Abstract
Chromium was proposed to be an essential element over 50 y ago and was shown to have therapeutic potential in treating the symptoms of type 2 diabetes; however, its mechanism of action at a molecular level is unknown. One chromium-binding biomolecule, low-molecular weight chromium-binding substance (LMWCr or chromodulin), has been found to be biologically active in in vitro assays and proposed as a potential candidate for the in vivo biologically active form of chromium. Characterization of the organic component of LMWCr has proven difficult. Treating bovine LMWCr with trifluoroacetic acid followed by purification on a graphite powder micro-column generates a heptapeptide fragment of LMWCr. The peptide sequence of the fragment was analyzed by MS and tandem MS (MS/MS and MS/MS/MS) using collision-induced dissociation and post-source decay. Two candidate sequences, pEEEEGDD and pEEEGEDD (where pE is pyroglutamate), were identified from the MS/MS experiments; additional tandem MS suggests the sequence is pEEEEGDD. The N-terminal glutamate residues explain the inability to sequence LMWCr by the Edman method. Langmuir isotherms and Hill plots were used to analyze the binding constants of chromic ions to synthetic peptides similar in composition to apoLMWCr. The sequence pEEEEGDD was found to bind 4 chromic ions per peptide with nearly identical cooperativity and binding constants to those of apoLMWCr. This work should lead to further studies elucidating or eliminating a potential role for LMWCr in treating the symptoms of type 2 diabetes and other conditions resulting from improper carbohydrate and lipid metabolism.
Introduction
Despite chromium being proposed as an essential trace element over 50 y ago and having been demonstrated to have potential as a adjuvant therapy to improve insulin resistance and related symptoms in rodent models of type 2 diabetes, the mode of action of chromium at a molecular level has not been elucidated (1). Two biomolecules are known to bind chromium: transferrin and low-molecular weight chromium-binding substance (LMWCr);4 the latter has been proposed to be biologically active and has the ability to potentiate the activity of insulin-activated insulin receptor in vitro (1).
Dietary chromium as trivalent chromium is absorbed by passive diffusion from the gastrointestinal tract and quickly passes to the bloodstream where transferrin is responsible for maintaining chromium supplies in the bloodstream and transporting chromium to the tissue (1). In the tissue, chromium ultimately binds to a small molecule(s), after which it is rapidly cleared and eliminated in the urine as the small molecule complex.
The identification of this molecule(s) and attempts at characterizing it and its complex with chromium were begun in the 1980s by Wada et al. (2–9). In 1981, a low-molecular weight chromium compound was identified by size exclusion chromatography of the cytosol of liver cells of male mice injected with a single dose of potassium chromate (2). A similar low-molecular weight compound was found in the feces and urine hours after the i.v. administration. These researchers suggested that LMWCr was formed in the liver and participates in retention and excretion of chromium in the body. The material from the livers of rabbits similarly treated with chromate was partially purified and found to apparently be an anionic organic-chromium complex containing amino acids (2). Also in 1981, Wu and Wada (3) reported additional studies on LMWCr from urine. LMWCr was found to occur in urine normally, although the amounts were greatly increased after rats were injected with chromate (3). Normal human and rat urine LMWCr was found not be saturated with chromium. The LMWCr was thought to be similar to that of the liver and other organs of rabbits and dogs and to be involved in removing excess chromium from the body. The distribution of LMWCr in mice 2 h after injection with potassium dichromate was also examined (4). LMWCr was found in liver, kidney, spleen, intestine, testicle, brain, and blood plasma, with the greatest amount in liver followed by kidney. Supernatants of homogenates of the organs were found to possess more chromium bound to LMWCr when dichromate was added to the homogenate than when the mice were injected with dichromate. The time course of chromium binding to LMWCr after injection of dichromate was investigated (5). Chromium was found to be associated with liver and kidney LMWCr only 2 min after injection and reached a maximum 1–2 h after treatment. In these studies, LMWCr was again identified by its elution behavior in size exclusion chromatography and its chromium-binding ability.
