regions of the cDNA that contribute to the yeast phenotype (Fig. 4A). In this method, GFP is fused in-frame to the C terminus of the target cDNA that is carried on a low-copy (CEN) plasmid, thereby providing a fluorescent signal that allows us to monitor expression of the full-length fusion protein. This method requires that expression of the fusion protein still causes growth interference, which we found is generally the case.
Figure 4.
Theory and application of a novel tool that combines random mutagenesis with the ability to select mutations required for activity and then screen for those that maintain full-length expression. (A) A yeast strain expressing a heterologous cDNA fused in-frame to GFP at the C terminus is able to grow under conditions when gene expression is not induced (glucose), but cannot grow when gene expression is induced (galactose); i.e., the tagged cDNA still confers the growth interference phenotype. The plasmid is then mutagenized in vitro, and a selection for colonies that grow is followed by a screen for those colonies that fluoresce (full-length expression; growing and green). Thus, the tool can map the residues (or domain) of the expressed protein that are necessary for the growth interference phenotype. (B) The mutagenesis tool is applied to a nuclear receptor, CPF, that causes growth interference when expressed in yeast. The total number of colonies of a strain bearing CPF on SC-His glucose plates (CPF expression not induced) shows that, over time, the plasmid is mutated to such a degree that it no longer supports transformation ability or propagation in yeast. However, at shorter exposures to the mutagen, there are a large number of viable colonies. The mutants were chosen at those times when there were viable colonies. (C) Further analysis of the colonies shows that at early times of exposure to mutagen, colonies can be identified that are able to grow and that are fluorescent when CPF is induced (SC-His galactose plates). Sequencing of these mutants reveals that these colonies contain alleles of CPF carrying mutations that restore growth to yeast by mutation of CPF's DNA-binding domain.
The GFP-tagged constructs were mutagenized chemically in vitro and then transformed into yeast under inducing conditions. Those carrying mutations that destroyed the growth interference phenotype resulted in two types of colonies: green fluorescent colonies (glowing) arising from missense mutations in the heterologous cDNA, and white colonies (nonglowing) resulting from stop codons (nonsense mutations). The glowing colonies were visualized on plates using a light source and filters as described by Cronin and Hampton (1999).
Using this technique, we mapped the domains of several different proteins (indicated in Table 2, last column [Valid.], as R; S. Tugendreich, unpubl.), including the nuclear receptor CPF (CYP7A promoter binding factor [Nitta et al. 1999]). Figure 4B shows that as the CPF-GFP fusion construct was subjected to increasing lengths of exposure to the mutagen, the total number of colonies growing on non-cDNA-inducing conditions (glucose) declines such that by 30 min few colonies were growing. We attribute the sharp decrease in colony number to the mutation of the auxotrophic markers and other key elements of the plasmid required for efficient transformation and selection. Conversely, Figure 4C shows that on cDNA-inducing media, the number of colonies increases on exposure to mutagen at early time points (5–25 min). This is the result of mutations that destroy the interference activity encoded by the CPF ORF. The colonies that are glowing are shown in Figure 4C to be a subset of the total colonies appearing on the inducing plates.
To determine the nature of the mutations, the plasmids in the glowing and nonglowing colonies were recovered into E. coli, the inserts were sequenced, and the mutations were mapped onto the full-length CPF protein. The mutant plasmids were also reintroduced into yeast to confirm the absence of growth interference and the gain of green fluorescence. As discussed below, the “glowing growers” have missense mutations in the CPF ORF, whereas the nonglowing colonies have stop codons in CPF.
In Figure 5A the domain structure for CPF is depicted. Many of the chemically induced single base pair mutations obtained map to the key cysteines that form the zinc finger DNA-binding domain (Fig. 5B). Thus, the growth interference observed in yeast from expression of CPF can be reversed by destruction of its DNA-binding function. We speculate that CPF causes growth interference in yeast by binding inappropriately to promoters of yeast genes essential for cell viability and somehow disrupting their transcription.
Figure 5.
Analysis of the mutants that “grow and glow” shows that they are expressing full-length alleles of the human CPF gene that contain mutations in the DNA-binding domain. (A) Schematic of the domain structure of the nuclear receptor, CPF. CPF contains a DNA-binding domain and a ligand-binding domain. The Cs indicate the location of the zinc fingers within the DNA-binding domain. (B) The selected mutations are mapped to the gene sequence and indicate that most of the mutations are in the zinc fingers. (Inset) Alleles of CPF carrying stop codons result in white colonies, whereas the fluorescent colonies encode missense mutations in the CPF ORF.
Further analysis indicates that the glowing CPF mutants are expressed stably as full-length proteins of the correct size as detected by immunoblotting with anti-GFP antibodies. As predicted, loss-of-function mutants deriving from stop codons (that result in truncation before the GFP moiety) are not visualized on the immunoblot (Fig. 6A, lanes 49 and 51). Importantly, the unmutagenized fusion protein is localized properly in the nucleus in yeast (Fig. 6B), as one would expect for a DNA-binding protein.
