Abstract
Two cellular retinol-binding proteins (CRBP I and II) with distinct tissue distributions and retinoid-binding properties have been recognized thus far in mammals. Here, we report the identification of a human retinol-binding protein resembling type I (55.6% identity) and type II (49.6% identity) CRBPs, but with a unique H residue in the retinoid-binding site and a distinctively different tissue distribution. Additionally, this binding protein (CRBP III) exhibits a remarkable sequence identity (62.2%) with the recently identified ι-crystallin/CRBP of the diurnal gecko Lygodactylus picturatus [Werten, P. J. L., Röll, B., van Alten, D. M. F. & de Jong, W. W. (2000) Proc. Natl. Acad. Sci. USA 97, 3282–3287 (First Published March 21, 2000; 10.1073/pnas.050500597)]. CRBP III and all-trans-retinol form a complex (Kd ≈ 60 nM), the absorption spectrum of which is characterized by the peculiar fine structure typical of the spectra of holo-CRBP I and II. As revealed by a 2.3-Å x-ray molecular model of apo-CRBP III, the amino acid residues that line the retinol-binding site in CRBP I and II are positioned nearly identically in the structure of CRBP III. At variance with the human CRBP I and II mRNAs, which are most abundant in ovary and intestine, respectively, the CRBP III mRNA is expressed at the highest levels in kidney and liver thus suggesting a prominent role for human CRBP III as an intracellular mediator of retinol metabolism in these tissues.
Vitamin A fulfills essential roles in the physiology of vertebrates, being involved in cell growth and differentiation, embryonic development, and vision. Plasma retinol-binding protein and cellular retinol-binding proteins (CRBPs) are specific carriers of the alcoholic form of vitamin A (retinol) in body fluids and within the cell, respectively. CRBPs are monomeric proteins of approximately 15.5 kDa belonging to a superfamily of small cytoplasmic proteins that specifically interact with hydrophobic ligands [intracellular lipid-binding proteins (iLBPs)]. Besides CRBPs, well characterized members of this superfamily are the cellular retinoic acid-binding proteins and the fatty acid-binding proteins (reviewed in refs. 1–3). Two mammalian CRBPs (rat CRBP type I and II) have been subjected previously to an extensive characterization. Their structures consist of 10 antiparallel β-strands, forming a β-barrel that accommodates the retinol molecule, and two α-helices that cover the open end of the barrel (4, 5). The NMR solution structures of apo-CRBP II and holo-CRBP II have revealed the occurrence of conformational changes, affecting discrete regions of the protein molecule, associated with retinol binding (6, 7). In different organisms, CRBP I exhibits rather distinct tissue distributions; in the adult rat, it is expressed mainly in liver, kidney, and the genital tract (8), whereas in humans it is expressed at high levels in ovary, adrenal and pituitary glands, and testis (9). Instead, in both rat and humans, CRBP II is confined essentially to the small intestine (8). Another remarkable difference between the two binding proteins is their affinity for all-trans-retinol. By two different methodologies, it has been found that the Kd of the retinol–CRBP I complex may be as low as 0.1 nM (10, 11), whereas the binding of retinol to CRBP II is ≈100-fold weaker (10). This property, along with the different tissue distribution, points to functionally distinct roles for the two carrier proteins. A number of in vitro studies have suggested that mammalian CRBPs may play an important role in vitamin A metabolism (reviewed in ref. 1). However, despite the wealth of in vitro data obtained with purified proteins and/or subcellular fractions, the physiological functions of CRBPs are not well established yet. According to a recent in vivo study (12), retinol esterification and intercellular retinol transfer between liver cells are strongly impaired in CRBP I-null mice. These two processes thus seem to be main targets of CRBP I action.
Here we report on the identification and the functional and structural characterization of a human protein that exhibits several of the characteristic features of CRBPs and is expressed mainly in liver and kidney.
Materials and Methods
Materials.
All-trans- and 13-cis-retinol, all-trans-retinaldehyde, and all-trans-retinoic acid were purchased from Sigma. All-trans-3,4-didehydroretinol was produced by saponification of all-trans-3,4-didehydroretinyl acetate. All-trans-3,4-didehydroretinyl acetate and 9-cis-retinol were kind gifts of S. A. Tanumihardjo and D. Cavazzini, respectively. All other chemicals were of analytical grade.
Identification, Bacterial Expression, and Purification of Human CRBP III.
