Abstract
The intracellular degradation of many proteins is mediated in an ATP-dependent manner by large assemblies comprising a chaperone ring complex associated coaxially with a proteolytic cylinder, e.g., ClpAP, ClpXP, and HslUV in prokaryotes, and the 26S proteasome in eukaryotes. Recent studies of the chaperone ClpA indicate that it mediates ATP-dependent unfolding of substrate proteins and directs their ATP-dependent translocation into the ClpP protease. Because the axial passageway into the proteolytic chamber is narrow, it seems likely that unfolded substrate proteins are threaded from the chaperone into the protease, suggesting that translocation could be directional. We have investigated directionality in the ClpA/ClpP-mediated reaction by using two substrate proteins bearing the COOH-terminal ssrA recognition element, each labeled near the NH2 or COOH terminus with fluorescent probes. Time-dependent changes in both fluorescence anisotropy and fluorescence resonance energy transfer between donor fluorophores in the ClpP cavity and the substrate probes as acceptors were measured to monitor translocation of the substrates from ClpA into ClpP. We observed for both substrates that energy transfer occurs 2–4 s sooner with the COOH-terminally labeled molecules than with the NH2-terminally labeled ones, indicating that translocation is indeed directional, with the COOH terminus of the substrate protein entering ClpP first.
The ClpAP proteolytic machine of Escherichia coli is one member of a diverse group of energy-dependent chaperone/protease complexes, including ClpXP and HslUV in prokaryotes and the 26S proteasome complex in eukaryotes (1–4). These complexes share a common architecture, in which a hexameric chaperone ring complex (e.g., ClpA, ClpX, HslU, or the base of the 19S regulatory particle) binds coaxially to a protease ring complex (e.g., ClpP, HslV, or the 20S proteasome), whose proteolytic active sites are located in a central chamber, sequestered from the bulk solution (5–10). The axial channels into the proteolytic chambers generally appear to be narrow, measuring ≈10 Å in diameter in the case of ClpP (11), and thus may only be wide enough to accommodate unfolded proteins. Therefore, substrate proteins apparently must be unfolded by the chaperone component of these complexes to be threaded into the protease for degradation. Consistently, recent in vitro studies of ClpA and ClpX demonstrate that these chaperones, members of the Hsp100 family, can mediate ATP-dependent unfolding of substrate proteins in isolation (12–15).
ClpA and ClpX recognize substrate proteins through specific amino acid “tag” sequences, usually found at one terminus of the protein (3, 4). The best-studied of these is the 11-residue, COOH-terminal tag specified by the ssrA gene, added to translationally arrested proteins (16–18). In the presence of ATP, but not nonhydrolyzable analogues, ClpA and ClpX mediate unfolding of ssrA-tagged proteins, e.g., a green fluorescent protein (GFP)-ssrA fusion (12–15). When such reactions were carried out in the presence of proteolytically active ClpP, the GFPssrA protein was degraded in a process that was apparently committed, because addition of a “trap” molecule to the reaction mixture, able to bind but not release nonnative GFPssrA, could not interfere with the degradation process (12). This implied that the initially recognized and unfolded GFPssrA was directly translocated into the ClpP cylinder for proteolysis. Additional studies with proteolytically inactive versions of ClpP supported this, in that the GFPssrA protein could be isolated in a stable complex with inactive ClpP, sequestered inside the ClpP cavity (E.W.-B., unpublished observations; see also refs. 14 and 15). In the case of ClpA, the translocation step has also been analyzed in isolation from recognition and unfolding, as chemically unfolded proteins, even ones devoid of a terminal recognition element, can become bound to ClpA in the absence of ATP (13, 14). This enabled the determination that translocation into ClpP depends on the presence of ATP. Here also, the translocation step could not be supported by nonhydrolyzable analogues, suggesting that ATP hydrolysis may be critical to conformational changes of ClpA that enable both unfolding and translocation actions.