We also used this technique to investigate RCC1, which is a guanine nucleotide exchange factor for the small GTPase Ran (Gorlich et al. 1996; Carazo-Salas et al. 1999; Nemergut and Macara 2000). RCC1 has a symmetrical seven-bladed propeller structure (Fig. 7B; Renault et al. 1998), in which only one face interacts with Ran (Azuma et al. 1999). Interestingly, the mutations that reverse the growth interference in yeast depicted in Figure 7A map to the face of RCC1 known to interact with Ran (Fig. 7B). These data indicate that the RCC1 phenotype is due to appropriate interactions between the mammalian protein and endogenous yeast protein(s). Furthermore, the mutants were tested for nucleotide exchange activity and all showed reduced exchange when compared with wild-type RCC1 (M. Hetzer and I. Mattaj, pers. comm.). The one mutation that mapped away from the Ran-binding face (G364D) appears to result in instability of the protein as assessed by immunoblot analysis (S. Tang, unpubl.).
Figure 7.
Application of the random mutagenesis tool to yeast expressing the human guanine nucleotide exchange factor RCC1 identifies mutations in the seven-bladed propeller. (A) Shows the seven-bladed propeller sequence of the guanine nucleotide exchange factor RCC1 and maps the mutations to B1 (A), B2, B6, B7, and B1 (B). The mutation circled in the B7 region is one that destabilizes the RCC1 protein. (B) Crystal structure of RCC1 (Renault et al. 1998) from two different angles; the residues shown by space filling molecules indicate the location of the mutations that eliminate the growth interference phenotype caused by RCC1 expression. All the mutations, with the exception of one (see rightmost structure), map to a single surface of the protein. The exception is G364D, which we have found to destabilize the protein. The other mutations line the face of RCC1 that interacts with the small G protein Ran (Azuma et al. 1999).
DISCUSSION
Many of the new genes identified as a result of sequencing the human genome belong to established gene families with a successful history as drug targets (e.g., kinases, phosphatases, and GPCRs). Thus, there is a pressing need to establish a high-throughput and rapid means for assay development to enable screening for small molecule inhibitors of these new family members. Indeed, the kinase family alone has approximately 600 members (Venter et al. 2001). To meet this demand, we have developed a scheme that allows the uniform and parallel processing of mammalian cDNAs to obtain HTS compatible yeast cell-based assays.
Our ability to use the in vivo biochemical activity of mammalian or viral proteins to generate a generic growth interference assay in yeast whose reversal allows the identification of small molecule inhibitors is highlighted in this paper. Although the use of growth interference for this purpose has been published in the past, such a process was applied to a few specific targets on a “case by case” basis (Kurtz et al. 1995). These low-throughput studies were performed in many different genetic backgrounds and were not optimized for responsiveness to small molecule inhibitors, or for expression levels. To adapt yeast to high-throughput screening, several issues had to be addressed: eliminating uncontrolled variations in the genetic background between strains in different screens, decreasing compound efflux, obtaining high-level expression of foreign proteins, stabilizing plasmids carrying the expressed gene, and correlating phenotype with the activity of the expressed protein. We have addressed these issues and developed a systematic and homogenous set of assays such that the inhibitors identified during the screening of each assay are cataloged and compared with prior screening results to determine selectivity. Importantly, as a screening tool, growth interference assays have an advantage over complementation assays in that hits restore growth, and therefore toxic compounds don't act as false positives.
Furthermore, to make use of the abundance of full-length cDNAs and partial Expressed Sequence Tags available in nonyeast host vectors (such as those in clone repositories like at Incyte Genomics: http://www.incyte.com), we developed a rapid means to transfer them into yeast expression vectors. This procedure, referred to as GRIPP recombinational cloning, is accomplished without PCR amplification of the ORFs. Instead, the ORF-containing insert is released from its host vector by restriction digestion and cotransformed into yeast with a PCR-amplified yeast vector. The vector has been modified during the amplification to contain short tails of sequence identical to the ends of the heterologous ORF. Interestingly, we have found that the restriction digest is not absolutely required, but rather improves the efficiency of obtaining correct recombinants (S. Tugendreich, unpubl.). The GRIPP method is a highly scalable and parallel process because the same vector is PCR-amplified for each ORF. That is, the PCR conditions are uniform and result in one product size, that of the template vector. An E. coli variation of this process was published after completion of our work in yeast (Zhang et al. 2000).