The expressed sequence-tag (EST) database was searched with the BLAST program (13) by using the partial sequence of the bovine protein that copurified with CRBP I as a query. A highly similar (88% identity) human cDNA was found in the ATCC clone 454664 [containing the EST sequences H60427 (3′) and H60473 (5′) in plasmid pT7T3D-Pac] and sequenced on both strands by the dideoxy chain termination method (14). The region of plasmid pT7T3D-Pac corresponding to the entire human CRBP III coding sequence (405 bp) was PCR amplified by using a high-fidelity thermostable DNA polymerase (Pfu DNA polymerase, Stratagene) and two sequence-specific primers: an NdeI-tailed upstream primer (5′-CATCCACCATATGCCTCCCAACCTCACTGG-3′) and a BamHI-tailed downstream primer (5′-TAGGATCCCTATCTGACCTTCCTGAAGAC-3′). After the addition of 3′ terminal adenosine residues by incubation for 30 min at 72°C in the presence of Taq DNA polymerase (Perkin–Elmer), the amplification product was inserted into the pGEM vector (Promega) to generate the intermediate vector pGEM-CRBP III. The restriction fragment obtained from NdeI/BamHI digestion of plasmid pGEM-CRBP III was ligated then into the dephosphorylated NdeI and BamHI sites of the expression vector pET11b (Novagen), and the resulting plasmid (pET-CRBP III) was electroporated into Escherichia coli BL21 (DE3) cells. The expression of CRBP III was induced by adding 1 mM isopropyl-1-thio-β-D-galactopyranoside, and after a 3-h incubation at 28°C, cells were lysed by three cycles of freezing and thawing, followed by ten 15-sec bursts of sonication. CRBP III was purified to homogeneity by using a previously described three-step procedure (15) with a final yield of ≈2 mg/liter of cell culture. The ɛ280 nm (extinction coefficient) of CRBP III, calculated on the basis of its predicted amino acid sequence, was estimated to be 24,070 M−1⋅cm−1 (16). Phylogenetic analyses were performed with programs of the PHYLIP package (17); genetic distances were calculated with PROTDIST (PAM substitution matrix) and analyzed with the neighbor-joining algorithm; bootstrap was performed with SEQBOOT.
Ligand-Binding Assays.
Recombinant CRBP III in 50 mM K-phosphate, pH 7.3/150 mM NaCl was supplemented with all-trans-retinol or other retinoids dissolved in ethanol and stirred gently in the spectrophotometer cuvette. Absorption spectra were recorded with a Varian Cary 1E spectrophotometer. The interaction of retinol with CRBP III was investigated by fluorescence titrations carried out with a Perkin–Elmer LS-50B spectrofluorometer. Retinol binding to apo-CRBP III was monitored by measuring the quenching of protein fluorescence (excitation at 280 nm, emission at 330 nm). Aliquots (0.2- to 1-μl) of an ethanolic stock solution of all-trans-retinol (ɛ325 nm = 46,000 M−1⋅cm−1) were added to a cuvette containing CRBP III in 50 mM K-phosphate, pH 7.3/150 mM NaCl. After each addition, the protein solution was stirred gently and allowed to equilibrate at 20°C in the dark for a few minutes before spectrofluorometric analysis. Binding data were analyzed as described (15) by using SIGMA PLOT (Jandel, San Rafael, CA).
Crystallization, Data Collection, and Structure Determination.
Single crystals of apo-CRBP III were obtained in 1–2 weeks by sitting drop vapor-diffusion using PEG 8000 as a precipitant, at 21°C, in the presence of 25 mM potassium phosphate, pH 5.0. A single crystal of apo-CRBP III (0.3 × 0.1 × 0.03 mm3) was used to obtain a native data set at 2.3-Å resolution at the x-ray diffraction beam-line of the ELETTRA synchrotron facility (Trieste, Italy). By using a wavelength of 1 Å, 50 frames of 1° rotation each were measured. An image plate detector (MAR Research, Hamburg) with a diameter of 180 mm and a crystal-to-detector distance of 250 mm was used. The crystal structure was solved by the molecular replacement method with the software AMORE (18) by using the structure of the CRBP I as a template (4). Two independent molecules were positioned in the asymmetric unit. The structure was refined with the CNS program (19), imposing restraints on noncrystallographic symmetry. Data collection and refinement statistics are shown in Table 1.
Table 1.
Data collection and refinement statistics
| Measurement | Value |
|---|---|
| Space group | P212121 |
| Unit cell | a = 59.51 Å; b = 59.84 Å; c = 68.98 Å |
| Resolution, Å | 21–2.3 |
| Unique reflections | 9874 |
| Overall completeness, % | 87.1 |
| Last resolution shell completeness, % | 86.3 |
| Rsym, % | 12.7 |
| Multiplicity | 3.7 |
| 〈I/σ〉 | 3.6 |
| Reflections in working set | 8679 |
| Reflections in test set | 890 (8%) |
| Rcryst/Rfree, % | 22.9/28.9 |
| Protein atoms | 2222 |
| Water molecules | 144 |
| rms deviations on bond lengths, Å | 0.01 |
| rms deviations on bond angles, ° | 1.81 |
RNA Blot Analysis.