Insulin dose response studies using rat adipocytes have indicated a potential intrinsic biological function for LMWCr; the first detailed studies were reported by Yamamoto et al. (7) in 1988 in the Journal of Nutrition. Isolated rat adipocytes in the presence of LMWCr and insulin display a greater ability to metabolize glucose to produce carbon dioxide or total lipids; this increase occurs without a change in the insulin concentration required for half-maximal stimulation (7, 8, 10). This lack of change in half-maximal insulin concentration suggests a role for LMWCr inside the insulin-sensitive cells after insulin binds externally to the insulin receptor (10). The stimulation of glucose metabolism by LMWCr is proportional to the chromium content of the oligopeptide (6). A role has been proposed for LMWCr in the autoamplification of insulin signaling by binding to activated insulin receptor (11).
Efforts continued to isolate and characterize LMWCr. To date, LMWCr has been isolated and purified from several mammalian sources, including rabbit liver (9), bovine liver (12), and porcine kidney (13). The material from rabbit was loaded with chromium by injection of the animal with chromate, or Cr(III), which provides lower yields. For the materials from bovine liver and porcine kidney, chromate was added to the homogenized liver or kidney or suspended kidney powder. A chromium-loading procedure is required so that the material can be followed by its chromium content during the isolation and purification procedures. Amino acid analysis of the bovine LMWCr indicates the presence of 4 glutamate (and/or glutamine), 2 aspartate (and/or asparagine), 2 glycine, and 2 cysteine residues (12), and the rabbit LMWCr possesses an additional glycine residue (9). Thus, LMWCr appears to be a naturally occurring oligopeptide composed of glycine, cysteine, aspartate, and glutamate, with the carboxylates comprising more than one-half of the total amino acid residues. No amino acid sequence data has appeared despite attempts at sequencing by Edman degradation, NMR, and MS. The lack of additional characterization of the organic components of the materials is a matter of concern, particularly because the rabbit LMWCr amino acid composition was reported over 20 y ago. Herein are reported successful efforts to sequence the oligopeptide of LMWCr and the demonstration that a synthetic peptide of this sequence binds Cr in a similar fashion to LMWCr.
Experimental Procedures
Materials.
α-Cyano-4-hydroxycinnamic acid, 2,5-dihydroxybenzoic acid, trifluoroacetic acid (TFA), and activated charcoal (C-5510) were obtained from Sigma. Liquid chromatography(LC)-MS grade acetonitrile was obtained from Riedel-de Haën. LMWCr were purified from livers of alligator (14), bovine (12), and chicken (14) and human urine (15) utilizing methods previously described or were available from previous work. The peptides pEEEEGDD and pEEEGEDD were synthesized using standard Fmoc procedures (16) with an Advanced ChemTech Model 90 peptide synthesizer. 51CrCl3 was obtained from ICN, CrCl3 was from Fisher Scientific, and HEPES was from Research Organics, Inc.
MS.
Matrix-assisted laser desorption ionization time-of-flight MS (MALDI/TOF MS) was performed on a Bruker Daltonics Reflex III mass spectrometer with a 2-stage reflectron (17). Ionization used the 337 line of a Laser Science VSL-337ND-S nitrogen laser. Positive and negative ion spectra were obtained in linear and reflectron modes with an accelerating voltage of 20 kV. Post-source decay (PSD) spectra used precursor ion selection with a pulsed voltage that deflected matrix and contaminant ions from entering the flight tube. Product ions were detected in segments by stepping down the reflectron voltage as follows: −21.0, −19.55, −15.75, −11.82, −8.86, −6.64, −4.98, −3.74, and −2.80 kV. The MALDI matrix was generally α-cyano-4-hydroxycinnamic acid, although some experiments utilized 2,5-dihydroxybenzoic acid.
For LC-MS analysis, an Agilent 1200 series liquid chromatograph with a Zorbax (150 × 0.5 mm) 5B-C18 column was interfaced to a Bruker HCT ultra PTM discovery system high capacity quadrupole ion trap (QIT) mass spectrometer via electrospray ionization (ESI). The mobile phase involved doubly deionized water (ddH2O) and acetonitrile; gradient elution was employed. Direct infusion experiments used a syringe pump with a flow rate of ~140 μL/h. The ESI needle spray voltage was 4 kV, the capillary temperature was 300°C, and mass spectra were acquired over a range of m/z 100–2000. Low energy collision-induced dissociation (CID) used helium as the collision gas. The fragmentation amplitude was 1.0 V and the acquisition software’s smart fragmentation was on (the start Amplitude 30% and the end amplitude 200%).