The analysis of growth interference phenotypes elicited by the expression of heterologous cDNAs in yeast resulted in an average net success rate of about 30%. Notably, representatives of diverse classes of proteins yielded screening-competent phenotypes in yeast. For those cDNAs whose expression as an integrated copy did not result in measurable growth interference in the yeast cell-based assay, we found that, in some cases, engineering specific mutations into the yeast genetic background exacerbated the phenotype caused by expression of the heterologous cDNA. For example, although p38 expression in wild-type yeast causes a growth interference of ∼70% when expressed from a high-copy episome, it is almost negligible when expressed from a genomically integrated copy in the wild-type background. However, we found that deletion of PTP2 and PTP3 (Wurgler-Murphy et al. 1997) sensitized the yeast strain to the lower expression of p38. These results indicate that it might be possible to generally increase the success rate at which cDNAs cause interference phenotypes in yeast by manipulating the yeast genetic background (Guarente 1993). These types of synthetic lethal or synthetic dosage lethality effects have been observed previously to enhance the severity of phenotypes from overexpression of yeast genes (Kroll et al. 1996), and here we observe a similar synergistic effect for a heterologous gene. The entire collection of yeast nonessential genes has been deleted and “bar-coded” individually, providing a means to assay synthetic dosage lethality across the entire yeast genome in parallel (Winzeler et al. 1999).
For those cDNAs that we scored as positive for growth interference in yeast, we showed that the phenotype was due to the activity of the expressed gene product by analyzing the effect of mutations and/or known small molecule inhibitors. In several cases we used a random mutagenesis scheme as a proteomic tool both to correlate the protein's structure with its activity in yeast and to help validate the assay as a screen for small molecule inhibitors. In all cases the missense mutations mapped to recognizable functional domains of the proteins (S. Tugendreich, unpubl.).
Although random mutagenesis methods have been cited in the literature before the method reported here, we are not aware of any that simultaneously correlate the structure/activity of the protein with a cell-based phenotype and provide a method to screen for only those mutations that retain expression of the full-length gene (Belfort and Pedersen-Lane 1984; Patterson and Spudich 1995; Harms et al. 1999; Blaesing et al. 2000; Lanio et al. 2000).
Our findings confirm the potential use of yeast cell-based assays to characterize new genes and screen for small molecule inhibitors of genes involved in key cellular pathways. The assay development process described here is a valuable means to screen potential therapeutic targets homogeneously, rapidly, and on a genomic scale.
METHODS
Batch PCR Primer Design
A PERL script to design PCR primers automatically from text files containing an indefinite number of FASTA-formatted sequences is an important element of our process. The program's parameters were as follows: For fixed primers (primers that must start either with the ATG or the stop codons) the program started with a 19-mer and extended it in 1-nt increments until the predicted Tm (Tm = 81.5 + 0.41[%GC] − 675/N − %mismatch [N = primer length]) was at least 60°C. The program also required that the 3′ end be G or C but not a row of five or more G/Cs. For nonfixed primers, the program scanned in an inchworm-like fashion for primers with lengths between 20 and 25 nt or as specified. These primers also followed the requirements listed above for fixed primers.
Depending on whether our source material was genomic cDNA or a previous plasmid construct, we either PCR amplified the coding region or the entire expression vector using a method referred to as GRIPP (patent pending). In the first case, our PERL script designed two primers that yielded the following PCR product: tgaagcaagcctcctgaaagATG … (CDS) … TAAgagatctatgaatcgtagatac (TAA could be replaced by TGA or TAG depending on the gene).
This product was then adapted for recombination by extending the tails to at least 45 nt using universal primer adapters or “extendamers”, such that the final product ready for recombinational insertion was as follows: atcaacaaaaaattgttaatatacc tctatactttaacgtcaaggtgaagcaagcc tcctgaaagATG … (CDS) … TAAgag atctatgaatcgtagatactgaaaaaccccgc aagttcacttc. The program was also used to design two other primers within the coding region for construct confirmation purposes.
For inverse PCR (GRIPP), the program was used to design primers that polymerized the entire expression plasmid. The primers included a vector-specific region (shown in lower-case letters below) as well as 45-nt tails that were homologous to the beginning and the end of the ORF such that the PCR product was as follows: CDS(45 nt)TAAgagatcta tgaatcgtagatactg … (vector) … gt taatatacctctatactttaacgtcaagga atataATG-CDS(45 nt). Similar primer batch design scripts were used for planning protein fusions.
Primers for GFP tagging were designed automatically as follows: The 5′ primer was a chimeric primer starting with the last 42 nt of the ORF (excluding the stop codon) and ending with the first 24 nt of GFP (atgagtaaaggagaagaactt ttc). The 3′ primer (gatgtataaatgaa agaaattgagatggtgcacgatgcaca gttgtggatggcggcgttagtatcg) anneals to the KanMX marker and has a 45-nt tail to recombine with the GAL4 terminator of the pARC yeast expression vector (see below).
