Abstract
The Drosophila mutant methuselah (mth) was identified from a screen for single gene mutations that extended average lifespan. Mth mutants have a 35% increase in average lifespan and increased resistance to several forms of stress, including heat, starvation, and oxidative damage. The protein affected by this mutation is related to G protein-coupled receptors of the secretin receptor family. Mth, like secretin receptor family members, has a large N-terminal ectodomain, which may constitute the ligand binding site. Here we report the 2.3-Å resolution crystal structure of the Mth extracellular region, revealing a folding topology in which three primarily β-structure-containing domains meet to form a shallow interdomain groove containing a solvent-exposed tryptophan that may represent a ligand binding site. The Mth structure is analyzed in relation to predicted Mth homologs and potential ligand binding features.
The sequence of the protein affected by the methuselah (mth) mutation predicts that it contains a 195-residue extracellular domain followed by seven transmembrane domains related to the membrane domains of G protein-coupled receptors (GPCRs) (1). GPCRs are key transducers of biological signals, and the critical roles they play in eukaryotic organisms are reflected in the diverse range of ligands that signal through GPCRs, including small molecules, peptides, proteins, and light (2). The membrane domains of Mth and Mth relatives identified in the Drosophila genome (3) are most closely related to membrane regions of the B family of GPCRs (also known as family-2 GPCRs), which includes peptide hormone receptors that transduce signals via activation of heterotrimeric G proteins (see Fig. 4, which is published as supplemental material on the PNAS web site, www.pnas.org). Phylogenetic analysis places Mth-related proteins in a separate subgroup of GPCR family B (Fig. 1A). Family B also includes GPCR-like proteins containing extracellular epidermal growth factor-like modules, Ig superfamily domains, or cadherin domains (cell adhesion family; Fig. 1A) (4).
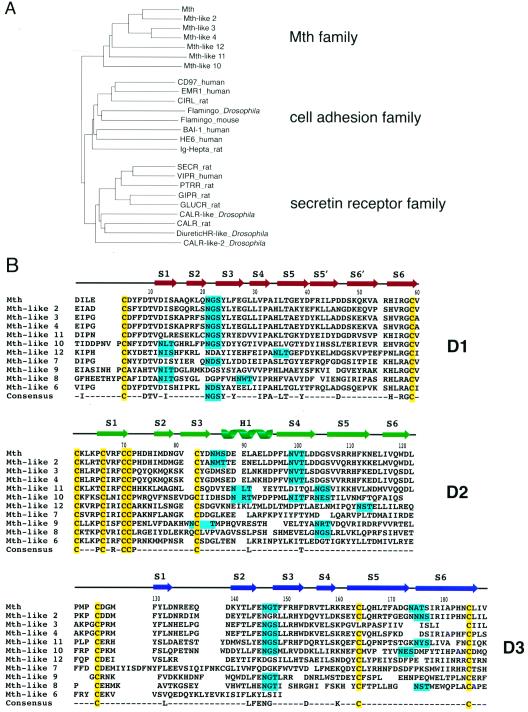
Figure 1.
Relation of Mth to family B GPCRs and other Mth homologs. (A) Phylogenetic relationship of transmembrane regions of GPCR family B proteins. A phylogenetic tree was generated by using the neighbor-joining method based on an alignment of transmembrane domains generated with the clustalw program (25). The human β-2 adrenergic receptor (not shown) served as the outgroup. A subfamily consisting of ≈10 Mth-like proteins identified from the Drosophila genome project clusters with Mth. CIRL, calcium-independent receptor of α-latrotoxin; BAI-1, brain-specific angiogenesis inhibitor 1; HE6, human epididymal gene product 6; SECR, secretin receptor; VIPR, vasoactive intestinal peptide receptor; PTRR, parathyroid hormone-related peptide receptor; GIPR, gastric inhibitory peptide receptor; GLUCR, glucagon receptor; CALR, calcitonin receptor. (B) Sequence alignment of the Mth ectodomain with predicted homologs from the Drosophila genome. Residue numbering begins at the first residue of the mature protein. Crystallographically determined secondary-structural elements are shown above the sequences. The 10 cysteines that form five disulfide bonds in the Mth ectodomain are highlighted in yellow, and potential N-linked glycosylation sites are highlighted in cyan.