Graphite powder microcolumn.
Custom-made chromatographic microcolumns were used for desalting and concentration of the peptide prior to MS analysis. Activated charcoal was packed in a constricted gel loader tip (Eppendorf). A 10-mL syringe was used to force liquid through the column by applying gentle air pressure. The columns were equilibrated with 10 μL of 0.1% TFA. An aliquot of the LMWCr after purification by Sephadex G-15 column chromatography was diluted to 30 μL in 0.1% TFA and loaded onto the column using gentle syringe air pressure. The column was washed with 60 μL 0.1% TFA. The resulting samples were mixed with ~2 μL of 4HCCA in 70% acetonitrile/0.1% TFA and spotted onto the MALDI target (plate) with a micropipettor.
To generate more purified samples, ESI/MS samples were prepared with a modified method in which activated charcoal powder was packed into a microconcentrator instead of the gel loader tip. A tabletop centrifuge was used to wash the sample and elute the sample through charcoal powder and filter membrane. A solvent of 70% acetonitrile/0.1%TFA was used for the final elution solution. Eluent was lyophilized and redissolved in ddH2O before LC-MS processing.
Chromium-binding studies.
A variation of the equilibrium dialysis method using an ultrafiltration device was utilized to examine the binding of chromium to the synthetic peptides. Aliquots of a mixture of CrCl3 and 51CrCl3 were combined to generate different concentrations of Cr(III) while maintaining the synthetic peptide in solution at a constant volume. Known amounts (~0.46 μmol) of peptide and 200 mL of 0.1 mol/L HEPES buffer (pH 7.4) were slowly stirred in an Amicon 8400 ultrafiltration unit (with a YC05 membrane) at 4°C temperature for at least 12 h to achieve equilibration. The ultrafiltration unit was then pressurized and effluent was collected. The content of free chromic ion in the effluent was determined by gamma counting using a Packard Cobra II auto-gamma counter. Chromium-binding experiments were performed in triplicate and the synthetic peptide used in at least 1 of the 3 sets of triplicates was from a different synthesis. As a control for the chromium-binding experiments, the ultrafiltration procedure was performed without peptide to establish the amount of chromium that adhered to the ultrafiltration unit; all experiments were corrected for this background. Linear regression analyses of the Langmuir isotherms and Hill plots were performed using SigmaPlot 11.0. The chromium concentration of solutions of isolated peptides were determined by graphite furnace atomic absorption spectroscopy using a PerkinElmer Analyst 400 atomic absorption spectrometer equipped with an HGA-900 graphite furnace and an AS-800 autosampler using a chromium hollow cathode lamp operating at 10 mA; a spectral bandwidth of 0.8 nm was selected to isolate the light at 353.7 nm. Chromium standard solution obtained from Perkin Elmer was utilized to generate a standard curve.
Miscellaneous.
Errors are presented throughout as SD of the triplicate analyses. ddH2O was used throughout. Amino acid analyses of samples were performed by the Protein and Separation Analysis Laboratory at Purdue University. Protein concentrations were determined by the fluorescamine method (12). Fluorescence measurements were obtained on a Jobin Yvon FluoroMax-3 fluorescence spectrophotometer. UV-visible spectra were obtained using a Hewlett-Packard 8451A or a Beckman Coulter DU 800 spectrophotometer.
Results
Production of apo-oligopeptide of LMWCr.
