Abstract
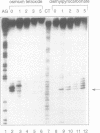
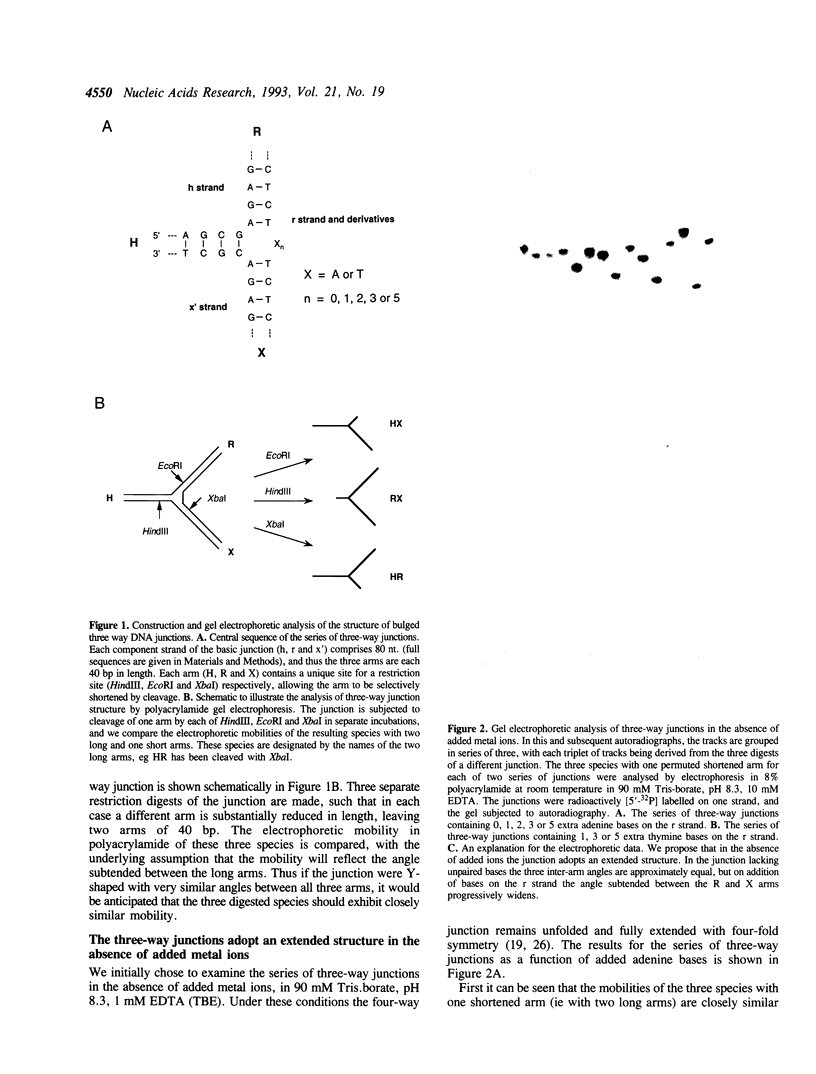
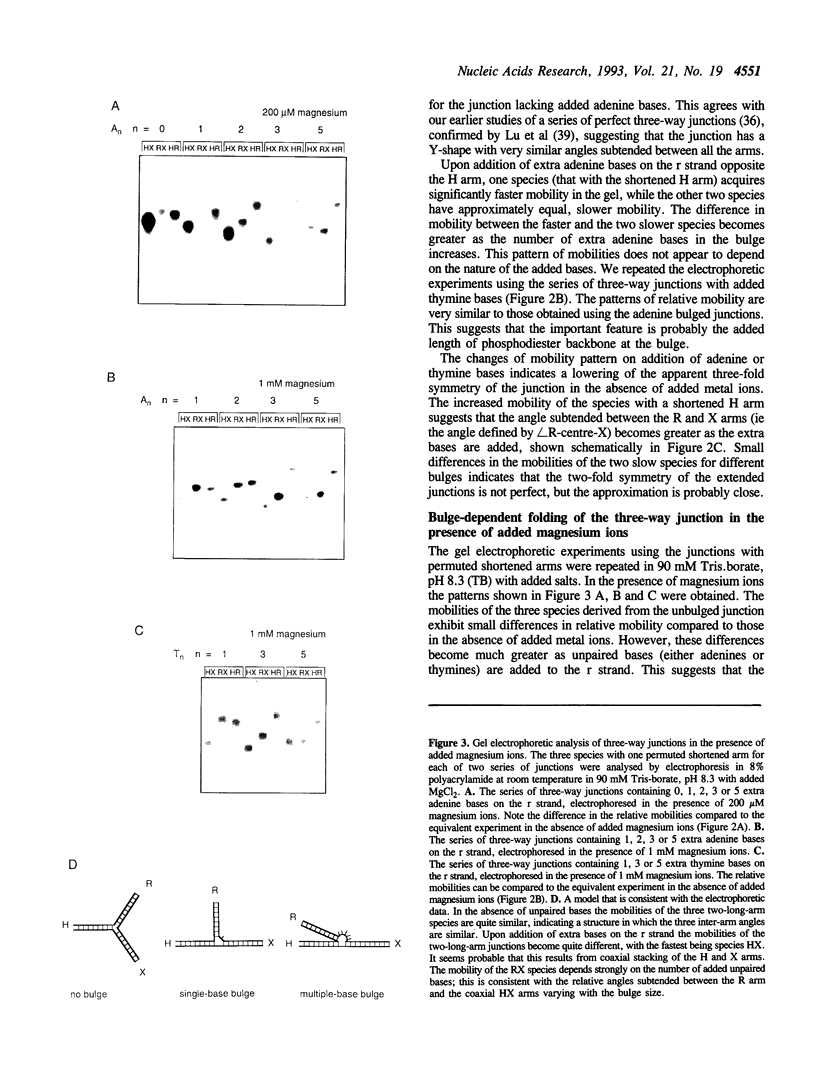
We have studied a series of three-way DNA junctions containing unpaired bases on one strand at the branch-point of the junctions. The global conformation of the arms of the junctions has been analysed by means of polyacrylamide gel electrophoresis, as a function of conditions. We find that in the absence of added metal ions, all the results for all the junctions can be accounted for by extended structures, with the largest angle being that between the arms defined by the strand containing the extra bases. Upon addition of magnesium (II) or hexamine cobalt (III) ions, the electrophoretic patterns change markedly, indicative of ion-dependent folding transitions for some of the junctions. For the junction lacking the unpaired bases, the three inter-arm angles appear to be quite similar, suggesting an extended structure. However, the addition of unpaired bases permits the three-way junction to adopt a significantly different structure, in which one angle becomes smaller than the other two. These species also exhibit marked protection against osmium addition to thymine bases at the point of strand exchange. These results are consistent with a model in which two of the helical arms undergo coaxial stacking in the presence of magnesium ions, with the third arm defining an angle that depends upon the number of unpaired bases.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhattacharyya A., Lilley D. M. The contrasting structures of mismatched DNA sequences containing looped-out bases (bulges) and multiple mismatches (bubbles). Nucleic Acids Res. 1989 Sep 12;17(17):6821–6840. doi: 10.1093/nar/17.17.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A., Murchie A. I., Lilley D. M. RNA bulges and the helical periodicity of double-stranded RNA. Nature. 1990 Feb 1;343(6257):484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- Broker T. R., Lehman I. R. Branched DNA molecules: intermediates in T4 recombination. J Mol Biol. 1971 Aug 28;60(1):131–149. doi: 10.1016/0022-2836(71)90453-0. [DOI] [PubMed] [Google Scholar]
- Chen J. H., Churchill M. E., Tullius T. D., Kallenbach N. R., Seeman N. C. Construction and analysis of monomobile DNA junctions. Biochemistry. 1988 Aug 9;27(16):6032–6038. doi: 10.1021/bi00416a031. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Tullius T. D., Kallenbach N. R., Seeman N. C. A Holliday recombination intermediate is twofold symmetric. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4653–4656. doi: 10.1073/pnas.85.13.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg R. M., Murchie A. I., Zechel A., Carlberg C., Diekmann S., Lilley D. M. Fluorescence resonance energy transfer analysis of the structure of the four-way DNA junction. Biochemistry. 1992 May 26;31(20):4846–4856. doi: 10.1021/bi00135a016. [DOI] [PubMed] [Google Scholar]
- Cooper J. P., Hagerman P. J. Gel electrophoretic analysis of the geometry of a DNA four-way junction. J Mol Biol. 1987 Dec 20;198(4):711–719. doi: 10.1016/0022-2836(87)90212-9. [DOI] [PubMed] [Google Scholar]