The observations that a terminal peptide provides the recognition element for ClpA and that translocation occurs through a narrow passageway suggest that translocation could proceed in a preferred direction, that is, with either the NH2 or COOH terminus of the protein moving into the proteolytic chamber first. Here, we have examined translocation of ssrA-tagged substrate proteins through the ClpAP complex by using two independent fluorescence methods that permit real-time observation of substrate dynamics. Fluorescence anisotropy of substrates labeled with fluorescein near either terminus and fluorescence resonance energy transfer (FRET) between donor fluorophores in the ClpP cavity and the fluorescein-labeled substrates show that translocation indeed occurs in a directional manner.
Experimental Procedures
Proteins.
ClpA, ClpP, and miClpP were overproduced and purified as described (12). Cys-substituted variants of miClpP (F31C, L62C, and L139C), λ repressor (1–93) (16) (A21C and S93C), and cysteine-less T4 lysozyme (19) (E5C and T151C) were produced by oligonucleotide-directed mutagenesis, confirmed by DNA sequencing, overexpressed, and purified similarly to the parental proteins.
Gel Filtration.
Gel filtration was carried out on a Superose 12 column (Amersham Pharmacia) in Clp reaction buffer [50 mM N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonate), pH 7.5/0.3 M NaCl/20 mM MgCl2/10% glycerol]; 0.5 mM adenosine 5′-O-(3-thiotriphosphate) (ATPγS) or 1 mM ATP was added to the buffer where indicated.
Fluorescence.
Stopped-flow fluorescence anisotropy and FRET measurements were carried out with a Biologique 4-syringe stopped-flow device and two channel detection with instrumentation previously described (20, 21). The stopped-flow syringes and cuvette were thermostated at 25°C. For anisotropy experiments, the excitation wavelength was 485 nm, and emission was recorded in both the parallel (Iparallel) and perpendicular (Iperpendicular) orientations through long-pass (>500 nm) filters, as described. For FRET experiments, excitation was at 336 nm, and emission was recorded in both donor (nominally 400–500 nm) and acceptor (nominally >500 nm) emission regions, using a set of separation filters as described (21). Only the donor data are reported, but the acceptor data gave essentially the same results. Both studies were carried out in Clp reaction buffer, as above, with 1 mM ATPγS and 10 mM ATP, where indicated.
Anisotropy (r) was calculated according to the formula r = (Iparallel − Iperpendicular)/(Iparallel + 2 Iperpendicular). G factor correction and data processing were carried out as previously described (22). FRET efficiency was calculated from the ratio of the change in donor-side fluorescence when using an acceptor-labeled Fl-λRssrA or Fl-LYssrA (emissionD-A) to the fluorescence change in an otherwise identical reaction with unlabeled λRssrA or LYssrA (emissionD), according to the formula % efficiency = [1 − (emissionD-A/emissionD)] × 100. The donor-only changes were 10–30% positive, depending on the ClpP variant used.
Results
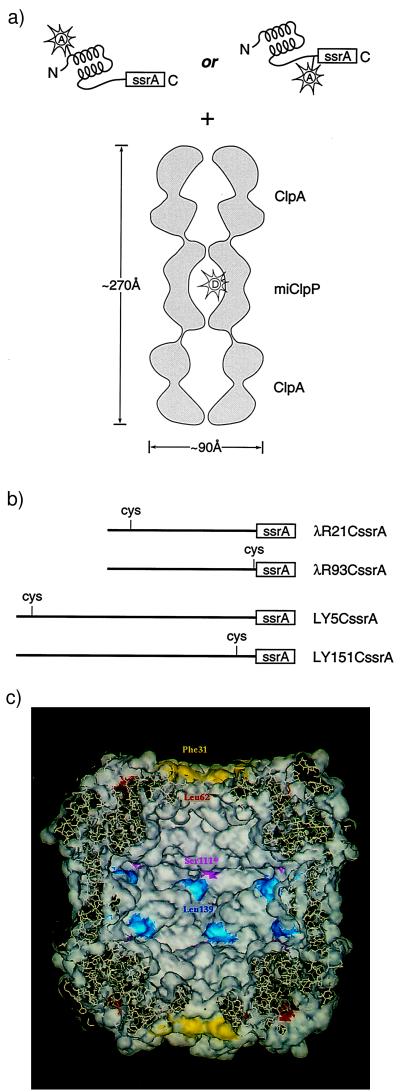
To generate substrates that could be recognized by the ClpA/ClpP system, sequences encoding the 11-residue ssrA tag were added at the 3′-end of the coding sequence for the DNA-binding domain (amino acids 1–93) of λ repressor (16) and at the 3′-end of the coding sequence for a T4 lysozyme variant with no native cysteines (19), producing λRssrA and LYssrA, respectively. To monitor the behavior of the termini of these ClpA substrates, we produced two versions of each, with a single cysteine near either the NH2 terminus or the COOH-terminal ssrA tag (Fig. 1). These sites were then modified with fluorescein-5-maleimide (Molecular Probes) to an extent of >90%.