Construction of the pARC25B Vector
A brief summary will be presented in lieu of a detailed description of the steps used to construct the pARC series of vectors. The vectors are derived from the pRS series of vectors (Sikorski and Hieter 1989; Christianson et al. 1992). The basic elements of the pARC plasmids include a 713-bp SphI/BamHI fragment containing the S. cerevisiae GAL1 promoter and a 243-bp BglII/HindIII fragment containing the GAL4 terminator region. This region surrounds a 45-bp polylinker that contains unique sites for PstI, SalI, SpeI, XhoI, and AvaI and replaces the pRS polylinker: The base vector and polylinker of the pRS vectors is pBluescript II. Both CEN- and 2μ-based vectors were generated with 44 bp of the 5′ end of the LYS2 gene (bp 8 –52 of the LYS2 ORF) and 43 bp of the 3′ end of the LYS2 gene (bp 4133–4176 of the LYS2 ORF) flanking the CEN-ARSH4 element or the 2μ replication origin. In addition, SfiI sites flank the CEN-ARSH4 or 2μ elements such that digesting with this restriction enzyme liberates the elements and allows the subsequent integration into the endogenous LYS2 locus via a γ deletion mechanism (Sikorski and Hieter 1989). For this work, integrants were selected for α-aminoadipate resistance and by marker prototrophy, that is, Leu+ or Ura+. Integration was confirmed by PCR. The sequence for the 2μ version of the vector (pARC25B), which is the only one actually used in this work, has been deposited into GenBank with accession number AF359244.
GRIPP Recombination
Approximately 10 ng of vector was PCR amplified with the Bio-X-Act polymerase mixture (Bioline) as per the manufacturer's instructions. After an initial denaturing step of 2 min at 94°C, a cycle of 20 sec at 94°C, 35 sec at 55°C, and 10 min at 68 °C was repeated 32 times. Recombination was performed by first mixing ∼100–300 ng of vector PCR product (5 μL of the reaction) with ∼500–1000 ng of digested insert/host vector (10 μL of a 15 μL digest). This mixture was then transformed into competent EIS20-2B yeast by the standard LiAc procedure (Ito et al. 1983), followed by growth on SC-Leu plates containing 2% glucose. The genotype of the W303-derived EIS-20-2B is MATa, ade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 pdr5Δ snq2Δ. Typically about 100 colonies appeared on transformation with the PCR amplified vector alone (presumably because of reclosure of the PCR-amplified vector and carryover from the intact vector template), whereas transformation with the mixture of the digested insert and the PCR-amplified vector resulted in ∼500–5000 colonies.
Cloning from cDNA Libraries
ORFs were cloned with PCR using automatically designed primers and “extendamers” as described above. Reactions were performed in an MJ-research PTC-200 thermocycler, in 25 μL containing 600 nM extendamer primers, 6 nM ORF-specific primers, 200 μM each dNTP, and 0.1 units/μL Taq-plus precision polymerase (Stratagene) under the buffer conditions recommended by the supplier. The normal cycling conditions were as follows: (2 min/94°C), 23×(30 sec/94°C, 30 sec/59°C, 1 min/72°C), (10 min/72°C), (4°C) with the exception of the extension step at 72°C that varied depending on the length of the template (∼1 min/1000 nt). Long ORFs were often polymerized in two overlapping segments. PCR products were added directly to gapped expression vectors for yeast transformation. Recombination into the vector was highly efficient as described by others (Raymond et al. 1999), either as a double recombination for single PCR products or triple recombination for overlapping PCR products.
Whole-Cell PCR to Check Recombinants
Yeast recombinants were analyzed by transferring colonies into 100 μL of SC-Leu 2% glucose in 96-well dishes and allowing the cells to grow to saturation overnight at 30°C without shaking. Twenty-five microliters of this culture was pipeted into 100 μL of water in a 48-well PCR reaction plate (MJ Research) and centrifuged in a swinging platform centrifuge at 2000 rpm for 5 min. The supernatant was then removed by a vigorous shake of the hand and blotted inverted on a paper towel. Twenty-five microliters of Bio-X-Act PCR cocktail containing the primers IPI_178 (GTTAAAGTGGTTATGCAGCTTTTCC) and IPI_179 (CGATGCACAGTTGAAGTGAACTT GCG), which flank the site of the insertion, was added to the cells. The thermocycling program was as for GRIPP (see above) except the extension time was 2.5 min and was performed for 40 cycles total.
Plasmid Rescue into E. coli
To rescue insert-containing recombinant plasmids from yeast into E. coli for amplification, the yeast was patched on one sixth of an SC-Leu 2% glucose plate and grown 24–48 h at 30°C and scraped into 1 mL of water. The cells were centrifuged briefly, then washed again. The cell pellet was then resuspended in 100 μL of buffer containing 2% Triton X-100, 1% SDS, 100 mM NaCl, 10 mM Tris-HCl (pH 8), 1 mM EDTA. One hundred microliters of 425- to 600-μm glass beads (Sigma G-8772) and 100 μL of phenol:chloroform was added, and the mixture was vortexed for 10 min on an Eppendorf vortexer and centrifuged for 5 min at 15,000 rpm in a microfuge. Approximately 40 μL of the supernatant was then removed to a fresh tube, and 10 μL of this supernatant was float dialyzed against 10% glycerol for 20 min on either Millipore VS filter discs (cat#: VSWP 025 00, pore size: 0.025 μm) or 96-well Multiscreen-VM plates (Millipore, cat# MAVMN0510). Three to five microliters of the dialysate (containing the yeast DNA) was then electroporated into 50–75 μL of DH5α electrocompetent cells by standard methods and selected on LB ampicillin plates.