- Cooper J. P., Hagerman P. J. Geometry of a branched DNA structure in solution. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7336–7340. doi: 10.1073/pnas.86.19.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R., Fujiwara T., Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990 Aug 24;62(4):829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- Delihas N., Andersen J., Singhal R. P. Structure, function and evolution of 5-S ribosomal RNAs. Prog Nucleic Acid Res Mol Biol. 1984;31:161–190. doi: 10.1016/s0079-6603(08)60377-3. [DOI] [PubMed] [Google Scholar]
- Diekmann S., Lilley D. M. The anomalous gel migration of a stable cruciform: temperature and salt dependence, and some comparisons with curved DNA. Nucleic Acids Res. 1987 Jul 24;15(14):5765–5774. doi: 10.1093/nar/15.14.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett D. R., Lilley D. M. Effects of base mismatches on the structure of the four-way DNA junction. J Mol Biol. 1991 Sep 5;221(1):147–161. doi: 10.1016/0022-2836(91)80211-c. [DOI] [PubMed] [Google Scholar]
- Duckett D. R., Lilley D. M. The three-way DNA junction is a Y-shaped molecule in which there is no helix-helix stacking. EMBO J. 1990 May;9(5):1659–1664. doi: 10.1002/j.1460-2075.1990.tb08286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Diekmann S., von Kitzing E., Kemper B., Lilley D. M. The structure of the Holliday junction, and its resolution. Cell. 1988 Oct 7;55(1):79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Lilley D. M. The role of metal ions in the conformation of the four-way DNA junction. EMBO J. 1990 Feb;9(2):583–590. doi: 10.1002/j.1460-2075.1990.tb08146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Furlong J. C., Lilley D. M. Highly selective chemical modification of cruciform loops by diethyl pyrocarbonate. Nucleic Acids Res. 1986 May 27;14(10):3995–4007. doi: 10.1093/nar/14.10.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough G. W., Lilley D. M. DNA bending induced by cruciform formation. Nature. 1985 Jan 10;313(5998):154–156. doi: 10.1038/313154a0. [DOI] [PubMed] [Google Scholar]
- Guo Q., Lu M., Churchill M. E., Tullius T. D., Kallenbach N. R. Asymmetric structure of a three-arm DNA junction. Biochemistry. 1990 Dec 11;29(49):10927–10934. doi: 10.1021/bi00501a010. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Herr W. Diethyl pyrocarbonate: a chemical probe for secondary structure in negatively supercoiled DNA. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8009–8013. doi: 10.1073/pnas.82.23.8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess R., Wierzbicki A., Abremski K. Isolation and characterization of intermediates in site-specific recombination. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6840–6844. doi: 10.1073/pnas.84.19.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C. H., Griffith J. D. Deletions of bases in one strand of duplex DNA, in contrast to single-base mismatches, produce highly kinked molecules: possible relevance to the folding of single-stranded nucleic acids. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4833–4837. doi: 10.1073/pnas.86.13.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Klug A. Crystallographic refinement of yeast phenylalanine transfer RNA at 2-5A resolution. J Mol Biol. 1976 Dec 25;108(4):619–649. doi: 10.1016/s0022-2836(76)80109-x. [DOI] [PubMed] [Google Scholar]
