Abstract
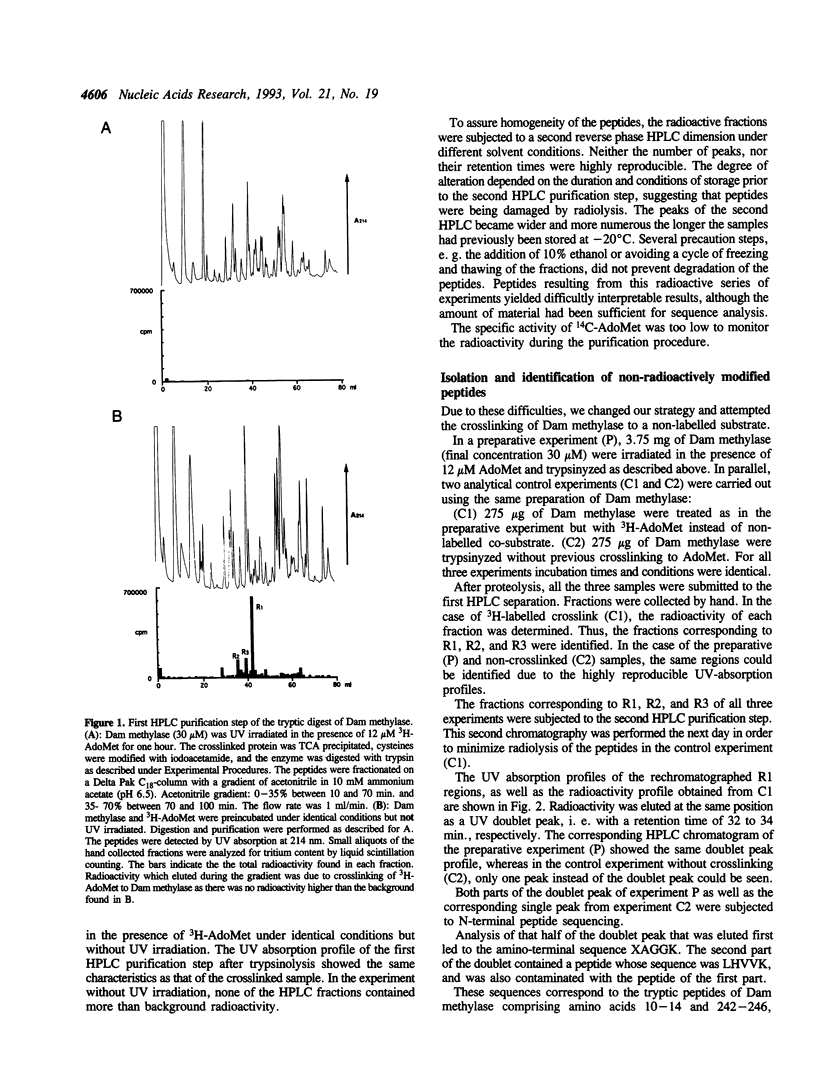
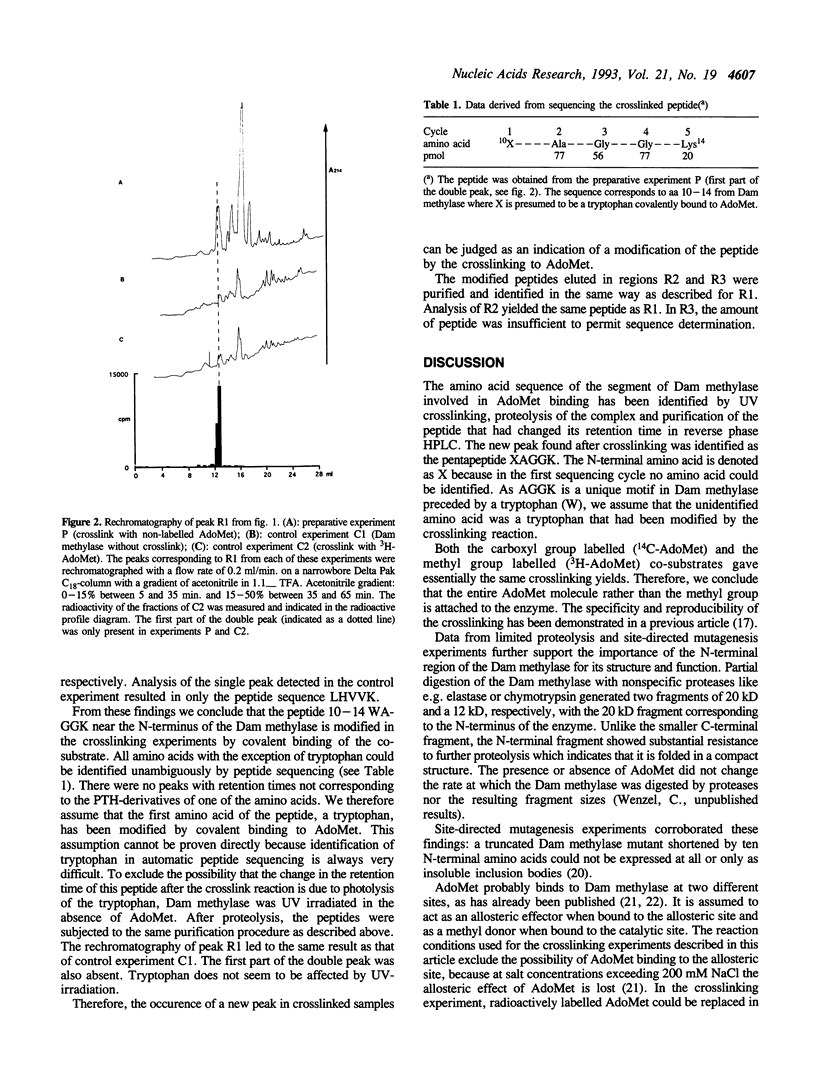
DNA adenine methyltransferase (Dam methylase) has been crosslinked with its cofactor S-adenosyl methionine (AdoMet) by UV irradiation. About 3% of the enzyme was radioactively labelled after the crosslinking reaction performed either with (methyl-3H)-AdoMet or with (carboxy-14C)-AdoMet. Radiolabelled peptides were purified after trypsinolysis by high performance liquid chromatography in two steps. They could not be sequenced due to radiolysis. Therefore we performed the same experiment using non-radioactive AdoMet and were able to identify the peptide modified by the crosslinking reaction by comparison of the separation profiles obtained from two analytical control experiments performed with 3H-AdoMet and Dam methylase without crosslink, respectively. This approach was possible due to the high reproducibility of the chromatography profiles. In these three experiments only one radioactively labelled peptide was present in the tryptic digestions of the crosslinked enzyme. Its sequence was found to be XA-GGK, corresponding to amino acids 10-14 of Dam methylase. The non-identified amino acid in the first sequence cycle should be a tryptophan, which is presumably modified by the crosslinking reaction. The importance of this region near the N-terminus for the structure and function of the enzyme was also demonstrated by proteolysis and site-directed mutagenesis experiments.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barras F., Marinus M. G. Arrangement of Dam methylation sites (GATC) in the Escherichia coli chromosome. Nucleic Acids Res. 1988 Oct 25;16(20):9821–9838. doi: 10.1093/nar/16.20.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barras F., Marinus M. G. The great GATC: DNA methylation in E. coli. Trends Genet. 1989 May;5(5):139–143. doi: 10.1016/0168-9525(89)90054-1. [DOI] [PubMed] [Google Scholar]
- Bergerat A., Guschlbauer W., Fazakerley G. V. Allosteric and catalytic binding of S-adenosylmethionine to Escherichia coli DNA adenine methyltransferase monitored by 3H NMR. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6394–6397. doi: 10.1073/pnas.88.15.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat A., Guschlbauer W. The double role of methyl donor and allosteric effector of S-adenosyl-methionine for Dam methylase of E. coli. Nucleic Acids Res. 1990 Aug 11;18(15):4369–4375. doi: 10.1093/nar/18.15.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye E. A turnstile for initiation of DNA replication. Trends Cell Biol. 1991 Nov;1(5):107–109. doi: 10.1016/0962-8924(91)90100-n. [DOI] [PubMed] [Google Scholar]
- Braun R. E., Wright A. DNA methylation differentially enhances the expression of one of the two E. coli dnaA promoters in vivo and in vitro. Mol Gen Genet. 1986 Feb;202(2):246–250. doi: 10.1007/BF00331644. [DOI] [PubMed] [Google Scholar]