Liquid Growth Assays
Individual transformants and controls transformed with vector alone were inoculated into 200 μL of selective medium (SC-Leu 2% glucose) in 96-well microtiter dishes (Greiner LaborTechnik #655198/plates and #656197/lids). They were allowed to grow to saturation overnight at 30°C. The cultures were then diluted 1:400 in triplicate into new 96-well microtiter dishes containing 100 μL of four different types of SC-Leu media containing increasing amounts of galactose inducer (2% glucose, 2% raffinose + 0.02% galactose, 2% raffinose + 0.2% galactose, and 2% galactose) using a Beckman Multimek 96 Automated 96-channel pipetter. Plates were incubated at 30°C wrapped in plastic to prevent edge effects caused by evaporation. Growth was monitored at approximately 24 h, 36 h, and 48 h, although the ideal time depended on each particular strain and vector type. At each of these time points plates were shaken vigorously for at least 1 min using a titer plate shaker (LAB-LINE Instruments) at speed setting 8, to disperse the cells uniformly. They were then analyzed immediately in a Molecular Devices Spectra Max 340 plate reader set to measure absorbance at 600 nm. Results were shown as percent inhibition relative to the vector control provided that the vector control itself grew to an OD600 of at least 0.1. The % inhibition = 100 (1 − SICu/CISu) where SI = sample induced, Su = sample uninduced, CI = control induced, and Cu = control uninduced.
Determining the Robustness and Sensitivity of the Yeast Cell-Based Assay for Testing of Small Molecule Inhibitors
Those yeast strains expressing cDNAs that generated an interference phenotype of 55% or greater (using the semiautomated liquid microtiter assay; mentioned above) were optimized further for screening of small molecule inhibitors. This was accomplished by varying the starting inoculum and growth end point to produce a signal-to-noise ratio of 3.0 or higher. Achieving such a ratio was an indication that the phenotype was qualified to screen with small molecule inhibitors. Signal-to-noise is defined by the ratio of the final OD600 of yeast containing integrated vector control to yeast expressing the heterologous cDNA. For example, a signal-to-noise ratio of 3.0 translates to an OD value of 0.075 for yeast expressing the integrated cDNA and a value of 0.225 for those with the integrated vector control.
To determine whether known inhibitors of p38 were able to restore growth to yeast expressing p38, 5 mL of cells was grown overnight at 30°C in SC-Leu with 2% glucose (p38 not induced) in a cell rotator. Cells were centrifuged, washed in SC-Leu medium containing no carbon source, and resuspended to a calculated OD600 of 0.0025 in SC-Leu with 2% galactose (p38 induced). The known p38 inhibitor PD169316 was tested from 0 to 4 μM and at 0 to 128 μM for SB203580. The inhibitors were diluted into a final volume of 100 μL of SC-Leu with 2% galactose, 2 mM HEPES buffer (pH 7.2), and 1% DMSO. Under these conditions the signal-to-noise ratio was 4.8 at 48 h. The effect of the compounds was measured as percent of growth restoration using the following equation: percent growth restoration = (TEST − MEDarc)/(MEDvec − MEDarc) × 100, where TEST is OD600 of the well with test compound, MEDarc is the median value of OD600 of the cells without compound, and MEDvec is the median value of OD600 of vector-containing cells. The EC50 values were calculated using KaleidaGraph software to fit the data to a logistic equation using a four-parameter fit.
Hydroxylamine Mutagenesis
Four micrograms of CEN-plasmid DNA carrying both the HIS3 auxotrophic marker and a GAL1 promoter-driven fusion of the foreign heterologous cDNA fused in-frame to GFP (see below) was incubated in 100 μL of a freshly prepared solution of 1 M hydroxylamine (Sigma), 50 mM sodium pyrophosphate (pH 7.0), 100 mM NaCl, and 2 mM EDTA at 75°C. At various intervals (see Results), 20-μL aliquots were moved to fresh tubes on ice and then float dialyzed for 30 min versus TE (see above). The dialyzed lysate was then transformed into EIS20-2B yeast by the standard LiAc procedure, split into two aliquots, and plated to SC-His plates with either 2% glucose or a mixture of 2% galactose and 0.005% glucose. The glucose plate was used to count total colonies, whereas the galactose plate (containing trace amounts of glucose) was used to select for mutations that destroyed the toxic effect of the expressed human cDNA. The trace glucose was added because we found that it increased the transformation efficiency on galactose plates, possibly by helping the cells convert to galactose use. Green colonies were identified by visual inspection using a slide projector light source and filter set as described by Cronin and Hampton (1999).
Analysis by Immunoblot for GFP Fusion Proteins
To monitor the expression of the heterologous cDNAs in yeast, strains carrying the integrated cDNA constructs were tagged initially at the 3′ end of each ORF using the GFP (S65T)-KanMX (which confers G418 resistance) construct as described previously (Longtine et al. 1998). Four G418-resistant transformants were streaked to 2% galactose plates to induce expression of the GFP fusion protein and were checked for fluorescence as described previously (Cronin and Hampton 1999). In some cases a fluorescent signal was not observed, possibly because of low expression level and/or toxicity of the fusion protein. In all cases we confirmed the tagging event by whole-cell PCR amplification using primers that anneal to the noncoding strand of the GFP (gcatcaccttcaccc tctccactg) ORF and a cDNA-specific primer approximately 500 bp from the 3′ end of the cDNAs. Transformants showing the predicted-size PCR product were selected for immunoblot analysis.