Treatment of LMWCr from a variety of sources (alligator liver, chicken liver, bovine liver, and human urine) with 0.1% TFA resulted in no visible precipitation. Separation of the products was attempted by graphite powder (GP) microcolumn. GP has been utilized to effectively retain small and hydrophilic peptides, which could readily be eluted for mass spectral analysis (18). The primary organic product eluting from the column contained no detectable chromium by the diphenylcarbazide method (12). Attempts to assay for protein content via reactions with a free primary amine group with fluorescamine indicated that the isolated material was either not a protein or that the amino terminus was blocked. Amino acid analysis of the component of bovine LMWCr eluting from the GP column produced the composition 1.0 glycine:4.5 glutamate (and/or glutamine):2.2 aspartate (and/or asparagine):0 cysteine, indicating that the LMWCr lost some of its amino acids during the TFA treatment and/or GP microcolumn processing. No other amino acids were detected above trace quantities. If the amino acid ratio is calculated assuming 2.0 aspartate residues, then the ratio becomes 0.91 glycine:4.1 glutamate:2.0 aspartate, indicating the loss of 1 glycine residue and 2 cysteine residues compared with the original composition of bovine LMWCr (2 glycine:4 glutamate:2 aspartate:2 cysteine) (12).
MALDI/TOF MS studies.
A molecular ion (m/z 804) was observed for all treated LMWCr samples with GP column under negative mode MALDI/TOF MS (Supplemental Fig. 1). No corresponding m/z peak was found under positive mode. The lack of a positive signal, [M+H]+, is consistent with the highly acidic nature of LMWCr. The intensities for the ions of interest are only a few hundred detector counts, which is very low; a more typical value would be ~10 times higher. The low signal intensity possibly resulted from poor binding capacity of the microcolumn, inefficient elution using 70% acetonitrile/0.1% TFA, or the samples not being ionized well.
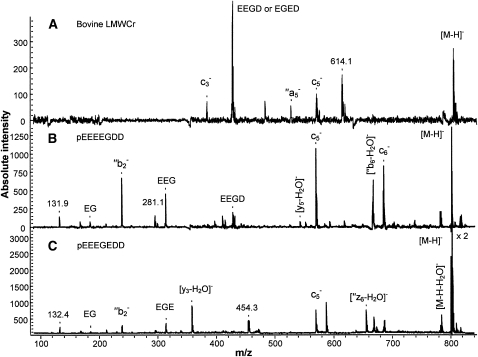
PSD of the m/z 804 ion of bovine LMWCr was performed (Fig. 1) and 2 sequences were proposed based on this data: pEEEEGDD and pEEEGEDD (where pE is pyroglutamate). Peptide backbone cleavage ions were identified and are denoted in Figure 1 with Roepstorffand Fohlman nomenclature (19). All assigned product ions match the m/z of the predicted ions to within m/z ± 1, which is within accepted accuracy of PSD. There are several unassigned peaks of appreciable intensity in the spectra that are not standard peptide cleavage fragments. Past work in the Cassady group (T. Yalcin and C. J. Cassady, unpublished data) indicated that nonstandard cleavages frequently appear in negative mode tandem MS (MS/MS) when adjacent acidic residues exist in a peptide. The precursor ion, [M-H]−, at m/z 804, is 2 Da higher in mass than expected. The reason for this is unclear, but it occurs consistently in the MALDI/TOF experiments of the LMWCr samples. This extra 2 Da may indicate that the biological peptides have been modified by the addition of 2 hydrogen atoms (e.g. hydrogenation across the double bond of a carbonyl) during the MALDI process; perhaps a trace component in the biological samples is facilitating the process. Sample modification during MALDI is unusual, but not without precedent. Tanaka et al. (20) observed hydrogenation during MALDI on oligosaccharides tagged with 2-aminopyridine, whereas Yamaguchi et al. (21) found that MALDI induced dehydrogenation for a peptide derizatized with 2-hydrazino-2-imidazoline.
FIGURE 1.
Negative mode MALDI/TOF PSD spectra of m/z 804 of bovine liver LMWCr peptide (A) and m/z 802, [M-H]− of synthetic peptide pEEEEGDD (B) and synthetic peptide pEEEGEDD (C).