- Campbell J. L., Kleckner N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990 Sep 7;62(5):967–979. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991 Nov 19;30(46):11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- Geier G. E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J Biol Chem. 1979 Feb 25;254(4):1408–1413. [PubMed] [Google Scholar]
- Guyot J. B., Grassi J., Hahn U., Guschlbauer W. The role of the preserved sequences of Dam methylase. Nucleic Acids Res. 1993 Jul 11;21(14):3183–3190. doi: 10.1093/nar/21.14.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Brooks J. E., Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978 Dec 15;126(3):367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- Herman G. E., Modrich P. Escherichia coli dam methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1982 Mar 10;257(5):2605–2612. [PubMed] [Google Scholar]
- Ho D. K., Wu J. C., Santi D. V., Floss H. G. Stereochemical studies of the C-methylation of deoxycytidine catalyzed by HhaI methylase and the N-methylation of deoxyadenosine catalyzed by EcoRI methylase. Arch Biochem Biophys. 1991 Feb 1;284(2):264–269. doi: 10.1016/0003-9861(91)90294-s. [DOI] [PubMed] [Google Scholar]
- Hülsmann K. H., Quaas R., Georgalis Y., Saenger W., Hahn U. High-level expression of a semisynthetic dam gene in Escherichia coli. Gene. 1991 Feb 1;98(1):83–88. doi: 10.1016/0378-1119(91)90107-m. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Timinskas A., Menkevicius S., Butkienè D., Butkus V., Janulaitis A. Sequence motifs characteristic of DNA[cytosine-N4]methyltransferases: similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res. 1989 Dec 11;17(23):9823–9832. doi: 10.1093/nar/17.23.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Cheng X., Pflugrath J. W., Roberts R. J. Purification, crystallization, and preliminary X-ray diffraction analysis of an M.HhaI-AdoMet complex. Biochemistry. 1992 Sep 15;31(36):8648–8653. doi: 10.1021/bi00151a035. [DOI] [PubMed] [Google Scholar]
- Lahue R. S., Modrich P. Methyl-directed DNA mismatch repair in Escherichia coli. Mutat Res. 1988 Mar;198(1):37–43. doi: 10.1016/0027-5107(88)90037-1. [DOI] [PubMed] [Google Scholar]
- Landoulsi A., Hughes P., Kern R., Kohiyama M. dam methylation and the initiation of DNA replication on oriC plasmids. Mol Gen Genet. 1989 Apr;216(2-3):217–223. doi: 10.1007/BF00334359. [DOI] [PubMed] [Google Scholar]
- Landoulsi A., Malki A., Kern R., Kohiyama M., Hughes P. The E. coli cell surface specifically prevents the initiation of DNA replication at oriC on hemimethylated DNA templates. Cell. 1990 Nov 30;63(5):1053–1060. doi: 10.1016/0092-8674(90)90508-c. [DOI] [PubMed] [Google Scholar]
- Lauster R. Evolution of type II DNA methyltransferases. A gene duplication model. J Mol Biol. 1989 Mar 20;206(2):313–321. doi: 10.1016/0022-2836(89)90481-6. [DOI] [PubMed] [Google Scholar]