GPCR family B members that have been experimentally characterized fall into two categories: (i) those that bind and transmit signals from peptides (e.g., secretin, insect diuretic hormone, calcitonin), and (ii) those that appear to have cell adhesion roles (CD97 and Flamingo) (4). In both cases, the extracellular domains of these receptors have been implicated in their function, either providing hormone ligand specificity or facilitating cell adhesion (e.g., the cadherin domains in Flamingo). Although the ectodomains of both the hormone receptor GPCRs and Mth proteins show some similarities in that they are about the same size and each contain their own pattern of conserved cysteines, the Mth extracellular domain does not share significant sequence similarity to any GPCR family B members or other known proteins. Because the Mth ectodomain does not include the multidomain stalk structure common to the GPCRs with known cell adhesion roles (Flamingo, CD97), a cell adhesion function seems unlikely. Instead, the Mth ectodomain could represent a ligand binding region, by analogy to several hormone receptor GPCRs whose isolated ectodomains can function in ligand binding. For example, recombinant forms of the ectodomains of the secretin receptor (5), the glucagon-like peptide-1 receptor (6), and the pituitary adenylate cyclase activating peptide receptor (7) form high-affinity peptide hormone binding sites and/or inhibit activation of the full-length form of the corresponding receptor.
As a first step in characterizing the potential ligand binding properties of Mth and in understanding the role of Mth and its relatives in affecting aging and stress resistance, we solved the crystal structure of the Mth ectodomain. The structure reveals a folding topology likely to be conserved in Mth-related proteins and a potential ligand binding site.
Materials and Methods
Protein Expression and Purification.
A construct encoding a soluble portion of Mth (including the Mth signal sequence and residues 1–195 of the mature protein) with a C-terminal His tag was subcloned into the baculovirus transfer vector pVL1392 (PharMingen). Mth was purified from supernatants of baculovirus-infected High 5 cells, buffer exchanged to 50 mM phosphate at pH 8.0, followed by Ni-NTA chromatography (Ni-NTA superflow, Qiagen, Chatsworth, CA). Protein from an imidazole elution was further purified by gel filtration chromatography. NH2-terminal protein sequencing yielded the sequence DILEXDYFDTV.
Crystallization and Data Collection.
Crystals (space group P212121, a = 68.9 Å, b = 73.0 Å, c = 115.1 Å; two molecules per asymmetric unit) of Mth were grown at 22°C in hanging drops by combining 4 μl of protein solution [Mth (20 mg/ml) and 20 mM Tris at pH 7.4] with 2 μl of precipitant solution (1.6 M Li2SO4 and 0.1 M Hepes at pH 7.5). Before data collection, crystals were transferred to a cryoprotectant solution (1.6 M Li2SO4, 0.1 M Hepes at pH 7.5, and 12.5% glycerol). Initial native data to 2.6 Å were collected at −150°C from a single crystal at Stanford Synchrotron Radiation Laboratory (SSRL, Stanford, CA) beamline 7–1. A selenomethionine derivative data set was collected at SSRL beamline 9–2. A multiwavelength anomalous dispersion (MAD) data set to 2.3 Å was collected on the lead derivative at Brookhaven beamline X8C.
Structure Determination, Phasing, and Refinement.
Data were processed and scaled with denzo and scalepack (8). Heavy atom refinement of four lead and four gadolinium sites was done with sharp (9), yielding phases to 3.0 Å with a mean figure of merit of 0.43. A solvent-flattened map calculated from multiple isomorphous replacement including anomalous scattering phases was skeletonized and used to calculate a noncrystallographic symmetry (NCS) operator. A 3.0-Å NCS-averaged, solvent-flattened map was used for initial model building with o (10). Map interpretation was aided by a native anomalous Fourier map to locate disulfides and an anomalous Fourier by using the selenomethionine derivative to locate the four methionine residues. Refinement was carried out on the lead λ1 dataset (“native” II) in place of the native data (native I) by using the simulated annealing and minimization protocols in cns (11) with bulk solvent and anisotropy corrections, refinement of individual B factors, and tight NCS restraints (200 kcal/mol⋅ Å2). The NCS operator was broken into two operators covering D1 (residues 1–41 and 55–63) and D2D3 (residues 86–185). The model includes residues 1–188, four lead atoms, disulfide bonds between cysteine residues 5 and 59, 61 and 66, 70 and 164, 71 and 82, and 126 and 185, and ordered N-linked carbohydrates attached to Asn-21, Asn-99, and Asn-146. Ramachandran plot statistics are as defined by procheck (12). Figures were made with molscript (13) and RASTER-3D (14).
