Abstract
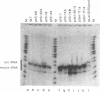
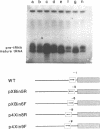
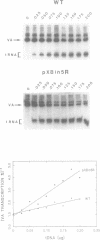
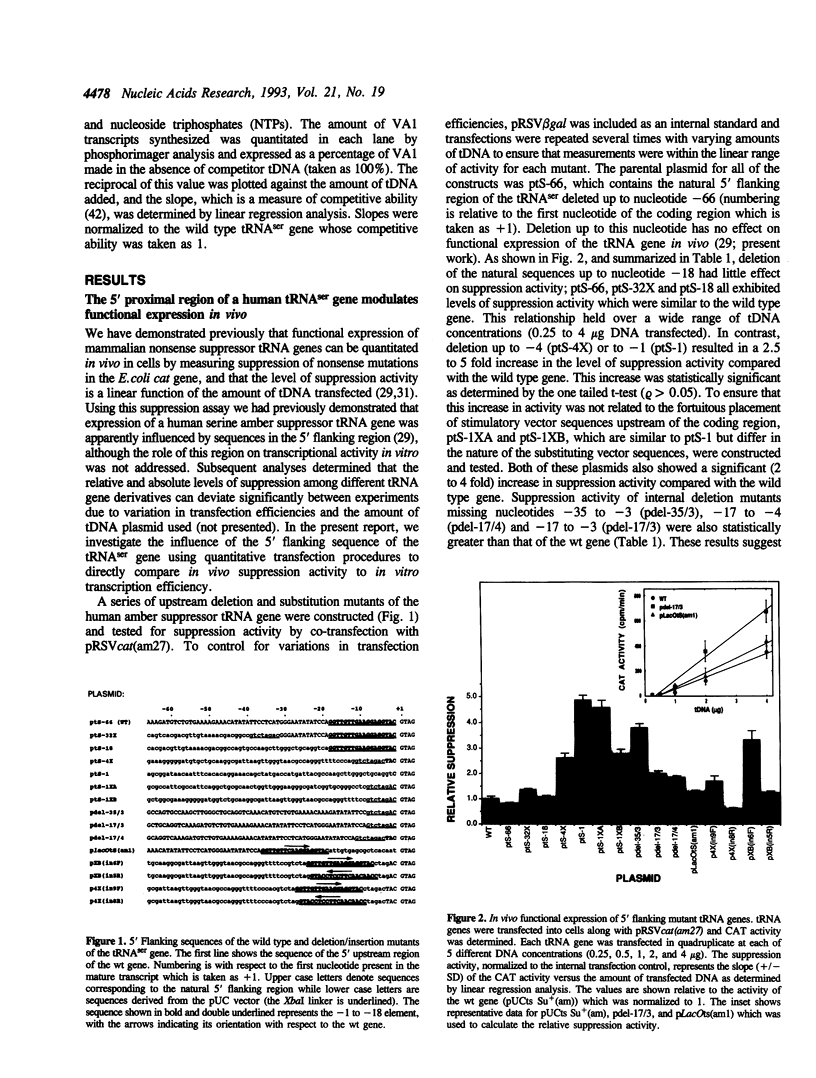
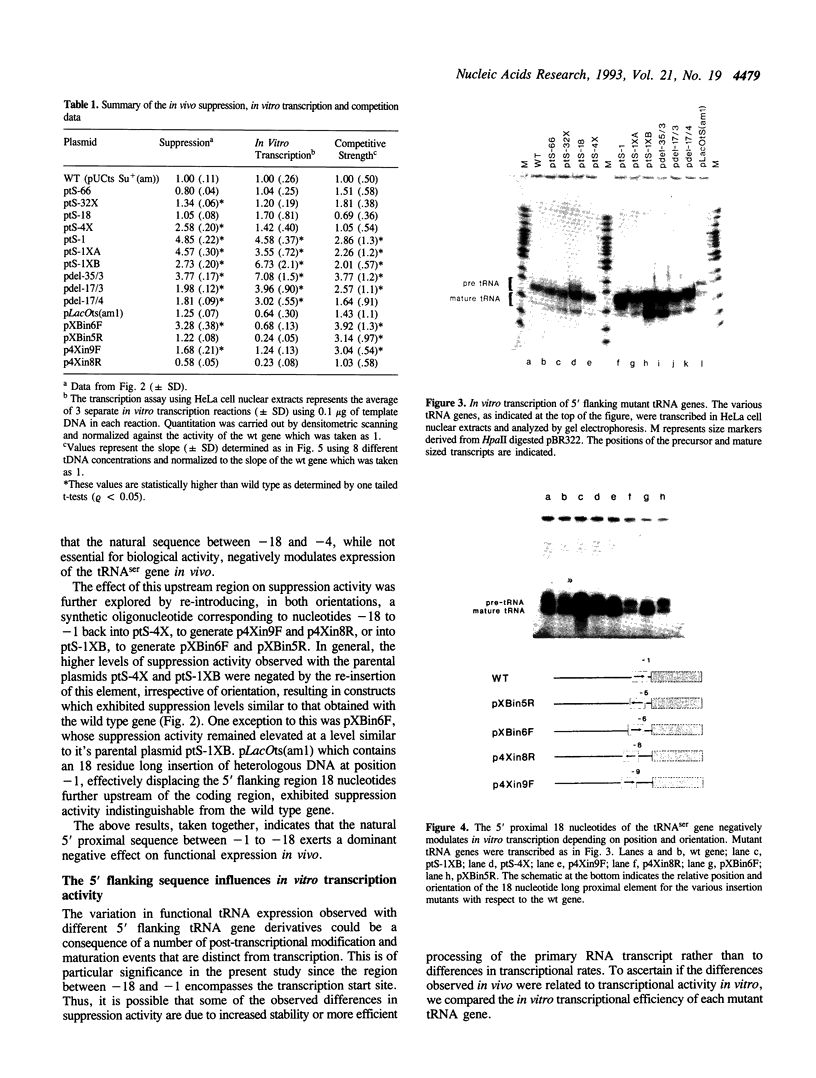
The consequences of altering the 5' flanking region of a human amber suppressor tRNA(ser) gene on phenotypic expression in vivo and transcription in vitro was examined by constructing a series of upstream deletion and substitution mutants. The resulting tDNA variants were examined for functional tRNA expression in vivo, by measuring suppression of a nonsense mutation in the Escherichia coli chloramphenicol acetyltransferase (cat) gene in co-transfection assays, and for transcriptional activity in vitro using HeLa cell nuclear extracts. Mutant genes in which the 18 nucleotides 5' proximal to the coding region were deleted and replaced with heterologous sequences were 2 to 5 fold more active in vivo in comparison to the wild type gene. There was a strong, but not exclusive, correlation between the levels of nonsense suppression observed in vivo and transcriptional activity in vitro. In certain cases, introduction of an oligonucleotide encompassing this 18 nucleotide element upstream of more active tRNA genes reduced both the levels of suppression and template activity. These results indicate that the immediate 5' contiguous sequence of this tRNA gene negatively modulates expression both in vivo and in vitro.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnold G. J., Gross H. J. Unrelated leader sequences can efficiently promote human tRNA gene transcription. Gene. 1987;51(2-3):237–246. doi: 10.1016/0378-1119(87)90312-x. [DOI] [PubMed] [Google Scholar]
- Arnold G. J., Schmutzler C., Thomann U., van Tol H., Gross H. J. The human tRNAVal gene family: organization, nucleotide sequences and homologous transcription of three single-copy genes. Gene. 1986;44(2-3):287–297. doi: 10.1016/0378-1119(86)90193-9. [DOI] [PubMed] [Google Scholar]
- Bull P., Thorikay M., Moenne A., Wilkens M., Sánchez H., Valenzuela P., Venegas A. The yeast tRNA(Phe) gene family: structures and transcriptional activities reveal member differences not explained by intragenic promoters. DNA. 1987 Aug;6(4):353–362. doi: 10.1089/dna.1987.6.353. [DOI] [PubMed] [Google Scholar]
- Capone J. P. Modulation of the phenotypic expression of a human serine tRNA gene by 5'-flanking sequences. DNA. 1988 Sep;7(7):459–468. doi: 10.1089/dna.1.1988.7.459. [DOI] [PubMed] [Google Scholar]