Figure 1.
The ClpAP machine and the substrate proteins used in this study. (a) Schematic of the ssrA-tagged substrate proteins and the molecular architecture of the ClpAP complex, based on a cryo-electron microscopy reconstruction (5). The ClpA hexamer binds to one or both ends of the ClpP tetradecamer. The donor fluorophore (EDANS) used in FRET studies is represented in the ClpP cavity by the letter “D.” The acceptor fluorophores are represented on the native, ssrA-tagged substrates by the letter “A.” (b) Substrate proteins. Single cysteine versions of two different ssrA-tagged proteins were generated for this study. λR21CssrA and λR93CssrA are the NH2- and COOH-terminal single-Cys versions of the 93-residue DNA-binding domain of lambda repressor, fused at the COOH terminus to the ssrA tag. LY5CssrA and LY151CssrA are the NH2- and COOH-terminal single-Cys variants of T4 lysozyme, fused at the COOH terminus to a Ser-Gly-Gly linker and the ssrA tag. (c) A cut-away view of the inside surface of ClpP, showing the positions of the cysteine substitutions and the active site mutation. The surface rendering was produced by Insight II (Molecular Simulations, Waltham, MA), based on the ClpP crystal structure (Protein Data Bank: 1TYF) (11). The surface of the residues substituted with cysteine are colored yellow (Phe-31), red (Leu-62), and blue (Leu-139); the active site residue (Ser-111) is purple.
λRssrA Forms a Stable Complex with ClpA in ATPγS.
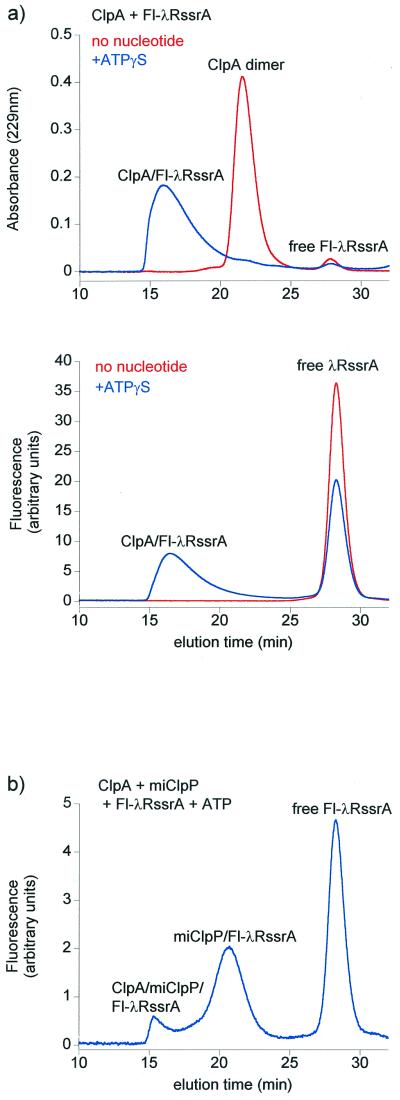
The modified λRssrA proteins, termed Fl-λR21CssrA and Fl-λR93CssrA, exhibited the same CD spectra as the unmodified parent molecule (not shown), suggesting that the structure of this α-helical DNA binding domain had not been perturbed by amino acid substitution and modification with the fluorophore. Both proteins exhibited the same kinetics of ClpA/ClpP-mediated proteolytic degradation as unlabeled λRssrA, as monitored by SDS/PAGE. Importantly, like the parental λRssrA, the modified proteins formed stable binary complexes with ClpA in the presence of the nonhydrolyzable ATP analogue, ATPγS, which supports the assembly of the ClpA hexamer but is unable to promote unfolding or translocation of substrate proteins (Fig. 2a). This enabled examination of the kinetic phases of translocation and degradation upon addition of ATP and ClpP, independent of a recognition step.