- Jayaram M., Crain K. L., Parsons R. L., Harshey R. M. Holliday junctions in FLP recombination: resolution by step-arrest mutants of FLP protein. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7902–7906. doi: 10.1073/pnas.85.21.7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper B., Janz E. Function of gene 49 of bacteriophage T4. I. Isolation and biochemical characterization of very fast-sedimenting DNA. J Virol. 1976 Jun;18(3):992–999. doi: 10.1128/jvi.18.3.992-999.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Quigley G. J., Suddath F. L., McPherson A., Sneden D., Kim J. J., Weinzierl J., Rich A. Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain. Science. 1973 Jan 19;179(4070):285–288. doi: 10.1126/science.179.4070.285. [DOI] [PubMed] [Google Scholar]
- Kimball A., Guo Q., Lu M., Cunningham R. P., Kallenbach N. R., Seeman N. C., Tullius T. D. Construction and analysis of parallel and antiparallel Holliday junctions. J Biol Chem. 1990 Apr 25;265(12):6544–6547. [PubMed] [Google Scholar]
- Kitts P. A., Nash H. A. Homology-dependent interactions in phage lambda site-specific recombination. Nature. 1987 Sep 24;329(6137):346–348. doi: 10.1038/329346a0. [DOI] [PubMed] [Google Scholar]
- Leontis N. B., Kwok W., Newman J. S. Stability and structure of three-way DNA junctions containing unpaired nucleotides. Nucleic Acids Res. 1991 Feb 25;19(4):759–766. doi: 10.1093/nar/19.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Clegg R. M. The structure of the four-way junction in DNA. Annu Rev Biophys Biomol Struct. 1993;22:299–328. doi: 10.1146/annurev.bb.22.060193.001503. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Palecek E. The supercoil-stabilised cruciform of ColE1 is hyper-reactive to osmium tetroxide. EMBO J. 1984 May;3(5):1187–1192. doi: 10.1002/j.1460-2075.1984.tb01949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Guo Q., Kallenbach N. R. Effect of sequence on the structure of three-arm DNA junctions. Biochemistry. 1991 Jun 18;30(24):5815–5820. doi: 10.1021/bi00238a001. [DOI] [PubMed] [Google Scholar]
- Lumpkin O. J. Mobility of DNA in gel electrophoresis. Biopolymers. 1982 Nov;21(11):2315–2316. doi: 10.1002/bip.360211116. [DOI] [PubMed] [Google Scholar]
- Ma R. I., Kallenbach N. R., Sheardy R. D., Petrillo M. L., Seeman N. C. Three-arm nucleic acid junctions are flexible. Nucleic Acids Res. 1986 Dec 22;14(24):9745–9753. doi: 10.1093/nar/14.24.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moras D., Comarmond M. B., Fischer J., Weiss R., Thierry J. C., Ebel J. P., Giegé R. Crystal structure of yeast tRNAAsp. Nature. 1980 Dec 25;288(5792):669–674. doi: 10.1038/288669a0. [DOI] [PubMed] [Google Scholar]