PSD spectra of synthetic peptides were generated and compared with those of the LMWCr ( . The synthetic peptides produced the expected [M-H]− at m/z 802. The PSD spectra for the biological LMWCr were produced from m/z 804 and the PSD spectra for the synthetic peptides were generated from m/z 802. (MALDI/TOF MS analysis of mixtures of biological and synthetic peptides yielded both m/z 802 and 804, showing that these were distinct ions.) This mass discrepancy may prevent a strong match between the PSD spectra for the biological peptides and the model peptides. The spectra of the LMWCr from different biological sources shared common features at m/z 384, 428, 482, and 570, which suggests a similarity in sequence. However, the PSD spectra of peptides pEEEEGDD and pEEEGEDD both showed only a few similar features at m/z 428, 482, and 570 to those of the LMWCr. Neither is a sufficient match to positively identify the biological peptide.
Analysis of LMWCr using ESI/QIT MS.
Larger samples of LMWCr were isolated by the modified GP column with the application of the microconcentrator. The ability of reverse phase column (Zorbax 5B-C18 on LC/MS) to retain LMWCr was tested; experiments revealed that the products of the TFA treatment of LMWCr elute during the first 5 min when washing the column with 2% acetonitrile; these were detected by obvious UV absorbances at 260 nm. No ESI response (m/z 802) could be observed, because ionization interferences occur when an extract from a biological specimen, LMWCr in this case, is loaded into the LC portion of the instrument. Suppression of the signal at the time point that corresponds to the void volume of the column is common (22). Consequently, for the experiments described below, LMWCr samples were introduced into the ESI source by infusion with a syringe pump rather than by LC.
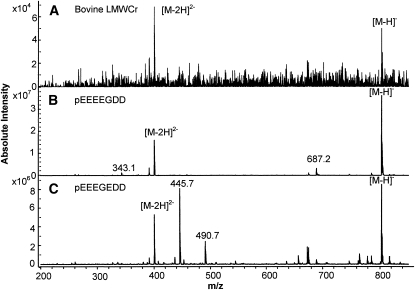
As was the case for MALDI, no positive ion signal was observed when LMWCr samples were ionized by ESI. The negative mode ESI spectrum of bovine LMWCr (Fig. 2) shows 1 peak at m/z 802 and another at m/z 401 corresponding to the singly and doubly charged species, [M-H]− and [M-2H]2−, respectively. Unlike the ions generated by MALDI, these ions generated by ESI (which is a much gentler ionization technique) exactly conform to the molecular weight of pEEEEGDD or pEEEGEDD. Low-energy CID MS/MS (MS2) on m/z 802 ions, [M-H]−, was carried out to elucidate the sequence. The synthetic peptides pEEEEGDD and pEEEGEDD were dissociated under the same conditions. The MS/MS spectra of m/z 802, [M-H]−, from the bovine sample and the 2 synthetic peptides (not shown) were dominated by a very intense water elimination ion at m/z 784, [M-H-H2O]−. Water loss during low-energy CID is common and abundant in negative mode when a peptide has adjacent acidic residues (T. Yalcin and C. J. Cassady, unpublished data). Relative to this large m/z 784, other CID products were only a few percent relative intensity (or less), and no obvious differences existed among these low-intensity ions. That is, CID on [M-H]− (MS/MS) cannot distinguish between these 2 synthetic peptides, and both model spectra are a good match for the biological sample.
FIGURE 2.
Negative mode ESI/QIT MS spectra of bovine liver LMWCr (A), synthetic peptide pEEEEGDD (B), and synthetic peptide pEEEGEDD (C).