cDNA fragments corresponding to the 3′ untranslated regions of the human CRBP I, CRBP II, and CRBP III messengers were generated by PCR and used as transcript-specific CRBP probes. The CRBP I (182 bp) and CRBP III (239-bp) probes were amplified under standard PCR conditions by using plasmids containing the corresponding full-length cDNAs as templates. A human infant intestine library (Invitrogen) was used as a template for the amplification of the CRBP II probe (192 bp). The sequences of the oligonucleotide primers used for these amplifications were: CRBP I upstream primer, 5′-TGAGGCCCAAGCAGACAACC-3′; CRBP I downstream primer, 5′-ACAGGTCACTTTATTGGCATGG-3′; CRBP II upstream primer, 5′-ATGATGGCGACGTGGGAGGC-3′; CRBP II downstream primer, 5′-ATGCTAGGTTTCAAAGGAGGGG-3′; CRBP III upstream primer, 5′-CGGAGAGGAGCCAAGATCCC-3′; and CRBP III downstream primer, 5′-TGGGACGCCAGACCTTGACC-3′. Radioactively labeled probes of comparable specific activities [≈150 μCi/pmol (1 Ci = 37 GBq)] were prepared with the Megaprime labeling kit (Amersham Pharmacia) by using [α-32P]dCTP as the labeling precursor; probe specificity was verified by cross-hybridization experiments. A multiple tissue RNA dot blot (CLONTECH) containing poly(A)+ RNAs from 50 different human tissues arrayed on a nylon membrane was used to analyze the expression pattern of the human CRBP III mRNA; after stripping, the same blot was used for the analysis of human CRBP I and II expression. Prehybridizations and hybridizations were carried out according to the manufacturer's instructions. Approximately 2 × 107-cpm equivalents of each probe were used for hybridization. The first four washes were done with a 2× SSC (0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/1% SDS solution (at 65°C for 20 min under continuous agitation), and a 0.1× SSC/0.5% SDS solution was used for the final two washes (55°C, 20 min). Hybridization signals were quantified from phosphorimages recorded with a Personal Molecular Imager FX (Bio-Rad) by using the computer program QUANTITY-ONE.
Results
Identification and Sequence Analysis of a Putative CRBP.
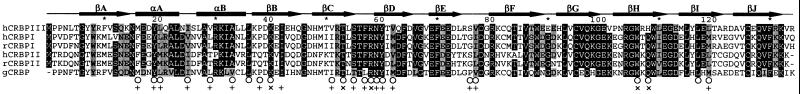
A previously unidentified protein copurifying with CRBP I during chromatographic fractionation of bovine kidney extracts was found to possess an N-terminal amino acid sequence (PPNLTGYYRFVSQKNLEDYLQALNVNMALRKIALLLKPDKEI) similar to that of CRBP I (unpublished data). The isolation of only minute amounts of this protein, contaminated by CRBP I, precluded the possibility of identifying its natural ligand and characterizing it functionally and structurally. When used as a query for a similarity search conducted against the entire expressed sequence tag data bank, however, the sequence of the above peptide led to the identification of a highly similar full-length cDNA randomly sequenced from both human liver and spleen. The deduced amino acid sequence of such cDNA is shown in Fig. 1. Five amino acid residues of a total of 42 N-terminal residues differ between the bovine and the human sequence. This putative CRBP, designated CRBP III, shares a considerable sequence identity with iLBPs and has an estimated molecular mass (15.9 kDa) typical of this protein superfamily. Because it exhibits a remarkably higher sequence identity with CRBPs (55.6 and 49,6% identity with human CRBP I and II, respectively) than with the other iLBPs thus far characterized (less than 40% identity), it presumably belongs to the CRBP family. Interestingly, CRBP III shares a high sequence identity (62.2%) with the ι-crystallin/housekeeping CRBP of the diurnal gecko L. picturatus, a “moonlighting” protein expressed in the eye lens, in which it is complexed with 3,4-didehydroretinol (vitamin A2), and in the liver, in which the physiological ligand is presumably all-trans-retinol (20, 21). Human CRBP I is very similar also to the gecko ι-crystallin/CRBP (59.3% sequence identity), whereas a substantially lower sequence identity (40.6%) is shared by human CRBP II and the gecko protein. Indeed, as revealed by a molecular phylogenetic analysis (data not shown) conducted according to ref. 17, gecko CRBP seems related more closely to CRBP III than to CRBP I and II.
Figure 1.
Multiple sequence alignment between human CRBP III and previously characterized CRBPs. Residues that are identical or similar in at least five of the six sequences are boxed in black or gray, respectively. The symbols below amino acid letter residues denote: ○, residues that in rat retinol–CRBP I change their accessible surface area by more than 1 Å2 after ligand removal (4); +, residues that have an atom within a distance of 3.6–5.1 Å from bound retinol in rat retinol–CRBP II (5); ×, residues that have an atom within 3.6 Å of bound retinol in rat retinol–CRBP II (5). The positions of β-strands A-J (arrows) and α-helices A and B (boxes) are indicated according to ref. 4. Conservative substitutions are defined as follows: A/I/L/M/V, S/T, F/Y/W, D/E, N/Q, and K/R. GenBank accession nos. are: P09455 and P50120, human (h) CRBP I and II, respectively; P02696 and P06768, rat (r) CRBP I and II, respectively; CA885608, gecko (g) (Lygodactylus picturatus) ι-crystallin/CRBP.
Retinoid Binding to Human Recombinant CRBP III.