Figure 2.
Fluorescently labeled lambda repressor bearing the COOH-terminal 11-residue ssrA tag (Fl-λRssrA) forms a stable complex with ClpA hexamers in ATPγS and associates stably with mature, inactive ClpP (miClpP) following ATP-induced translocation. (a) Gel-filtration profiles of Fl-λR21CssrA and ClpA in the presence and absence of 0.5 mM ATPγS, monitored by absorbance at 229 nm (Upper) and by fluorescein fluorescence (Lower). ClpA (3 μM hexamer) was incubated with 2 μM Fl-λR21CssrA in Clp reaction buffer plus 1 mM ATPγS for 1 h. Reactions were analyzed by gel filtration on a Superose 12 column. (b) Gel-filtration profile of Fl-λR21CssrA encapsulated in miClpP after ATP-driven translocation from ClpA, monitored by fluorescein fluorescence. ClpA (2 μM), Fl-λR21CssrA (2 μM), and miClpP (8 μM) were incubated for 15 min at room temperature in Clp reaction buffer plus 10 mM ATP before Superose 12 chromatography in reaction buffer containing 1 mM ATP. The elution position of miClpP/Fl-λRssrA was confirmed by SDS/PAGE analysis of column fractions (not shown). The majority of ClpA was found in the ternary complex with miClpP and Fl-λRssrA (ClpA/miClpP/Fl-λRssrA).
Fluorescence Anisotropy Changes upon Translocation of Fl-λRssrA into ClpP.
In an initial experiment, when binary complexes between ClpA and the Fl-λRssrA proteins in ATPγS were rapidly mixed with ATP and ClpP, a transient rise in fluorescence anisotropy (r) from 0.21 to 0.24 was observed, followed by a steep fall to 0.10 (21C) or 0.07 (93C) (not shown). The latter phase was presumed to reflect proteolysis with the release of fluorescein-bearing peptides, a conclusion supported by the observation that incubation of either labeled substrate alone in solution with proteinase K produced a drop in anisotropy from 0.14 to 0.04.
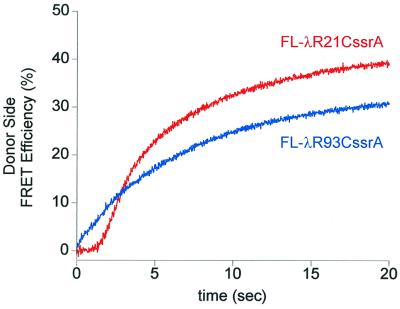
To remove the complicating effects of proteolysis and to limit observation to the unfolding and translocation phases, a proteolytically defective mature version of ClpP, miClpP, was produced, in which the active site serine (Ser-111) was changed to alanine, inactivating the protease, and the NH2-terminal 14-residue propeptide was deleted to avoid blockage of the ClpP channel (23). This mutant ClpP has been shown to associate normally with ClpA and to accept unfolded substrate proteins from it in the presence of ATP. For example, a GFPssrA fusion protein was recognized by ClpA and could be translocated into miClpP, where it remained trapped in an intact and nonfluorescent, apparently nonnative, conformation (E. W.-B., unpublished observations; see also refs. 14 and 15). Likewise, Fl-λR21CssrA was translocated into and stably maintained within the miClpP cavity (Fig. 2b). Thus, because translocation from ClpA into ClpP apparently does not depend on proteolysis, translocation could be examined in isolation. Upon addition of ATP and miClpP to the Fl-λRssrA/ClpA/ATPγS complexes, the fluorescence anisotropy of Fl-λR21CssrA or Fl-λR93CssrA was observed, as before, to rise from 0.22 or 0.23 to 0.25 or 0.26, respectively (Fig. 3). The anisotropy plateaued at the latter value, further indicating that the subsequent fall seen with intact ClpP had been because of proteolysis. Rapid kinetic analysis with stopped-flow mixing showed that there was a much faster rise of anisotropy for Fl-λR93CssrA than for Fl-λR21CssrA, with a t1/2 of 0.2 s for the COOH-terminal variant compared with 2.5 s for the NH2-terminal one (Fig. 3). It seems likely that the rise of anisotropy reports an altered environment of the fluorophores during the reaction. The slower rate of change of the anisotropy of the NH2-terminal probe suggests that this portion of the substrate is arriving more slowly and implies a COOH-terminal to NH2-terminal directionality of translocation.