- Capone J. P., Sedivy J. M., Sharp P. A., RajBhandary U. L. Introduction of UAG, UAA, and UGA nonsense mutations at a specific site in the Escherichia coli chloramphenicol acetyltransferase gene: use in measurement of amber, ochre, and opal suppression in mammalian cells. Mol Cell Biol. 1986 Sep;6(9):3059–3067. doi: 10.1128/mcb.6.9.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G., Duimio L., Coda-Zabetta F., Sprague K. U., Ottonello S. A novel RNA polymerase III transcription factor fraction that is not required for template commitment. J Biol Chem. 1993 May 25;268(15):11199–11207. [PubMed] [Google Scholar]
- Dingermann T., Sharp S., Schaack J., Söll D. Stable transcription complex formation of eukaryotic tRNA genes is dependent on a limited separation of the two intragenic control regions. J Biol Chem. 1983 Sep 10;258(17):10395–10402. [PubMed] [Google Scholar]
- Francis M. A., Rajbhandary U. L. Expression and function of a human initiator tRNA gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1990 Sep;10(9):4486–4494. doi: 10.1128/mcb.10.9.4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Hipskind R. A., Clarkson S. G. 5'-flanking sequences that inhibit in vitro transcription of a xenopus laevis tRNA gene. Cell. 1983 Oct;34(3):881–890. doi: 10.1016/0092-8674(83)90545-7. [DOI] [PubMed] [Google Scholar]
- Hollon T., Yoshimura F. K. Variation in enzymatic transient gene expression assays. Anal Biochem. 1989 Nov 1;182(2):411–418. doi: 10.1016/0003-2697(89)90616-7. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Braun B. R., Nguyen L. H., Geiduschek E. P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990 Jan 26;60(2):235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- Kondo K., Hodgkin J., Waterston R. H. Differential expression of five tRNA(UAGTrp) amber suppressors in Caenorhabditis elegans. Mol Cell Biol. 1988 Sep;8(9):3627–3635. doi: 10.1128/mcb.8.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K., Makovec B., Waterston R. H., Hodgkin J. Genetic and molecular analysis of eight tRNA(Trp) amber suppressors in Caenorhabditis elegans. J Mol Biol. 1990 Sep 5;215(1):7–19. doi: 10.1016/S0022-2836(05)80090-7. [DOI] [PubMed] [Google Scholar]
- Larson D., Bradford-Wilcox J., Young L. S., Sprague K. U. A short 5' flanking region containing conserved sequences is required for silkworm alanine tRNA gene activity. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3416–3420. doi: 10.1073/pnas.80.11.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopata M. A., Cleveland D. W., Sollner-Webb B. High level transient expression of a chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 1984 Jul 25;12(14):5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschalek R., Dingermann T. Identification of a protein factor binding to the 5'-flanking region of a tRNA gene and being involved in modulation of tRNA gene transcription in vivo in Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Jul 25;16(14B):6737–6752. doi: 10.1093/nar/16.14.