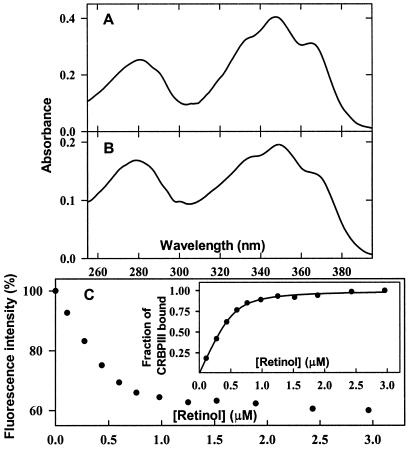
After overexpression in bacteria transformed with the pET-CRBPIII plasmid (see Materials and Methods for details), a protein with the molecular mass expected for CRBP III became detectable by SDS/PAGE analysis and was purified to homogeneity by ammonium sulfate fractionation, gel filtration, and ion-exchange chromatography (data not shown). By using UV absorption and fluorescence spectroscopy, various retinoids (all-trans-9-cis-, and 13-cis-retinol, all-trans-retinaldehyde, all-trans-retinoic acid, and all-trans-3,4-didehydroretinol) then were assayed for their ability to bind recombinant CRBP III. After incubation with CRBP III, all-trans retinol gave rise to an intense UV-visible absorption spectrum quite different from that of the unbound compound and similar to those of the all-trans retinol–CRBP I (22) and all-trans-retinol–CRBP II (23) complexes (see below). An absorption spectrum closely resembling the spectrum of the same retinoid added to CRBP I, albeit weaker and less defined, was produced also by all-trans-3,4-didehydroretinol (data not shown). The spectra obtained with all of the other retinoids added to CRBP III were very close to those of the unbound compounds, and as expected, they disappeared gradually after prolonged incubation. In contrast, the spectra produced by all-trans-retinol and all-trans-3,4-didehydroretinol essentially remained constant after incubation in the dark at room temperature for several hours, indicating the formation of relatively stable retinoid–CRBP III complexes. As shown in Fig. 2 A and B, which compare the absorption spectra of the complexes of all-trans-retinol with recombinant rat apo-CRBPI and human apo-CRBP III, respectively, the two spectra exhibit a remarkably similar fine structure in the retinol-absorption region. This similarity indicates that the ligand interacts with CRBP III in a specific manner and that such interaction is similar to that established between retinol and CRBP I. In the case of the retinol–CRBP III complex, however, the A350/A280 ratio (1.15) is lower than that of the retinol–CRBP I complex (1.6), both determined after the addition of a slight molar excess of all-trans-retinol to either binding protein. By assuming that the molar extinction coefficient of the complex of all-trans-retinol with CRBP III is the same as that of the complex with CRBP I (ɛmax = 50,200 ± 500 M−1⋅cm−1; ref. 22), it can be calculated that in the case of CRBP III, approximately 0.6 binding sites per protein molecule are occupied by retinol.
Figure 2.
Retinol binding to human apo-CRBP III. The absorption spectra of the retinol–CRBP I (A) and retinol–CRBP III (B) complexes were recorded after the addition of a slight molar excess of all-trans-retinol to recombinant rat apo-CRBP I and human apo-CRBP III (9 and 7 μM, respectively), followed by equilibration in the dark for a few minutes at 20°C. (C) Spectrofluorometric titration of human CRBP III with retinol. Intrinsic protein fluorescence (%) of apo-CRBP III (0.8 μM) is plotted as a function of retinol concentration. (Inset) Fitting of the experimentally determined fractional saturation to a theoretical curve corresponding to the binding of ≈0.66 mol of ligand per mol of CRBP III, with an apparent Kd of 60 nM.
A partial saturation of CRBP III by all-trans-retinol was evidenced also by spectrofluorometric titrations exploiting the marked decrease in intrinsic protein fluorescence that is brought about by energy transfer from excited tryptophans to protein-bound retinol. A typical fluorescence titration curve of humanCRBP III with all-trans-retinol is shown in Fig. 2C. Consistent with the partial saturation observed with all-trans-retinol, the extent of CRBP III fluorescence quenching at saturation (about 40%) was significantly lower than that normally observed with CRBP I and II (about 90%). A binding curve corresponding to an apparent Kd of ≈60 nM and a binding stoichiometry of ≈0.66 retinol-occupied sites per CRBP III molecule was found to fit the experimental data best (Fig. 2C Inset).
Three-Dimensional Structure of CRBP III.
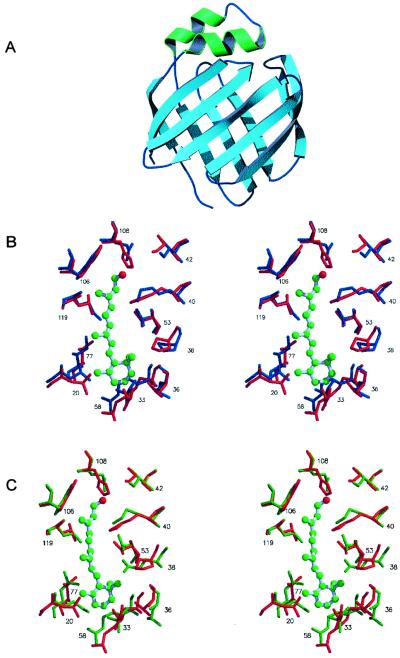
X-ray diffraction data are consistent with the presence of two CRBP III molecules in the asymmetric unit. Presumably, the two interacting molecules in the asymmetric unit do not form a real dimer. In fact, the buried area between them is relatively small (585 Å2), and only two hydrogen bonds are present at the protein–protein interface, a rather limited degree of interaction comparable to that involved in crystal packing. Because the two molecules are virtually identical to each other except for small differences in a few loops participating in intermolecular contacts, only one of such molecules will be described. The molecular model of CRBP III at 2.3-Å resolution closely resembles the CRBP I and II x-ray models (Fig. 3A). With the sole exception of strands D and E, which are separated by a gap, all β-strands are connected to each other by main–chain hydrogen bonds, as is the case for all the other structurally characterized iLBPs. As revealed by the superposition of the α-carbon atoms with the corresponding atoms of other iLBPs, CRBP III clearly belongs to the CRBP family. The overall rms deviations between equivalent α-carbons of CRBP I, II, and III in fact are rather small, and significantly increase when CRBP III is compared with iLBPs that bind negatively charged ligands such as cellular retinoic acid-binding proteins and fatty acid-binding proteins (Table 2). The crystals used for x-ray diffraction analysis were obtained from bacterially expressed CRBP III not supplemented with any ligand during purification and crystallization. However, a continuous, albeit not very long, electron density was visible in the central cavity of the β-barrel. This density might be attributed to a nonspecific, possibly hydrophobic ligand to which CRBP III became exposed during heterologous expression.