Figure 3.
The rise in fluorescence anisotropy of substrate fluorescein probes occurs more rapidly when the probe is adjacent to the COOH-terminal ssrA tag. Binary complexes of each Fl-λRssrA and ClpA in ATPγS were purified by gel-filtration chromatography as in Fig. 2a, and reactions were initiated in the stopped-flow apparatus by addition of ClpP (1 μM tetradecamer) and ATP (10 mM) to 400 nM complex. Each trace is the sum of 10 runs. The final anisotropy value (0.25–0.26) is the same as the anisotropy of miClpP/Fl-λRssrA complexes purified by gel-filtration chromatography as in Fig. 2b (data not shown). The total fluorescence intensity change during this reaction was ≈5% positive. In controls without miClpP, the anisotropy decreased with time to ≈0.14, indicating the release of Fl-λRssrA from ClpA.
Kinetics of FRET Development Support Directional Translocation.
To directly observe arrival of portions of the substrate protein inside ClpP, stopped-flow FRET studies were conducted, using the fluorescein-labeled λRssrA proteins as acceptors for donor 5-(2-(acetamido)ethylamino)naphthalene-1-sulfonic acid (EDANS) probes inside miClpP. Labeling of ClpP was accomplished by substituting a new cysteine residue in the ClpP subunit at one of several solvent-accessible sites in the ClpP cavity, based on inspection of the crystallographic model (11) (Fig. 1c). Although ClpP already contains two native cysteines in each of its subunits, they were found to be inaccessible to exogenous labeling with iodo-EDANS (not shown). One such substitution, F31C, was made at the “neck” of ClpP, just below the point of entry of polypeptide into the protease. Two additional individual substitutions, L62C and L139C, were made in the central cavity of the protease, one (L139C) near the position of the active site. The substitutions were programmed into the miClpP mutant, the protein was expressed, and the purified molecules were labeled with iodo-EDANS (1,5-IAEDANS; Molecular Probes) to a level of 50–80% of the available sites.
When Fl-λR21CssrA or Fl-λR93CssrA was incubated with complexes of ClpA and EDANS-miClpP139C in ATPγS, no FRET could be detected, as measured by fluorescence intensity change in the donor emission channel. This is consistent with localization of the ClpA-bound Fl-λRssrA at a sufficient distance from the EDANS probe so that significant energy transfer could not occur. Assuming substrate is bound at the terminal end of the ClpA in a ClpA–ClpP cylinder, the distance between the probes is likely to be at least 80 Å (5) (see Fig. 1a). Upon addition of ATP to these complexes, the donor signal decreased substantially over a time course of 20–30 s and remained reduced thereafter. This reflects FRET produced by translocation of the acceptor-labeled substrate protein into the ClpP cavity in proximity to the donor EDANS probe and its stable association there. Kinetic analysis of the ATP-triggered Fl-λR21CssrA and Fl-λR93CssrA reactions showed that Fl-λR93CssrA produced an increase in FRET efficiency immediately (within the mixing time of <50 ms), whereas a lag phase of ≈1.5–2 s was observed before the Fl-λR21CssrA molecule produced a FRET signal (Fig. 4). This apparently reflects the later arrival of the NH2-terminal fluorophore to a position within ClpP that is near enough to the EDANS to produce significant energy transfer. The same experiments were conducted with ClpP having an EDANS probe at position 31 (EDANS-miClpP31C). The NH2-terminally labeled substrate again exhibited a lag (≈1.5 s) in acquisition of FRET, whereas the COOH-terminally labeled substrate produced an immediate signal (not shown). Together, these data strongly support that translocation from ClpA into ClpP is directional, with the COOH-terminal, recognition-tagged end being translocated first.