To verify the structural difference between NCS molecules, the final model was subjected to simulated annealing and refinement with a single NCS operator. Both Rcryst and Rfree were 0.8% higher with a single NCS operator instead of two operators. The NCS-related regions are very similar (rms deviations of 0.04 Å for D1 and 0.06 Å for D2D3). When the two molecules are aligned by using the D2D3 NCS operator, the D1 domains have an rms deviation of 1.1 Å.
Analysis of Mth Homologs.
The predicted homologs Mth-like 1 and Mth-like 5 possess significant sequence similarity to Mth only in their transmembrane domains. One homolog, Mth-like 6, consists only of a truncated form of the Mth ectodomain. We identified two additional full-length Mth homologs in the Drosophila genome: Mth-like 11 (map position 3R;86A, in GenBank accession no. AE003687 between positions 16240 and 17991) and Mth-like 12 (map position 3R;87E, in GenBank accession no. AE003699 between positions 207030 and 208827). The transmembrane regions of Mth-like 3 and Mth-like 4 were not included in the first release of predicted protein sequences from the Drosophila genome (3) and were added to Fig. 1B after manual inspection revealed additional exons with homology to the Mth transmembrane region.
Results and Discussion
Overall Structure and Domain Organization.
The Mth ectodomain was expressed with a baculovirus expression system and purified from the supernatants of infected insect cells. The crystal structure was determined to 2.3 Å by multiple isomorphous replacement including anomalous scattering and MAD (Tables 1 and 2). The Mth ectodomain has a relatively compact, β-sheet-rich fold with five disulfide bonds (Fig. 2) that does not closely resemble that of any protein of known structure [i.e., no similar folds were found by using the dali (15) or vast (16) database search programs]. The structure can be divided into three domains. The first domain (D1; residues 1–60) and third domain (D3; residues 123–188) share a similar fold consisting of two three-stranded β-sheets, whereas the middle domain (D2, residues 61–122), which contains three two-stranded β-sheets and a short α-helix, wraps around the third domain to form a superdomain (D2D3). There are no backbone hydrogen bonds between D1 and the rest of the protein and the buried surface area between D1 and D2D3 [1,460 Å2, calculated with cns (11) using a 1.4-Å probe radius] is significantly smaller than that between D3 and D1-D2 (2,750 Å2), consistent with the assignment of D2D3 as a superdomain. Comparison of the two Mth ectodomains in the crystallographic asymmetric unit cell suggests the potential for flexibility at the interface between D1 and the D2D3 superdomain. A small, but measurable, difference is observed between the relative positions of the D1 and D2D3 domains in the two Mth ectodomains. The structural difference is verified by an improvement in the crystallographic R factors when the interdomain angle is allowed to vary compared with when the two Mth molecules are restrained to the same domain arrangement during refinement. The flexibility between D1 and D2D3 raises the possibility that ligand binding induces a similar conformational change, which may be relevant for the signaling mechanism of this receptor.
Table 1.
Data collection and heavy-atom phasing for Mth
| Data set | λ (Å) | Resolution (Å) | Complete (%)* | Rmerge† (%) | I/σI | rms fh/E‡ |
|---|---|---|---|---|---|---|
| Native I | 1.54 | 2.6 | 100.0 (99.9) | 5.6 (34.1) | 23.6 (3.5) | |
| Pb(CH3)3OAc | 1.54 | 2.8 | 98.8 (93.0) | 6.9 (46.3) | 13.3 (2.4) | 1.4 |
| GdCl3 | 1.54 | 3.4 | 98.9 (99.5) | 13.0 (43.4) | 6.5 (1.8) | 0.9 |
| SeMet | 0.979 | 2.3 | 99.4 (99.9) | 7.7 (32.8) | 20.9 (4.2) | N.D. |
| Pb(CH3)3OAc MAD data | ||||||
| λ1 peak/Nat. II | 0.9490 | 2.3 | 99.5 (98.6) | 6.9 (33.8) | 15.5 (2.8) | 2.3 |
| λ2 inflection | 0.9504 | 2.3 | 99.5 (99.3) | 6.7 (36.8) | 15.5 (2.6) | 1.9 |
| λ3 remote | 0.8610 | 2.3 | 98.1 (95.3) | 6.8 (41.5) | 13.4 (2.1) | 2.1 |
Complete represents (number of independent reflections)/total theoretical number.