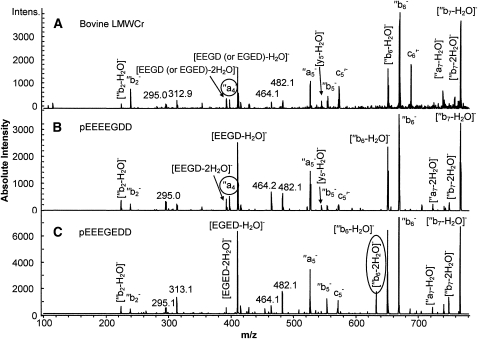
The intense peak at m/z 784, [M-H-H2O]−, which dominated the MS/MS spectra for both synthetic peptides and for bovine LMWCr, was subjected to a further stage of CID. The resulting MS/MS/MS (or MS3) spectra are shown in Figure 3. Again, very similar spectral features were shared by LMWCr and the 2 synthetic peptides. A few notable differences were observed in MS/MS/MS spectra of m/z 784 ions between the 2 synthetic peptides. A peak at m/z 397 (Fig. 3, circle) is found in the spectrum from pEEEEGDD; this corresponds to an ′′a4−. Because ′′a4− incorporates only the first 4 residues of the peptides (starting at the N terminus), it will not form at the same m/z in the spectrum of pEEEGEDD. The CID spectrum from the fragment of bovine LMWCr also contains a peak at m/z 397 in roughly the same abundance as in the spectrum for pEEEEGDD. In addition, a peak at m/z 632, corresponding to [′′b6 - 2H2O]−, is found only in the spectra from pEEEGEDD, but not the spectra from pEEEEGDD or bovine LMWCr. In negative mode CID of peptides, adjacent acidic residues (aspartic acid or gluatmic acid) promote water loss, and this is much more prevalent when one of the residues is aspartic acid (T. Yalcin and C. J. Cassady, unpublished data). Of the 2 model peptides, only pEEEGEDD has an aspartic acid residue (D at the 6th position) adjacent to another acidic residue (E at the 5th position) within the first 6 residues of the sequence, which comprise [′′b6 - 2H2O]−. Taking into account the 2 spectral features discussed here, the sequence of the fragment from bovine LMWCr is assigned as pEEEEGDD.
FIGURE 3.
Low-energy CID MS/MS/MS spectra of [M-H-H2O]− from bovine liver LMWCR peptide (A), synthetic peptide pEEEEGDD (B), and synthetic peptide pEEEGEDD (C).
Bioinformatics.
In the body, peptides, including numerous peptides with bioactivity, originate from the processing of proteins. Thus, the heptapeptide isolated from LMWCr should have at a point in its history been be part of a larger protein. A genomic search against the databases of the National Center for Biotechnology Information using the sequence EEEEGDD was performed to identify proteins containing this sequence motif. Multiple 100% hits were found due to the short sequence and low complexity: 7 sequences in Homo sapiens, 2 in Bos Taurus, 2 in Gallus gallus, and 1 in Mus musculus. Unfortunately, very little of the American alligator genome has been sequenced. None of the hits contain glycine and cysteine residues flanking the EEEEGDD sequence, suggesting that these residues are not part of a contiguous peptide and are attached to the heptapeptide in a nonstandard fashion. This is consistent with NMR studies on holoLMWCr and acid-hydrolyzed LMWCr that suggested the presence of some other organic moiety (12).
Curiously, several sequence hits correspond to genes for proteins involved in modifying chromatin. None of these hits correspond to a responsive gene in chromium treatment of mice (isolated adipocytes) or mouse testis (TM4 Sertoli-like) cells (23, 24). This is an area worthy of continued investigation.
Chromium binding.
Because chromium binding to the oligopeptide LMWCr is thought to be only through carboxylate residues and the heptapeptide pEEEEGDD retains all the aspartate and glutamate residues in LMWCr, the question arises as to whether the proposed heptapeptide can bind chromium in a similar fashion to LMWCr. The binding of chromium to bovine apoLMWCr prepared by the low-pH EDTA method (12), the heptapeptide pEEEEGDD, and 2 other acidic peptides was probed. The 2 peptides EDGEECDCGE and DGEECDCGEE were chosen, because they possess the same amino acid composition as bovine LMWCr, and searches of the human genome reveal these to be the only 2 sequences in this genome with this composition (25). The number of Cr3+ ions binding to the peptides was estimated using Langmuir isotherms. For the Langmuir isotherms of Cr3+ binding to all the synthetic peptides, chromium binding to apoLMWCr can be represented by 2 intersecting straight lines with different slopes (Supplemental Fig. 2). This biphasic behavior indicates that each peptide has 2 types of Cr3+ binding sites: tight binding sites and weak binding sites. Negative values of the Langmuir parameters Bt and K were found for each peptide: apoLMWCr, −2.69 mmol/g and −109 L/mmol, respectively; EDGEECDCGE, −6.97 mmol/g and −354 L/mmol; DGEECDCGEE, −62.9 mmol/g and −9.94 L/mmol; and pEEEEGDD, −0.73 mmol/g and −151 L/mmol. The appearance of negative Bt values for all peptides demonstrates the limitation of using the simple Langmuir model in cases of tight binding. The negative value of Bt indicates that most of the sorption sites have a high affinity for Cr3+ ions, especially at low Cr3+concentrations (26).