Figure 4.
FRET between donor-labeled EDANS-miClpP and acceptor-labeled Fl-λRssrA initiates earlier during a ClpA-mediated translocation reaction for a substrate probe that is adjacent to the COOH-terminal ssrA tag. ClpA (2 μM hexamer), miClpP (2 μM tetradecamer), either Fl-λRssrA (2 μM), and ATPγS (1 mM) were incubated together in reaction buffer at 25°C for 45 min, then rapidly mixed with an excess of ATP (10 mM) in the stopped-flow apparatus. Each trace is the sum of four runs. The difference in the apparent rate of acquisition of FRET between the two Fl-λRssrA molecules may reflect an early step in the overall reaction that is largely complete during the lag phase for the NH2-terminally labeled substrate, but that contributes to the apparent rate of the COOH-terminal one. The difference in the final FRET efficiency reached with the two molecules (42% vs. 34%, respectively) likely results from asymmetric binding of the tagged substrate within the ClpP chamber and, hence, different distances and relative orientations between the probes.
SsrA-Tagged T4 Lysozyme also Undergoes Directional Translocation.
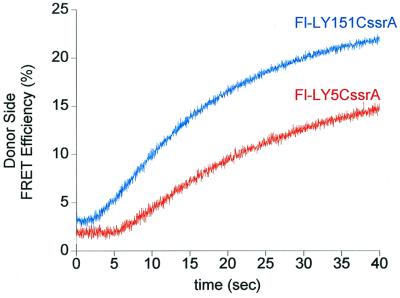
To establish that the directionality observed was not simply intrinsic to λR, we examined a second ssrA-tagged substrate protein, T4 lysozyme (LYssrA). This protein, like other ssrA fusions, was efficiently degraded by ClpA/ClpP in the presence of ATP (not shown). NH2- and COOH-terminal cysteine-substituted versions of LYssrA (Fig. 1b) were fluorescein-labeled and examined in the same energy transfer system, here using EDANS-miClpP62C as the FRET donor. Both substrates showed a lag before acquisition of FRET, but the lag with the NH2-terminally labeled substrate (Fl-LY5CssrA) was much longer (≈6 s) than with the COOH-terminally labeled one (Fl-LY151CssrA) (≈2 s) (Fig. 5). Once again, these data are consistent with the COOH terminus of an ssrA-tagged substrate protein being translocated first.
Figure 5.
FRET between donor-labeled EDANS-miClpP and acceptor-labeled Fl-LYssrA initiates earlier during a ClpA-mediated translocation reaction for a substrate probe that is adjacent to the COOH-terminal ssrA tag. ClpA (3 μM hexamer), miClpP (1.5 μM tetradecamer), either Fl-LYssrA (1.5 μM), and ATPγS (1 mM) were incubated together in reaction buffer at 25°C for 45 min, then rapidly mixed with an excess of ATP (10 mM) in the stopped-flow apparatus. Each trace is the sum of 10 runs. The difference in the final FRET efficiency reached with the two molecules (18% vs. 24.5%, respectively) likely results from asymmetric binding of the tagged substrate within the ClpP chamber and, hence, different distances and relative orientations between the probes.
The observation of a delay in acquiring a FRET signal even with the COOH-terminal LYssrA substrate probably reflects the occurrence of kinetically significant phases of binding and unfolding. Unlike λRssrA, LYssrA does not form a stable complex with ClpA in ATPγS (not shown); thus, the step of binding to form an initial LYssrA/ClpA/ClpP complex might contribute to the observed kinetics. Perhaps more importantly, T4 lysozyme is stably folded in its native state, whereas λ repressor is reported to be in an equilibrium between folded and unfolded states, with a time constant of about 75 ms for the unfolding reaction (24). Thus, the form of λRssrA bound to ClpA may already be unfolded or poised to unfold rapidly, whereas LYssrA must undergo an unfolding step after recognition and before translocation into ClpP. Consistent with this interpretation, it has been reported recently that, in the presence of ATPγS, ClpA can bind unfolded proteins that lack a recognition tag and subsequently deliver them to ClpP when ATP is added (13). Thus, a large fraction of the lag observed with Fl-LY151CssrA may reflect unfolding at ClpA before directional translocation.