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morry M. J., Harding J. D. Modulation of transcriptional activity and stable complex formation by 5'-flanking regions of mouse tRNAHis genes. Mol Cell Biol. 1986 Jan;6(1):105–115. doi: 10.1128/mcb.6.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W. Three in one and one in three: it all depends on TBP. Cell. 1993 Jan 15;72(1):7–10. doi: 10.1016/0092-8674(93)90042-o. [DOI] [PubMed] [Google Scholar]
- Rooney R. J., Harding J. D. Transcriptional activity and factor binding are stimulated by separate and distinct sequences in the 5' flanking region of a mouse tRNAAsp gene. Nucleic Acids Res. 1988 Mar 25;16(6):2509–2521. doi: 10.1093/nar/16.6.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutzler C., Gross H. J. Genes, variant genes, and pseudogenes of the human tRNA(Val) gene family are differentially expressed in HeLa cells and in human placenta. Nucleic Acids Res. 1990 Sep 11;18(17):5001–5008. doi: 10.1093/nar/18.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague K. U., Larson D., Morton D. 5' flanking sequence signals are required for activity of silkworm alanine tRNA genes in homologous in vitro transcription systems. Cell. 1980 Nov;22(1 Pt 1):171–178. doi: 10.1016/0092-8674(80)90165-8. [DOI] [PubMed] [Google Scholar]
- Stutz F., Gouilloud E., Clarkson S. G. Oocyte and somatic tyrosine tRNA genes in Xenopus laevis. Genes Dev. 1989 Aug;3(8):1190–1198. doi: 10.1101/gad.3.8.1190. [DOI] [PubMed] [Google Scholar]
- Syroid D. E., Tapping R. I., Capone J. P. Regulated expression of a mammalian nonsense suppressor tRNA gene in vivo and in vitro using the lac operator/repressor system. Mol Cell Biol. 1992 Oct;12(10):4271–4278. doi: 10.1128/mcb.12.10.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N. J., Green S., Jin J. R., Chambon P. The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell. 1988 Jul 15;54(2):199–207. doi: 10.1016/0092-8674(88)90552-1. [DOI] [PubMed] [Google Scholar]
- White R. J., Jackson S. P. The TATA-binding protein: a central role in transcription by RNA polymerases I, II and III. Trends Genet. 1992 Aug;8(8):284–288. doi: 10.1016/0168-9525(92)90255-3. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P. RNA polymerase III transcription. Curr Opin Cell Biol. 1991 Jun;3(3):461–466. doi: 10.1016/0955-0674(91)90074-9. [DOI] [PubMed] [Google Scholar]
- Wormington W. M., Bogenhagen D. F., Jordan E., Brown D. D. A quantitative assay for Xenopus 5S RNA gene transcription in vitro. Cell. 1981 Jun;24(3):809–817. doi: 10.1016/0092-8674(81)90106-9. [DOI] [PubMed] [Google Scholar]