Figure 3.
(A) Ribbon drawing of the molecular model of CRBP III. Barrel forming β-strands are shown in light blue and the two α-helices are shown in green. (B) Stereoview showing the superposition of residues in proximity to the bound retinol in rat holo-CRBP I (blue colored, PDB ID code 1CRB) with the corresponding residues in human apo-CRBP III (red colored, this work). (C) Stereoview showing the superposition of residues in proximity to the bound retinol in rat holo-CRBP II (green colored, PDB ID code 1OPB) with the corresponding residues in human apo-CRBP III (red colored, this work). Vitamin A bound to CRBP I and II (ball-stick model) is shown also.
Table 2.
Overall rms deviations between α-carbon atoms of CRBP III chain A and corresponding atoms of CRBP III chain B and of other iLBPs
| CRBP III chain A vs. | rms deviation, Å |
|---|---|
| CRBP III chain B | 0.4 |
| CRBP I | 0.7 |
| CRBP II | 0.7 |
| Cellular retinoic acid-binding protein | 1.6 |
| Muscle fatty acid-binding protein | 1.5 |
| Intestinal fatty acid-binding protein | 1.8 |
The 0.4-Å rms deviation between the two chains of CRBP III drops to 0.2 Å if eight residues pertaining to two loops are excluded from the calculations. CRBP I, PDB ID code 1CRP; CRBP II, PDB ID code 1OPB; cellular retinoic acid-binding protein II, PDB ID code 2CBS; muscle fatty acid-binding protein, PDB ID code 2HMB; intestinal fatty acid-binding protein, PDB ID code 2IFB.
To evaluate the structural similarity between CRBP III and type I and II CRBPs further, the amino acid residues that are in close proximity to the bound retinol in rat holo-CRBP I (4) and II (5) have been superimposed on the corresponding residues of apo-CRBP III. Indeed, the relative position of such residues in CRBP III is nearly the same as in CRBP I and II. Moreover, the residues lining the retinol-binding site that are either identical or chemically conserved in CRBP I and II are identical or conserved in CRBP III as well (Fig. 1). The only exception is the amino acid substitution that involves Q108, a residue whose amide group hydrogen bonds the alcoholic group of retinol in CRBP I and II, which is replaced by an H residue in CRBP III. The superpositions of the side chains of some of the residues that line the retinol-binding site of holo-CRBP I and II with the corresponding residues in apo-CRBP III are shown in Fig. 3 B and C, respectively.
Tissue Distribution of the Human CRBP III mRNA.
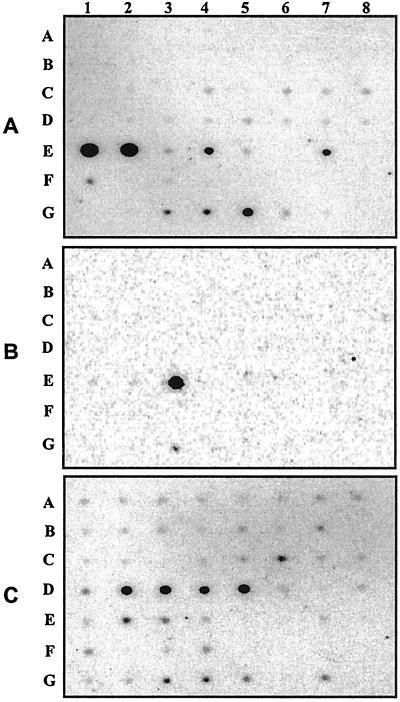
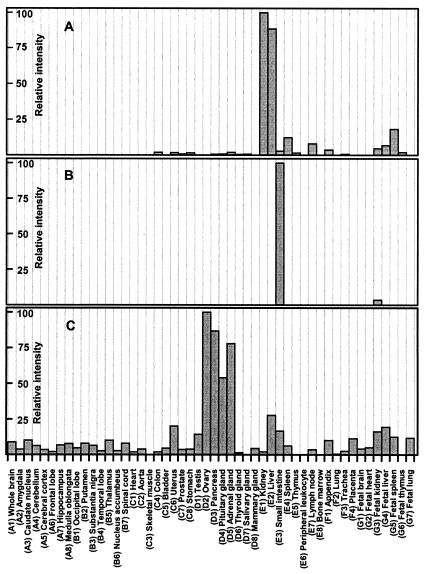
By using gene-specific DNA probes and a normalized dot blot containing poly(A)+ RNAs from 50 different human tissues, we next examined the tissue distribution of the human CRBP III mRNA (Fig. 4A) and compared it with the corresponding distributions of the CRBP II (Fig. 4B) and CRBP I (Fig. 4C) mRNAs. By comparing the intensities of hybridization signals produced by each CRBP probe, we determined the expression patterns of the three CRBP mRNAs in different tissues. As shown in Fig. 5A, the CRBP III mRNA has a fairly distinctive expression pattern. Consistent with the original isolation of the CRBP III protein from kidney extracts, the CRBP III mRNA is most abundant in adult kidney and liver tissues, whereas it is expressed at lower levels in spleen, lymph nodes, and appendix. This expression pattern is reversed somehow in the corresponding fetal tissues with a maximal accumulation in spleen and lower levels in both liver and kidney. Under identical hybridization conditions, the CRBP I messenger was detected in nearly all human tissues with the highest abundance in adult ovary, pancreas, pituitary and adrenal glands, and fetal liver (Fig. 5C). As expected, the CRBP II mRNA was confined to the small intestine in the adult, whereas among fetal tissues, it could be detected only in the kidney, albeit at very low levels (Fig. 5B). Finally, pairwise comparisons of the hybridization signals for CRBP III and I revealed that the levels of the CRBP III mRNA in human kidney and liver exceed those of the CRBP I messenger by approximately 200 and 13 times, respectively.