Initially, yeast cells were grown under conditions that no longer repressed transcription from the GAL1 promoter (SC-Leu medium containing 3%glycerol, 2% ethanol, 0.2% glucose to approximately 2 × 106 to 5 × 106 cells/mL). Cells were washed one time in dH2O and resuspended in an equal volume of 2% galactose medium to induce the expression of an in-frame carboxy-terminal fusion protein of a heterologous gene with GFP. Induction was allowed to proceed for approximately 6–8 h. At the end of the induction period, cells were harvested by centrifugation and washed once in dH2O. The resulting pellets were quick-frozen at −20°C in an ethanol/dry ice bath and stored at −80°C for later processing.
The samples were first thawed at 4°C and quickly resuspended in 10 μL of protein lysis buffer (100 mM sodium phosphate at pH 7.0, 1 mM EDTA [in 1× protease inhibitor solution], 0.1% SDS, and a mix of 1× protease inhibitors [Complete Protease Inhibitor Cocktail tablets, Boehringer Mannheim]). Protein concentrations of the resulting cell extracts were determined with the Bio-Rad Protein Assay Kit. In cases in which different samples were analyzed on the same immunoblot, they were normalized by adding the same total number of cells per lane on the SDS–polyacrylamide gel.
SDS-PAGE was performed using the mini-PROTEAN system and Ready Cast Gels (Bio-Rad) according to the manufacturer's specifications. Electrotransfer of proteins from the polyacrylamide gels to Immobilon PVDF membranes (Millipore) was performed using the Hoeffer SemiPhor TE70 transfer apparatus according to the manufacturer (Amersham Pharmacia Biotech). The Immobilon sheet was probed with anti-GFP antibodies to detect the fusion proteins. For detection, the Amersham Lumigen kit was used that contained a chemiluminescent substrate and horseradish peroxidase conjugated to goat anti-mouse secondary antibody.
Deletion of PTP2/PTP3
Overexpression of HOG1, the yeast homolog of p38, is lethal in yeast in the absence of the phosphatases PTP2 and PTP3. To enhance the growth interference phenotype observed with p38 expression we constructed a yeast strain with a ptp2 ptp3 background.
Deletion of the yeast PTP3 gene was accomplished using the oligos 5ptp53prs (ATGaaggacagtgtagactgcccaagcattc tacccaccgaccgcCAGAGCAGATTGTACTGAGAGTGCACC) and 3ptp33prs (CTAttgtggcaattctttcaacttatcatccacgaatgtct gcaaCGCATCTGTGCGGTATTTCACACCGC). These oligos were designed to PCR amplify the yeast URA3 gene from the plasmid pRS305 (Sikorski and Hieter 1989) followed by subsequent yeast transformation and selection for uracil prototrophs. Following purification of URA3-containing colonies, the Δptp3 allele was confirmed by whole-cell PCR amplification using the oligos Ptp3chk1 (GATCTACTTATCATATAGAA CATGAAGGACAGTG) and Ptp3chk2 (aaaatagagatcaaatac attcatattagcctaaC).
Using the strain carrying the Δptp3 mutation, deletion of the yeast PTP2 gene was constructed with the KanMX G418 resistance cassette as described previously (Wach et al. 1994) using the oligos 5ptp2kan (ATGgatcgcatagcacagcaa tatcgtaatggcaaaagagacaatCGTACGCTGCAGGTCGAC) and 3ptp2kan (CTAttaacaaggtaacgcgttctttatctgcttttgcagg gcaaaATCGATGAATTCGAGCTCG). After purification of G418 resistance colonies, the Δptp2 allele was confirmed by subsequent whole-cell PCR using the oligos Ptp2kanchk (Cga gatcactcaacctgacagacccg) and Kanfirm (Cctgtacataaccttcggg catggc) to identify the correct recombination junction.
Acknowledgments
We thank Ken Zaret, Phil Hieter, Kurt Jarnagin, and Keith Bostian for critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL tmelese@cc.ucsf.edu; FAX (415) 502-6779.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.191601.