Rmerge (I) = [Σ|I(i) − 〈I(h)〉 |/ΣI(i)], where I(i) is the ith observation of the intensity of the hkl reflection and 〈I〉 is the mean intensity from multiple measurements of the hkl reflection.
rms fh/E represents phasing power, where fh is the heavy-atom structure factor amplitude and E is the residual lack of closure error.
Table 2.
Refinement statistics for Mth
| Resolution (Å) | 20.0–2.3 |
| Theoretical refl. in resol. range | 26,430 |
| Reflections in working set |F| > 0 | 25,022 |
| Reflections in test set |F| > 0 | 1,283 (5%) |
| Rfree (%)* | 22.7 |
| Rcryst (%)† | 21.2 |
| rms deviations from ideality | |
| Bond lengths (Å) | 0.008 |
| Bond angles (deg) | 1.40 |
| Number of nonhydrogen atoms: | |
| Protein | 3,058 |
| Water | 201 |
| Carbohydrate | 98 |
| Pb and sulfate | 14 |
| Ramachandran plot | |
| Most favored (%) | 88.5 |
| Additional allowed (%) | 11.2 |
| Generously allowed (%) | 0.3 |
| Disallowed (%) | 0.0 |
Rfree is calculated over reflections in a test set not included in atomic refinement.
Rcryst (F) = Σh ∥Fobs(h)| − |Fcalc(h)∥/Σh|Fobs(h)|, where |Fobs(h)| and |Fcalc(h)| are the observed and calculated structure factor amplitudes for the hkl reflection.
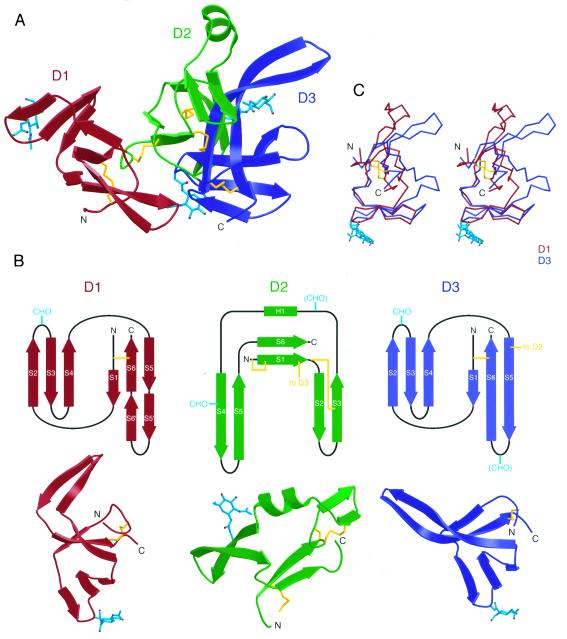
Figure 2.
Structure of the Mth ectodomain. (A) Ribbon diagram of the Mth structure (D1, red; D2, green; D3, blue). Ordered N-linked carbohydrates are shown in cyan in ball-and-stick representation and disulfide bonds are yellow. (B) (Upper) Topology diagrams for three extracellular domains of Mth. Potential N-linked glycosylation sites are labeled CHO, with parentheses denoting N-linked sites in which ordered carbohydrates were not observed. (Lower) Ribbon diagrams of the three domains of Mth. A disulfide bond (yellow) and an N-linked carbohydrate (cyan) are located at corresponding positions in D1 and D3. (C) Stereoview of a superposition of D1 (in red) and D3 (in blue) made by aligning the Cα atoms of the S2-S3-S4 β-sheet.