Figure 4.
Dot blot hybridization analysis of the CRBP III (A), CRBP II (B), and I (C) mRNAs in 50 different human tissues (see Materials and Methods for details). Each dot, identified by a letter (from A to G) and a number (from 1 to 8), corresponds to a distinct tissue. The identity of each tissue is specified in Fig. 5.
Figure 5.
Distribution of the CRBP III (A), II (B), and I (C) mRNAs in 50 different human tissues. The results are presented as hybridization intensities, derived from the dot blot experiments reported in Fig. 4, relative to the maximum hybridization intensity measured for each CRBP mRNA with each CRBP probe.
Discussion
A number of in vitro studies indicate that CRBP I and II might support retinol metabolism by facilitating enzymatic reactions involved in retinol storage and utilization. Additionally, the different tissue distribution and ligand-binding properties of the two proteins have suggested distinct functions for CRBP I and II. However, the precise functional roles of the two binding proteins have not been clarified yet. A human protein capable of binding retinol in a specific manner, CRBP III, has been shown here to resemble previously characterized CRBPs strongly, but with a distinctively different tissue distribution and a unique H residue in the retinoid-binding site.
The comparison of the amino acid sequence of CRBP III with those of other iLBPs clearly demonstrates that it is related most closely to members of the CRBP family including the moon-lighting protein ι-crystallin/CRBP of the diurnal gecko L. picturatus. The relatively low-yet-comparable sequence identities between human CRBP III and I (55.6%), CRBP III and II (49.6%), and CRBP I and II (53.7%) suggest that these proteins belong to distinct CRBP subfamilies.
The crystallization and structure determination of human apo-CRBP III have allowed a detailed comparison of the structural features of this protein with those of other members of the iLBP superfamily. The overall three-dimensional structure similarity of CRBP III is remarkably higher with CRBP I and II than with other members of the iLBP superfamily (Table 2). This structural similarity is significant particularly for the amino acid residues that are in close proximity to the retinol molecule in the retinol-binding sites of CRBP I and II. The residues that in rat holo-CRBP I change their accessible surface area by more than 1 Å2 after ligand removal (4) are nearly coincident with the residues that in rat holo-CRBP II have at least an atom within 5.1 Å of the bound retinol (ref. 5; Fig. 1). Remarkably, all such residues are positioned nearly identically in the three-dimensional structures of CRBP I, II, and III thus suggesting their involvement in the interaction with retinol also in the case of CRBP III. With regard to the five residues that in holo-CRBP II are within 3.6 Å of the retinol molecule (K40, T53, R58, W106, and Q108; ref. 5), they are identical in all of the CRBPs thus far characterized, except for Q108, which is replaced by an H residue in CRBP III. The side chain of Q108 hydrogen bonds the hydroxyl group of retinol in CRBP I and II. An H residue at the same position in CRBP III has the potential to hydrogen-bond retinol as well. It has been shown previously by site-directed mutagenesis that the replacement of Q108 by a positively charged residue like arginine remarkably lowers the affinity for retinol of both CRBP I and II (24–26). Accordingly, a loss of retinol binding is expected also to occur as a result of H108 protonation in CRBP III. The Q-H replacement at position 108 might have a functional significance. In fact, H108 protonation might take place physiologically after exposure of the protein to a weakly acidic microenvironment and/or as the consequence of a protein conformational change. A change in the ionization state of H108 in turn might impair the ability of this residue to hydrogen-bond the ligand and therefore might play a specific role in controlling retinoid binding to CRBP III and/or retinoid delivery to other protein acceptors of the vitamin.