Discussion
The fluorescence studies presented here indicate that the translocation of ssrA-tagged substrate proteins, recognized and unfolded by the ClpA chaperone, proceeds directionally into the ClpP protease, with the tagged COOH-terminal end entering first. This implies that, following recognition of the tag by ClpA, it must be displaced from its initial binding site to proceed through the narrow passageway into the protease. The ssrA tag itself might be specifically recognized at distal sites in the translocation pathway to reinforce this directionality, reminiscent of sequential recognition of mitochondrial targeting signals during translocation of precursor proteins into mitochondria (25). Additionally, unfolding itself might be a directional process, commencing at the initially recognized ssrA-tagged end of the substrate protein and freeing that end for translocation first. The rest of the protein would unfold and be translocated sequentially, with the distal, NH2-terminal end translocated last. Although such directional unfolding and degradation are attractive, recent experiments with the two mitochondrial inner membrane AAA proteases indicate that, in the case of membrane-spanning substrate proteins, domains at both sides of the membrane must be able to be unfolded for proteolysis by either enzyme, despite the fact that the proteases are localized to opposite faces of the membrane (26). On the other hand, in a similar system with FtsH, the bacterial inner membrane protease, a cytosolic domain has been observed to be selectively digested, whereas a tightly folded periplasmic domain remained intact (27). In the case of ClpA-mediated unfolding, an earlier deuterium exchange study with GFPssrA indicated that ClpA carried out global unfolding of this substrate in the presence of ATP (12); time-dependent studies of this and other ssrA-tagged substrate proteins should be able to resolve whether they are unfolded in a concerted or directional fashion.
It seems possible that the several ClpAP or ClpXP substrates reported to be recognized through NH2-terminal sequences (28–32) may exhibit a directionality of translocation opposite to that of the COOH-terminally ssrA-tagged proteins examined here, although this remains to be demonstrated. Whether specific directionality is involved for other chaperone-protease assemblies, such as the 26S proteasome, is unknown, although in the case of the partial proteolysis of the p105 precursor of the NF-κB p50 subunit, degradation by the proteasome seems to proceed from the COOH terminus (33).
Acknowledgments
We thank Hays Rye for assistance with the fluorescence measurements. E.U.W.-B. was a Jane Coffin Childs Foundation Fellow. This work was supported by the National Institutes of Health, the Howard Hughes Medical Institute, and the Nelson Fund.