The D3 portion of the D2D3 superdomain shares a similar folding topology and several common structural features with D1 (Fig. 2B). First, each domain includes two disulfide-linked cysteines at analogous positions (Cys-5 and Cys-59 in D1 and Cys-126 and Cys-185 in D3) that connect the beginning and end of each domain. Second, two of the five potential N-linked glycosylation sites occur at corresponding positions in D1 and D3 (Asn-21 and Asn-146) and ordered carbohydrate is visible at both sites. Although D1 and D3 share these structural features, they are related by <10% sequence identity (based on a structure-based sequence alignment) and superimpose well only over the S2-S3-S4 sheet (29 of 62 residues align with an rms deviation of 1.32 Å calculated with a 3.8-Å cutoff distance) (Fig. 2C). One significant difference between D1 and D3 is that the long S5-S6 β-stand pair in D3 is broken into two segments in D1 (near residues 40 and 53), which causes these strands to bend in such a way as to make contact with the D2D3 superdomain.
Potential Ligand Binding Site.
Between the D1 and D2D3 domains is a shallow groove containing a solvent exposed tryptophan, Trp-120, the only tryptophan residue in the Mth ectodomain (Fig. 3A). Receptor-ligand interfaces often bury otherwise exposed hydrophobic side chains, which contribute much of the free energy of binding (see discussion and references cited in ref. 17). For example, solvent exposed tryptophans in FcRn and HFE are critical for their interactions with binding partners (17, 18). Hence, the region near Mth Trp-120 may be suitable for binding a protein or peptide ligand or a hydrophobic small molecule. Localization of a ligand binding site in an interdomain groove also is not unprecedented: for example, the ligand binding domain of the ionotropic glutamate receptor, also a bilobed protein that bears a superficial resemblance to the Mth ectodomain structure, includes a shallow groove that serves as the ligand binding site (19).
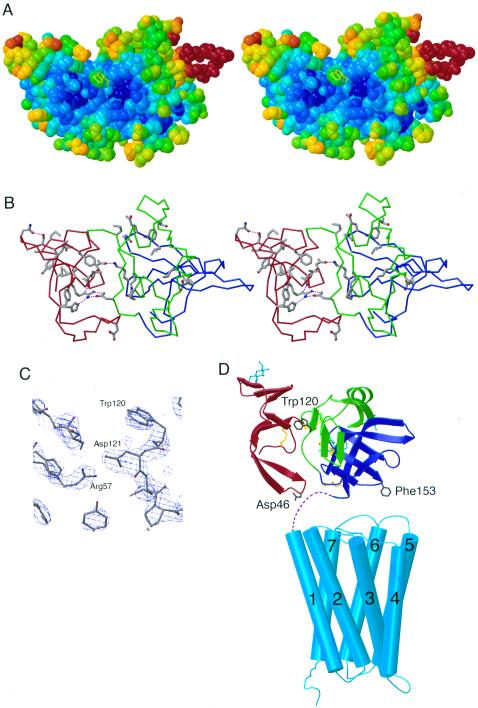
Figure 3.
Possible ligand binding site on Mth. (A) Stereoview of a space-filling model of Mth showing the shallow groove between D1 and D2D3. Atoms are colored according to their temperature factor, with blue and red corresponding to low (relatively well-ordered) and high temperature factors, respectively. Trp-120 (yellow), suggested to be at a ligand binding surface (see text), has a solvent accessible surface area of 126 Å2 [calculated by using a 1.4-Å probe radius with areaimol (26)] compared with 238 Å2 for a fully exposed tryptophan. Examples of protein–protein complexes including critical tryptophan residues at the interface include HFE/transferrin receptor (HFE Trp-81; 115 Å2; second most exposed tryptophan in HFE) (27) and FcRn/Fc (FcRn Trp-133; 136 Å2; third most exposed tryptophan in FcRn) (17). Other receptor-ligand interfaces that include surface-exposed hydrophobic side chains on one of the binding partners are discussed in ref. 17 and references therein. (B) Stereoview of Mth showing residues that are greater than 70% conserved in known Drosophila Mth homologs (excluding conserved cysteines). Asp-121 (conserved in five of the Mth-like proteins and conservatively substituted for glutamate in three others) is involved in an interdomain hydrogen bonds (purple dotted lines) with Arg-57 (conserved in all Mth homologs). (C) Experimental electron density map (MAD solvent-flattened map contoured at 2 σ) in the region near Trp-120. (D) Model for the structure of intact Mth showing relative sizes of Mth ectodomain and the transmembrane region. The Mth transmembrane region is represented by the structure of rhodopsin (21), with adjustments in loop regions to reflect differences in loop lengths between rhodopsin and Mth. Dashed lines represent the seven residues following the Mth ectodomain C terminus and the predicted start of the first transmembrane helix. The Mth ectodomain is oriented such that the C terminus of the model derived from the crystal structure is closest to the membrane. In this orientation, the interdomain groove and Trp-120 are accessible for ligand binding, and two regions containing residues (Asp-46 and Phe-153) suggested to interact with the extracellular face of the seven pass transmembrane domain are positioned near the interhelical loops.