The absorption spectrum of the all-trans-retinol-CRBP III complex exhibits a fine structure that closely resembles the fine structure of the holo-CRBP I and II spectra. This finding is consistent with the involvement of similar specific interactions between the chromophoric vitamin and the protein moiety in holo-CRBPs. Several retinoids have been tested for their ability to bind to apo-CRBP III, and only all-trans-3,4-didehydroretinol among them has been found to bind well to this protein. A tight binding of all-trans-3,4-didehydroretinol has been established already for mammalian CRBP I (27), II (28), and gecko ι-crystallin/CRBP (21). Thus, CRBP III seems to possess a high degree of ligand-binding specificity, and some of its binding properties are similar to those of the other CRBPs. With regard to the binding affinity of retinol for CRBP III (Kd ≈ 60 nM) it is significantly lower than that for CRBP I (Kd as low as 0.1 nM; refs. 10 and 11) and much closer to that for CRBP II (Kd ≈ 10 nM; ref. 29). However, recombinant CRBP III could be saturated only partially by all-trans-retinol. Several factors might be responsible for this undersaturated binding stoichiometry: for example, a structural alteration of the retinol-binding region caused by a partial misfolding of a fraction of protein molecules or the occupancy of a fraction of retinol-binding sites by nonspecific hydrophobic ligands, to which CRBP III became exposed during recombinant protein expression and purification. As mentioned above, the electron density detected in the retinoid-binding site of CRBP III might reflect the presence of a nonspecific ligand. However, various attempts to improve retinol-binding stoichiometry through organic solvent extraction of the protein were successful only partially thus indicating that the binding stoichiometry deficit of CRBP III probably is caused by the concomitant influence of different as-yet-unidentified factors.
The identification of a putative CRBP prompted us to analyze the distribution of the corresponding mRNA in human tissues. The CRBP III messenger is most abundant in kidney and liver, whereas drastically lower levels are present in several other tissues (Fig. 5A). Although CRBP III protein levels were not determined directly, previous studies have shown that the relative abundance of the CRBP I mRNAs in rat tissues (30, 31) is in fairly good agreement with the corresponding protein levels determined by radioimmunoassays (32, 33). As mentioned above, at variance with the tissue distribution in the rat of CRBP I, which is most abundant in liver, kidney, and the genital tract (8), the highest levels of CRBP I in humans have been found previously in the ovary; the next-richest tissues were the pituitary gland, testis, adrenal gland, and liver, with much lower levels in kidney and lung (9). Therefore, our determinations of CRBP I mRNA levels in human tissues (Fig. 5C) are in good agreement with the results of the aforementioned radioimmunoassays, and in addition they have revealed the presence of relatively high amounts of the CRBP I messenger in tissues such as the pancreas that had not been analyzed previously. The finding that human liver and kidney contain the highest levels of CRBP III mRNA, together with the relatively low abundance of the CRBP I messenger in the same tissues (especially the kidney), points to a role for CRBP III as a major human intracellular carrier of retinol in such tissues.
The observation that CRBP I-null mice fed with a vitamin A-enriched diet are healthy and fertile has been interpreted as an indication that CRBP I may not be an indispensable protein (12). However, this finding also may reflect the possible compensatory role of another CRBP distinct from CRBP I and II. CRBP III is possibly a good candidate for such a compensatory role.
Acknowledgments
We thank R. Percudani for his skillful assistance with sequence analysis and phylogenetic tree construction. We are grateful to D. Bellovino and D. Cavazzini for the human CRBP I full-length cDNA and for recombinant rat CRBP I, respectively. We thank S. Petrucco and E. Soragni for many valuable discussions. The technical assistance of A. Gandolfi, A. Grasselli, and D. Merli (University of Parma), of P. Arcidiaco (Centro Grandi Strumenti, University of Pavia), and of the staff of the x-ray diffraction beam line at the Elettra synchrotron (Trieste, Italy) is also gratefully acknowledged. This work was supported by the National Research Council of Italy, “Target Project on Biotechnology,” and the Ministry of University and of Scientific and Technological Research (Rome).
Abbreviations
- CRBP
cellular retinol-binding protein
- iLBP
intracellular lipid-binding protein
Note Added in Proof.
While this paper was being reviewed, the identification of a murine member of the iLBP family, also designated CRBP III but well distinct from the protein described here, was reported (34).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The human CRBP III cDNA sequence reported in this paper has been deposited in the GenBank database (accession no. AY007436).
Data deposition: The N-terminal sequence of bovine CRBP III reported in this paper has been deposited in the Swiss-Prot database (accession no. P82708).
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1GGL).