Abbreviations
- GFP
green fluorescent protein
- ATPγS
adenosine 5′-O-(3-thiotriphosphate)
- FRET
fluorescence resonance energy transfer
- EDANS
5-(2-(acetamido)ethylamino)naphthalene-1-sulfonic acid
References
- 1.Larsen C N, Finley D. Cell. 1997;91:431–434. doi: 10.1016/s0092-8674(00)80427-4. [DOI] [PubMed] [Google Scholar]
- 2.Baumeister W, Walz J, Zühl F, Seemüller E. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 3.Wickner S, Maurizi M R, Gottesman S. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 4.Horwich A L, Weber-Ban E U, Finley D. Proc Natl Acad Sci USA. 1999;96:11033–11040. doi: 10.1073/pnas.96.20.11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beuron F, Maurizi M R, Belnap D M, Kocsis E, Booy F P, Kessel M, Steven A C. J Struct Biol. 1998;123:248–259. doi: 10.1006/jsbi.1998.4039. [DOI] [PubMed] [Google Scholar]
- 6.Grimaud R, Kessel M, Beuron F, Steven A C, Maurizi M R. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- 7.Rohrwild M, Pfeifer G, Santarius U, Müller S A, Huang H-C, Engel A, Baumeister W, Goldberg A L. Nat Struct Biol. 1997;4:133–139. doi: 10.1038/nsb0297-133. [DOI] [PubMed] [Google Scholar]
- 8.Sousa M C, Trame C B, Tsuruta H, Wilbanks S M, Reddy V S, McKay D B. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- 9.Glickman M H, Rubin D M, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried V A, Finley D. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 10.Braun B C, Glickman M, Kraft R, Dahlmann B, Kloetzel P M, Finley D, Schmidt M. Nat Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Hartling J A, Flanagan J M. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 12.Weber-Ban E U, Reid B G, Miranker A D, Horwich A L. Nature (London) 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 13.Hoskins J R, Singh S K, Maurizi M R, Wickner S. Proc Natl Acad Sci USA. 2000;97:8892–8897. doi: 10.1073/pnas.97.16.8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh S K, Grimaud R, Hoskins J R, Wickner S, Maurizi M R. Proc Natl Acad Sci USA. 2000;97:8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y I, Burton R E, Burton B M, Sauer R T, Baker T A. Mol Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- 16.Keiler K C, Waller P R H, Sauer R T. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 17.Gottesman S, Roche E, Zhou Y, Sauer R T. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levchenko I, Seidel M, Sauer R T, Baker T A. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- 19.Matsumura M, Matthews B W. Science. 1989;243:792–794. doi: 10.1126/science.2916125. [DOI] [PubMed] [Google Scholar]
- 20.Rye H S, Burston S G, Fenton W A, Beechem J M, Xu A, Sigler P B, Horwich A L. Nature (London) 1997;388:792–798. doi: 10.1038/42047. [DOI] [PubMed] [Google Scholar]
- 21.Rye H S, Roseman A M, Chen S, Furtak K, Fenton W A, Saibil H R, Horwich A L. Cell. 1999;97:325–338. doi: 10.1016/s0092-8674(00)80742-4. [DOI] [PubMed] [Google Scholar]
- 22.Otto M R, Lillo M P, Beechem J M. Biophys J. 1994;67:2511–2521. doi: 10.1016/S0006-3495(94)80741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kessel M, Maurizi M R, Kim B, Kocsis E, Trus B L, Singh S K, Steven A C. J Mol Biol. 1995;250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- 24.Myers J K, Oas T G. Biochemistry. 1999;38:6761–6768. doi: 10.1021/bi990088x. [DOI] [PubMed] [Google Scholar]
- 25.Voos W, Martin H, Krimmer T, Pfanner N. Biochim Biophys Acta. 1999;1422:235–254. doi: 10.1016/s0304-4157(99)00007-6. [DOI] [PubMed] [Google Scholar]
- 26.Leonhard K, Guiard B, Pellecchia G, Tzagoloff A, Neupert W, Langer T. Mol Cell. 2000;5:629–638. doi: 10.1016/s1097-2765(00)80242-7. [DOI] [PubMed] [Google Scholar]
- 27.Kihara A, Akiyama Y, Ito K. EMBO J. 1999;18:2970–2981. doi: 10.1093/emboj/18.11.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoskins J R, Kim S Y, Wickner S H. J Biol Chem. 2000;275:35361–35367. doi: 10.1074/jbc.M006288200. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez M, Rasulova F, Maurizi M R, Woodgate R. EMBO J. 2000;19:5251–5258. doi: 10.1093/emboj/19.19.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tobias J W, Shrader T E, Rocap G, Varshavsky A. Science. 1991;254:1374–1347. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Elliott M, Elliott T. J Bacteriol. 1999;181:1211–1219. doi: 10.1128/jb.181.4.1211-1219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonciarz-Swiatek M, Wawrzynow A, Um S J, Learn B A, McMacken R, Kelley W L, Georgopoulos C, Sliekers O, Zylicz M. J Biol Chem. 1999;274:13999–14005. doi: 10.1074/jbc.274.20.13999. [DOI] [PubMed] [Google Scholar]
- 33.Palombella V J, Rando O J, Goldberg A L, Maniatis T. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]