REFERENCES
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Azuma Y, Renault L, Garcia-Ranea JA, Valencia A, Nishimoto T, Wittinghofer A. Model of the ran-RCC1 interaction using biochemical and docking experiments. J Mol Biol. 1999;289:1119–1130. doi: 10.1006/jmbi.1999.2820. [DOI] [PubMed] [Google Scholar]
- Becker DM, Fikes JD, Guarente L. A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc Natl Acad Sci. 1991;88:1968–1972. doi: 10.1073/pnas.88.5.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort M, Pedersen-Lane J. Genetic system for analyzing Escherichia coli thymidylate synthase. J Bact. 1984;160:371–378. doi: 10.1128/jb.160.1.371-378.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- Blaesing F, Weigel C, Welzeck M, Messer W. Analysis of the DNA-binding domain of Escherichia coli DnaA protein. Mol Microbiol. 2000;36:557–569. doi: 10.1046/j.1365-2958.2000.01881.x. [DOI] [PubMed] [Google Scholar]
- Botstein D, Fink GR. Yeast: An experimental organism for modern biology. Science. 1988;240:1439–1443. doi: 10.1126/science.3287619. [DOI] [PubMed] [Google Scholar]
- Broach JR, Thorner J. High-throughput screening for drug discovery. Nature. 1996;384:14–16. [PubMed] [Google Scholar]
- Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Cohen P. The development and therapeutic potential of protein kinase inhibitors. Curr Opin Chem Biol. 1999;3:459–465. doi: 10.1016/S1367-5931(99)80067-2. [DOI] [PubMed] [Google Scholar]
- Cronin S, Hampton RA. A genetics friendly GFP assay. Trends Cell Biol (technical tips) 1999;9:1:36. [Google Scholar]
- Cuenda A, Rouse J, Doza YN, Meier R, Cohen P, Gallagher TF, Young PR, Lee JC. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- Diehn M, Alizadeh AA, Brown PO. Examining the living genome in health and disease with DNA microarrays. JAMA. 2000;283:2298–2299. [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Greenhalf W, Stephan C, Chaudhuri B. Role of mitochondria and C-terminal membrane anchor of Bcl-2 in Bax induced growth arrest and mortality in Saccharomyces cerevisiae. FEBS Lett. 1996;380:169–175. doi: 10.1016/0014-5793(96)00044-0. [DOI] [PubMed] [Google Scholar]
- Greenhalf W, Lee J, Chaudhuri B. A selection system for human apoptosis inhibitors using yeast. Yeast. 1999;15:1307–1321. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1307::AID-YEA455>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Guarente L. Synthetic enhancement in gene interaction: A genetic tool come of age. Trends Genet. 1993;9:362–366. doi: 10.1016/0168-9525(93)90042-g. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Harms N, Oudhuis WC, Eppens EA, Valent QA, Koster M, Luirink J, Oudega B. Epitope tagging analysis of the outer membrane folding of the molecular usher FaeD involved in K88 fimbriae biosynthesis in Escherichia coli. J Mol Microbiol Biotechnol. 1999;1:319–325. [PubMed] [Google Scholar]
- Hertzberg RP. Whole cell assays in screening for biologically active substances. Curr Opin Biotechnol. 1993;4:80–84. doi: 10.1016/0958-1669(93)90037-w. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bact. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen T, Proft M, Randez-Gil F, Schultz JR, Marbois BN, Entian KD, Clarke CF. Yeast Clk-1 homologue (Coq7/Cat5) is a mitochondrial protein in coenzyme Q synthesis. J Biol Chem. 1998;273:3351–3357. doi: 10.1074/jbc.273.6.3351. [DOI] [PubMed] [Google Scholar]
- Kaur R, Bachhawat AK. The yeast multidrug resistance pump, Pdr5p, confers reduced drug resistance in erg mutants of Saccharomyces cerevisiae. Microbiology. 1999;145:809–818. doi: 10.1099/13500872-145-4-809. [DOI] [PubMed] [Google Scholar]
- Kirsch DR. Development of improved cell-based assays and screens in Saccharomyces through the combination of molecular and classical genetics. Curr Opin Biotechnol. 1993;4:543–552. doi: 10.1016/0958-1669(93)90075-8. [DOI] [PubMed] [Google Scholar]
- Klump H, Auer H, Liebig HD, Kuechler E, Skern T. Proteolytically active 2A proteinase of human rhinovirus 2 is toxic for Saccharomyces cerevisiae but does not cleave the homologues of eIF-4 γ in vivo or in vitro. Virology. 1996;220:109–118. doi: 10.1006/viro.1996.0291. [DOI] [PubMed] [Google Scholar]
- Kolaczkowski M, Kolaczowska A, Luczynski J, Witek S, Goffeau A. In vivo characterization of the drug resistance profile of the major ABC transporters and other components of the yeast pleiotropic drug resistance network. Microb Drug Resist. 1998;4:143–158. doi: 10.1089/mdr.1998.4.143. [DOI] [PubMed] [Google Scholar]
- Kowaluk EA, Jarvis MF. Therapeutic potential of adenosine kinase inhibitors. Expert Opin Investig Drugs. 2000;9:551–564. doi: 10.1517/13543784.9.3.551. [DOI] [PubMed] [Google Scholar]