References
- 1.Li E, Norris A W. Annu Rev Nutr. 1996;16:205–234. doi: 10.1146/annurev.nu.16.070196.001225. [DOI] [PubMed] [Google Scholar]
- 2.Bernlohr D A, Simpson M A, Hertzel A V, Banaszak L J. Annu Rev Nutr. 1997;17:277–303. doi: 10.1146/annurev.nutr.17.1.277. [DOI] [PubMed] [Google Scholar]
- 3.Newcomer M E, Jamison R S, Ong D E. Subcell Biochem. 1998;30:53–80. doi: 10.1007/978-1-4899-1789-8_3. [DOI] [PubMed] [Google Scholar]
- 4.Cowan S W, Newcomer M E, Jones T A. J Mol Biol. 1993;230:1225–1246. doi: 10.1006/jmbi.1993.1238. [DOI] [PubMed] [Google Scholar]
- 5.Winter N S, Bratt J M, Banaszak L J. J Mol Biol. 1993;230:1247–1259. doi: 10.1006/jmbi.1993.1239. [DOI] [PubMed] [Google Scholar]
- 6.Lu J Y, Lin C L, Tang C G, Ponder J W, Kao J L F, Cistola D P, Li E. J Mol Biol. 1999;286:1179–1195. doi: 10.1006/jmbi.1999.2544. [DOI] [PubMed] [Google Scholar]
- 7.Lu J Y, Lin C L, Tang C G, Ponder J W, Kao J L F, Cistola D P, Li E. J Mol Biol. 2000;300:619–632. doi: 10.1006/jmbi.2000.3883. [DOI] [PubMed] [Google Scholar]
- 8.Ong D E, Newcomer M E, Chytil F. In: The Retinoids: Biology, Chemistry and Medicine. Sporn M B, Roberts A B, Goodman D S, editors. New York: Raven; 1994. pp. 283–317. [Google Scholar]
- 9.Ong D E, Page D L. Am J Clin Nutr. 1986;44:425–430. doi: 10.1093/ajcn/44.3.425. [DOI] [PubMed] [Google Scholar]
- 10.Li E, Qian S J, Winter N S, d'Avignon A, Levin M S, Gordon J I. J Biol Chem. 1991;266:3622–3629. [PubMed] [Google Scholar]
- 11.Malpeli G, Stoppini M, Zapponi M C, Folli C, Berni R. Eur J Biochem. 1995;229:486–493. [PubMed] [Google Scholar]
- 12.Ghyselinck N B, Bavik C, Sapin V, Mark M, Bonnier D, Hindelang C, Dierich A, Nilsson C B, Hakansson H, Sauvant P, et al. EMBO J. 1999;18:4903–4914. doi: 10.1093/emboj/18.18.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul M, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malpeli G, Folli C, Cavazzini D, Sartori G, Berni R. In: Methods in Molecular Biology: Retinoids Protocols. Redfern C P F, editor. Vol. 89. Totowa, NJ: Humana; 1998. pp. 111–122. [DOI] [PubMed] [Google Scholar]
- 16.Gill S C, von-Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J. PHYLIP, Phylogeny Interference Package. WA: Seattle; 1993. , Version 3.5c. [Google Scholar]
- 18.Navaza J. Acta Crystallogr D. 1994;21:916–924. [Google Scholar]
- 19.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 20.Röll B, Amons R, de Jong W W. J BiolChem. 1996;271:10437–10440. doi: 10.1074/jbc.271.18.10437. [DOI] [PubMed] [Google Scholar]
- 21.Werten P J L, Röll B, van Alten D M F, de Jong W W. Proc Natl Acad Sci USA. 2000;97:3282–3287. doi: 10.1073/pnas.050500597. . (First Published March 21, 2000; 10.1073/pnas.050500597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ong D E, Chytil F. J Biol Chem. 1978;253:828–832. [PubMed] [Google Scholar]
- 23.Ong D E. J Biol Chem. 1984;259:1476–1482. [PubMed] [Google Scholar]
- 24.Stump D G, Lloyd R S, Chytil F. J Biol Chem. 1991;266:4622–4630. [PubMed] [Google Scholar]
- 25.Cheng L, Qian S J, Rothschild C, d'Avignon A, Lefkowith J B, Gordon J I, Li E. J Biol Chem. 1991;266:24404–24412. [PubMed] [Google Scholar]
- 26.Jakoby M G, Miller K R, Toner J J, Bowman A, Cheng L, Li E, Cistola D P. Biochemistry. 1993;32:872–878. doi: 10.1021/bi00054a019. [DOI] [PubMed] [Google Scholar]
- 27.Ong D E, Chytil F. Nature (London) 1975;255:74–75. doi: 10.1038/255074a0. [DOI] [PubMed] [Google Scholar]
- 28.MacDonald P N, Ong D E. J Biol Chem. 1987;262:10550–10556. [PubMed] [Google Scholar]
- 29.Levin M S, Locke B, Yang N C, Li E, Gordon J I. J Biol Chem. 1988;263:17715–17723. [PubMed] [Google Scholar]
- 30.Levin M S, Li E, Ong D E, Gordon J I. J Biol Chem. 1987;262:7118–7124. [PubMed] [Google Scholar]
- 31.Rajan N, Blaner W S, Soprano D R, Suhara A, Goodman D S. J Lipid Res. 1990;31:821–829. [PubMed] [Google Scholar]
- 32.Ong D E, Crow J A, Chityl F. J Biol Chem. 1982;257:13385–13389. [PubMed] [Google Scholar]
- 33.Kato M, Sung W K, Kato K, Goodman D S. Biol Reprod. 1985;32:173–189. doi: 10.1095/biolreprod32.1.173. [DOI] [PubMed] [Google Scholar]
- 34.Vogel S, Mendelsohn C L, Mertz J R, Piantedosi R, Waldburger C, Gottesman M E, Blaner W S. J Biol Chem. 2001;276:1353–1360. doi: 10.1074/jbc.M005118200. [DOI] [PubMed] [Google Scholar]