- Lauster R., Kriebardis A., Guschlbauer W. The GATATC-modification enzyme EcoRV is closely related to the GATC-recognizing methyltransferases DpnII and dam from E. coli and phage T4. FEBS Lett. 1987 Aug 10;220(1):167–176. doi: 10.1016/0014-5793(87)80897-9. [DOI] [PubMed] [Google Scholar]
- Marinus M. G. DNA methylation in Escherichia coli. Annu Rev Genet. 1987;21:113–131. doi: 10.1146/annurev.ge.21.120187.000553. [DOI] [PubMed] [Google Scholar]
- Messer W., Noyer-Weidner M. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell. 1988 Sep 9;54(6):735–737. doi: 10.1016/s0092-8674(88)90911-7. [DOI] [PubMed] [Google Scholar]
- Möller W., Amons R. Phosphate-binding sequences in nucleotide-binding proteins. FEBS Lett. 1985 Jul 1;186(1):1–7. doi: 10.1016/0014-5793(85)81326-0. [DOI] [PubMed] [Google Scholar]
- Ogden G. B., Pratt M. J., Schaechter M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell. 1988 Jul 1;54(1):127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- Pogolotti A. L., Jr, Ono A., Subramaniam R., Santi D. V. On the mechanism of DNA-adenine methylase. J Biol Chem. 1988 Jun 5;263(16):7461–7464. [PubMed] [Google Scholar]
- Reich N. O., Everett E. A. Identification of peptides involved in S-adenosylmethionine binding in the EcoRI DNA methylase. Photoaffinity laveling with 8-azido-S-adenosylmethionine. J Biol Chem. 1990 May 25;265(15):8929–8934. [PubMed] [Google Scholar]
- Reich N. O., Maegley K. A., Shoemaker D. D., Everett E. Structural and functional analysis of EcoRI DNA methyltransferase by proteolysis. Biochemistry. 1991 Mar 19;30(11):2940–2946. doi: 10.1021/bi00225a030. [DOI] [PubMed] [Google Scholar]
- Som S., Friedman S. Identification of a highly conserved domain in the EcoRII methyltransferase which can be photolabeled with S-adenosyl-L-[methyl-3H]methionine. Evidence for UV-induced transmethylation of cysteine 186. J Biol Chem. 1991 Feb 15;266(5):2937–2945. [PubMed] [Google Scholar]
- Sternberg N. Evidence that adenine methylation influences DNA-protein interactions in Escherichia coli. J Bacteriol. 1985 Oct;164(1):490–493. doi: 10.1128/jb.164.1.490-493.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaramaiah K., Simms S. A. Photolabeling of CheR methyltransferase with S-adenosyl-L-methionine (AdoMet). Studies on the AdoMet binding site. J Biol Chem. 1992 Apr 25;267(12):8636–8642. [PubMed] [Google Scholar]
- Takata Y., Fujioka M. Identification of a tyrosine residue in rat guanidinoacetate methyltransferase that is photolabeled with S-adenosyl-L-methionine. Biochemistry. 1992 May 5;31(17):4369–4374. doi: 10.1021/bi00132a030. [DOI] [PubMed] [Google Scholar]
- Wenzel C., Moulard M., Løbner-Olesen A., Guschlbauer W. Crosslinking of Dam methyltransferase with S-adenosyl-methionine. FEBS Lett. 1991 Mar 11;280(1):147–151. doi: 10.1016/0014-5793(91)80224-q. [DOI] [PubMed] [Google Scholar]
- Wu J. C., Santi D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4778–4786. [PubMed] [Google Scholar]