- Kroll ES, Hyland KM, Hieter P, Li JJ. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102. doi: 10.1093/genetics/143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Luo G, Hahnenberger KM, Brooks C, Gecha O, Ingalls K, Numata K, Krystal M. Growth impairment resulting from expression of influenza virus M2 protein in Saccharomyces cerevisiae: Identification of a novel inhibitor of influenza virus. Antimicrob Agents Chemother. 1995;39:2204–2209. doi: 10.1128/aac.39.10.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. International Human Genome Sequencing Consortium. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lanio T, Jeltsch A, Pingoud A. Automated purification of His6-tagged proteins allows exhaustive screening of libraries generated by random mutagenesis. Biotechniques. 2000;29:338–342. doi: 10.2144/00292rr01. [DOI] [PubMed] [Google Scholar]
- Lin W, Ame JC, Aboul-Ela N, Jacobson EL, Jacobson MK. Isolation and characterization of the cDNA encoding bovine poly (ADP-ribose) glycohydrolase. J Biol Chem. 1997;272:11895–11901. doi: 10.1074/jbc.272.18.11895. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Munder T, Hinnen A. Yeast cells as tools for target-oriented screening. Appl Microbiol Biotechnol. 1999;52:311–320. doi: 10.1007/s002530051525. [DOI] [PubMed] [Google Scholar]
- Nemergut ME, Macara IG. Nuclear import of the ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J Cell Biol. 2000;149:835–850. doi: 10.1083/jcb.149.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta M, Ku S, Brown C, Okamoto AY, Shan B. CPF: An orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7α-hydroxylase gene. Proc Natl Acad Sci. 1999;96:6660–6665. doi: 10.1073/pnas.96.12.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson B, Spudich JA. A novel positive selection for identifying cold-sensitive myosin II mutants in Dictyostelium. Genetics. 1995;140:505–515. doi: 10.1093/genetics/140.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Nash P. Protein–protein interactions define specificity in signal transduction. Genes & Dev. 2000;14:1027–1047. [PubMed] [Google Scholar]
- Perkins E, Sun D, Nguyen A, Tulac S, Francesco M, Tavana H, Nguyen H, Tugendreich S, Barthmaier P, Couto J, et al. Novel inhibitors of PARP1 and PARP2 identified using a cell-based screen in yeast. Cancer Res. 2001;61:4175–4183. [PubMed] [Google Scholar]
- Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Raymond CK, Mugford VR, Sexson SL. Plasmid topologies that enhance the transformation efficiency of yeast. Biotechniques. 1999;27:892–894. doi: 10.2144/99275bm03. , 896. [DOI] [PubMed] [Google Scholar]
- Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A. The 1.7 Å crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- Sato T, Hanada M, Bodrug S, Irie S, Iwama N, Boise LH, Thompson CB, Golemis E, Fong L, Wang HG, et al. Interactions among members of the Bcl-2 protein family analyzed with a yeast two-hybrid system. Proc Natl Acad Sci. 1994;91:9238–9242. doi: 10.1073/pnas.91.20.9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Heller RA, Theriault TP, Konrad K, Lachenmeier E, Davis RW. Microarrays: Biotechnology's discovery platform for functional genomics. Trends Biotechnol. 1998;16:301–306. doi: 10.1016/s0167-7799(98)01219-0. [DOI] [PubMed] [Google Scholar]
- Sgroi DC, Teng S, Robinson G, LeVangie R, Hudson JR, Elkahloun AG. In vivo gene expression profile analysis of human breast cancer progression. Cancer Res. 1999;59:5656–5661. [PubMed] [Google Scholar]
- Shaham S, Shuman MA, Herskowitz I. Death-defying yeast identify novel apoptosis genes. Cell. 1998;92:425–427. doi: 10.1016/s0092-8674(00)80934-4. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman L, Campbell R, Broach JR. New assay technologies for high-throughput screening. Curr Opin Chem Biol. 1998;2:397–403. doi: 10.1016/s1367-5931(98)80015-x. [DOI] [PubMed] [Google Scholar]
- Stein RC, Waterfield MD. PI3-kinase inhibition: A target for drug development? Mol Med Today. 2000;6:347–357. doi: 10.1016/s1357-4310(00)01770-6. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Tugendreich S, Bassett DE, McKusick VA, Boguski MS, Hieter P. Genes conserved in yeast and humans. Hum Mol Genet. 1994;3:1509–1517. doi: 10.1093/hmg/3.suppl_1.1509. [DOI] [PubMed] [Google Scholar]
- van Hille B, Hill BT. Yeast cells expressing differential levels of human or yeast DNA topoisomerase II: A potent tool for identification and characterization of topoisomerase II-targeting antitumour agents. Cancer Chemother Pharmacol. 1998;42:345–356. doi: 10.1007/s002800050828. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wurgler-Murphy SM, Maeda T, Witten EA, Saito H. Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol Cell Biol. 1997;17:1289–1297. doi: 10.1128/mcb.17.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Reed JC. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell. 1998;1:337–346. doi: 10.1016/s1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Muyrers JP, Testa G, Stewart AF. DNA cloning by homologous recombination in Escherichia coli. Nat Biotechnol. 2000;18:1314–1317. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
- Zweiger G. Knowledge discovery in gene-expression-microarray data: Mining the information output of the genome. Trends Biotechnol. 1999;17:429–436. doi: 10.1016/s0167-7799(99)01359-1. [DOI] [PubMed] [Google Scholar]