Ten homologs of Mth have been predicted from the Drosophila genome (3, 20). Most of these are organized in the same manner as Mth: an NH2-terminal ectodomain followed by a seven-pass transmembrane domain. The NH2-terminal domains of the Mth-related proteins share between 27% and 65% sequence identity with the Mth ectodomain, suggesting that they will fold into similar tertiary structures. The 10 cysteines that form five disulfide bonds in Mth are conserved in all but one Mth homolog (Mth-like 3, in which a leucine substitutes for the cysteine in the strand S5 region of D3). Several of the N-linked carbohydrate sites also are conserved in many of the Mth-related proteins, particularly the analogous sites in D1 and D3 located between strands S2 and S3. In Fig. 3B, the noncysteine residues that are well-conserved (present in at least 70% of the Mth homologs) are highlighted on the structure of the Mth ectodomain. A majority of these residues are in D1, likely reflecting the greater constraints required to maintain the fold of this small domain. In addition, some of the interdomain interactions that stabilize the Mth fold are conserved in the Mth homologs. For example, conserved residues (D2 Asp-121, D1 His-55, and D1 Thr-36) surround and form hydrogen bonds to D1 Arg-57. The interaction between the side chains of Arg-57 and Asp-121 is one of the most significant between D1 and D2D3 (Fig. 3 B and C), and conservation of this interaction suggests that D1 and D2D2 will have a common arrangement in the Mth homologs. Trp-120 is conserved in only one of the Mth homologs (Mth-like 2) (Fig. 1B), thus the proposal that this residue forms part of the ligand binding site implies that the Mth homologs bind different ligands.
In addition to a ligand interaction surface, another region of the Mth ectodomain that may be functionally important is the region contacting the extracellular face of the seven-pass transmembrane domain, potentially in such a way as to signal ligand binding. In the recently determined crystal structure of the GPCR rhodopsin (21), the extracellular NH2-terminal region (residues 1–35) makes contact with all three of the extracellular interhelical loops, thus the Mth ectodomain also may contact loops between transmembrane helices. Because there are only seven residues between the C terminus of the Mth ectodomain observed in the structure and the predicted beginning of the first hydrophobic transmembrane region, the region near the Mth ectodomain C terminus has the potential to interact with loop regions between the transmembrane regions. There are three loops in this region: between strands S3 and S4 in D3 (including Phe-153 and conserved residue Asp-154), between S5′ and S6′ in D1 (including conserved residue Asp-46), and preceding S1 in D3 (around Gly-128) (Fig. 3D). The occurrence of conserved residues within these loops suggests that these regions may be involved in positioning the ectodomain on the transmembrane region and/or signaling ligand binding to the transmembrane region. When the region of Mth containing these loops and the C terminus of the Mth ectodomain is oriented on a model of the seven-pass transmembrane portion, the ectodomain is positioned such that the interdomain cleft containing Trp-120 is oriented away from the membrane, consistent with the suggestion that it represents a ligand binding site (Fig. 3D).