- Murchie A. I., Carter W. A., Portugal J., Lilley D. M. The tertiary structure of the four-way DNA junction affords protection against DNase I cleavage. Nucleic Acids Res. 1990 May 11;18(9):2599–2606. doi: 10.1093/nar/18.9.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie A. I., Clegg R. M., von Kitzing E., Duckett D. R., Diekmann S., Lilley D. M. Fluorescence energy transfer shows that the four-way DNA junction is a right-handed cross of antiparallel molecules. Nature. 1989 Oct 26;341(6244):763–766. doi: 10.1038/341763a0. [DOI] [PubMed] [Google Scholar]
- Nunes-Düby S. E., Matsumoto L., Landy A. Site-specific recombination intermediates trapped with suicide substrates. Cell. 1987 Aug 28;50(5):779–788. doi: 10.1016/0092-8674(87)90336-9. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij C. W., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985 Mar 11;13(5):1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter H., Dressler D. In vitro system from Escherichia coli that catalyzes generalized genetic recombination. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3698–3702. doi: 10.1073/pnas.75.8.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter H., Dressler D. On the mechanism of genetic recombination: electron microscopic observation of recombination intermediates. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3000–3004. doi: 10.1073/pnas.73.9.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice J. A., Crothers D. M. DNA bending by the bulge defect. Biochemistry. 1989 May 16;28(10):4512–4516. doi: 10.1021/bi00436a058. [DOI] [PubMed] [Google Scholar]
- Riordan F. A., Bhattacharyya A., McAteer S., Lilley D. M. Kinking of RNA helices by bulged bases, and the structure of the human immunodeficiency virus transactivator response element. J Mol Biol. 1992 Jul 20;226(2):305–310. doi: 10.1016/0022-2836(92)90947-i. [DOI] [PubMed] [Google Scholar]
- Sinha N. D., Biernat J., McManus J., Köster H. Polymer support oligonucleotide synthesis XVIII: use of beta-cyanoethyl-N,N-dialkylamino-/N-morpholino phosphoramidite of deoxynucleosides for the synthesis of DNA fragments simplifying deprotection and isolation of the final product. Nucleic Acids Res. 1984 Jun 11;12(11):4539–4557. doi: 10.1093/nar/12.11.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J. L., Holbrook S. R., Warrant R. W., Church G. M., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J Mol Biol. 1978 Aug 25;123(4):607–630. doi: 10.1016/0022-2836(78)90209-7. [DOI] [PubMed] [Google Scholar]
- Tang R. S., Draper D. E. Bulge loops used to measure the helical twist of RNA in solution. Biochemistry. 1990 Jun 5;29(22):5232–5237. doi: 10.1021/bi00474a003. [DOI] [PubMed] [Google Scholar]
- Wyatt J. R., Puglisi J. D., Tinoco I., Jr RNA pseudoknots. Stability and loop size requirements. J Mol Biol. 1990 Jul 20;214(2):455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
- Zhong M., Kallenbach N. R. Conformation and thermodynamics of DNA "necks". Models for three-arm branch formation in a duplex. J Mol Biol. 1993 Apr 5;230(3):766–778. doi: 10.1006/jmbi.1993.1199. [DOI] [PubMed] [Google Scholar]
- von Kitzing E., Lilley D. M., Diekmann S. The stereochemistry of a four-way DNA junction: a theoretical study. Nucleic Acids Res. 1990 May 11;18(9):2671–2683. doi: 10.1093/nar/18.9.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]