Conclusions
The structure of the Mth ectodomain represents one of only a few available three-dimensional structures of GPCRs. Previous structural studies of GPCRs include the 2.8-Å crystal structure of rhodopsin (21), and NMR structures of the NH2 terminus (residues 1–47) of the cholecystokinin A receptor (22) and a 31-aa fragment of the extracellular domain of the parathyroid hormone receptor (23). There is no structural similarity of these extracellular domains to each other or to the Mth ectodomain. Further structural studies of GPCRs, combined with the structural characterizations of heterotrimeric G protein complexes (24), will facilitate obtaining a complete picture of the molecular events leading from ligand binding to G protein activation.
Supplementary Material
Acknowledgments
We thank Yi-Jyun Lin and Laurent Seroude for helpful discussions, Duilio Cascio for assistance in MAD data collection, and members of the Bjorkman lab for critical reading of the manuscript. This work is supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation, Grant DRG-1445 (A.P.W.), the Howard Hughes Medical Institute (P.J.B.), the Ellison Medical Foundation (S.B.), the National Science Foundation (S.B.), and the National Institutes of Health (S.B.).
Abbreviations
- GPCR
G protein-coupled receptor
- Mth
Methuselah
- MAD
multiwavelength anomalous dispersion
- NCS
noncrystallographic symmetry.
Note Added in Proof.
Also recently reported was the crystal structure of the extracellular domain of the metabotropic glutamate receptor (28).
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1FJR).
References
- 1.Lin Y-J, Seroude L, Benzer S. Science. 1998;282:943–946. doi: 10.1126/science.282.5390.943. [DOI] [PubMed] [Google Scholar]
- 2.Bockaert J, Pin J P. EMBO J. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 4.Stacey M, Lin H-H, Gordon S, McKnight A J. Trends Biochem Sci. 2000;25:284–289. doi: 10.1016/s0968-0004(00)01583-8. [DOI] [PubMed] [Google Scholar]
- 5.Chow B K C. Recept Signal Transduct. 1997;7:143–150. [PubMed] [Google Scholar]
- 6.Wilmen A, Goke B, Goke R. FEBS Lett. 1996;398:43–47. doi: 10.1016/s0014-5793(96)01214-8. [DOI] [PubMed] [Google Scholar]
- 7.Cao Y J, Gimpl G, Fahrenholz F. Biochem Biophys Res Commun. 1995;212:673–680. doi: 10.1006/bbrc.1995.2021. [DOI] [PubMed] [Google Scholar]
- 8.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 9.De La Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 10.Jones T A, Kjeldgaard M. Methods Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 11.Brünger A T, Adams P D, Clore G M, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, Read R J, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 12.Laskowski R A, McArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 13.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 14.Merritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 15.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 16.Gibrat J-F, Madej T, Bryant S H. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 17.Vaughn D E, Milburn C M, Penny D M, Martin W L, Johnson J L, Bjorkman P J. J Mol Biol. 1997;274:597–607. doi: 10.1006/jmbi.1997.1388. [DOI] [PubMed] [Google Scholar]
- 18.Lebrón J A, Bjorkman P J. J Mol Biol. 1999;289:1109–1118. doi: 10.1006/jmbi.1999.2842. [DOI] [PubMed] [Google Scholar]
- 19.Armstrong N, Sun Y, Chen G Q, Gouaux E. Nature (London) 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 20.Brody T, Cravchik A. J Cell Biol. 2000;150:F83–F88. doi: 10.1083/jcb.150.2.f83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palczewski K, Kumasaka T, Hori T, Behnke C A, Motoshima H, Fox B A, Trong I L, Teller D C, Okada T, Stenkamp R E, et al. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 22.Pellegrini M, Mierke D F. Biochemistry. 1999;38:14775–14783. doi: 10.1021/bi991272l. [DOI] [PubMed] [Google Scholar]
- 23.Pellegrini M, Bisello A, Rosenblatt M, Chorev M, Mierke D F. Biochemistry. 1998;37:12737–12743. doi: 10.1021/bi981265h. [DOI] [PubMed] [Google Scholar]
- 24.Sprang S R. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 25.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collaborative Computational Project No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 27.Bennett M J, Lebrón J A, Bjorkman P J. Nature (London) 2000;403:46–53. doi: 10.1038/47417. [DOI] [PubMed] [Google Scholar]
- 28.Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Nature (London) 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.