The intersection points in the isotherms allow the number of tightly binding ions to be estimated. For bovine apoLMWCr, the chromium:oligopeptide ratio at the intersection point is 3.6, which is very close to the average amount of chromium bound to isolated bovine LMWCr, which is 3.5 (12). Titration of bovine LMWCr with Cr3+ has previously shown that 4 Cr3+ ions are required to restore bioactivity (27, 28). For the synthetic peptides, the intersection points occurred at Cr:peptide ratios of ~2, 2, and 4 for EDGEECDCGE, DGEECDCGEE, and pEEEEGDD, respectively. This is consistent with the Cr:peptides binding ratios found after exposing the peptides to solutions of 10 equivalents of Cr3+ (CrCl3⋅6H2O in 0.1 mol/L HEPES buffer, pH 7.4 overnight at 4°C) and separating the peptides from the excess Cr3+ by G-10 size exclusion chromatography (EDGEECDCGE, 2.0 ±0.3; and DGEECDCGEE, 2.0 ± 0.3). A change in mode from specific coordinate covalent binding to just electrostatic absorption on the peptide surface is proposed. Thus, the inflection point of the isotherm for the peptide pEEEEGDD implies that it tightly binds 4 Cr3+ ions. The binding of 2 Cr3+ ions to the peptides EDGEECDCGE and DGEECDCGEE is consistent with mass spectrometric studies of chromium binding to these peptides (H. M. Watson, J. Gao, and C. J. Cassady, unpublished data). The electronic spectrum of Cr-loaded pEEEEGDD peptide has 2 visible maxima at ~411 and 577 nm and a broad shoulder in the UV region at ~270 nm. The 2 visible features are readily assigned to d→d transitions from the Cr3+ centers. The spectrum is very similar to that of bovine liver LMWCr (shoulder ~260 nm, 394 nm, 576 nm) (12).
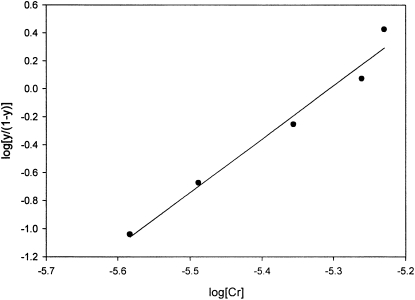
To further compare the binding properties between the synthetic peptides and apoLMWCr in CrCl3 solution, the method of Hill (29) was applied to establish the degree of cooperativity between the covalent binding sites with the initial low amounts of substrate in solution, assuming covalent binding sites are occupied before surface adsorption occurs. The total number of tight-binding sites established using the Langmuir isotherm was utilized. This method uses the binding number y defined as
where Kf is the binding of formation constant and n is the Hill constant, such that
Hill plots gave linear curves (Fig. 4). Kf and n were obtained as the value of the y-intercept and slope, respectively (Table 1). These data indicate a large degree of positive cooperativity such that the binding of the first and subsequent Cr3+ ions facilitates the binding of additional chromium, perhaps in a multinuclear assembly; the magnitudes suggest that essentially only apopeptide or peptide saturated with Cr3+ ions exist in solution. The Hill constants, Kf and n, of apoLMWCr measured in this study, 1.10 × 1021 (mol/L)−4 and 3.82, differ only slightly from published data, Kf = 1.54 × 1021 (mol/L)−4 and n = 3.47 (30). For EDGEECDCGE, the Hill constant is greater than the number of interacting sites, because only 2 Cr(III) binding sites are on the peptide. This suggests that the resulting Cr-peptide complex is actually a dimer of peptide (31). Because both apoLMWCr and pEEEEGDD bind 4 Cr3+ ions and the binding constants for apoLMWCr and pEEEEGDD are within an order of magnitude, whereas the Hill constants are identical, pEEEEGDD appears to contain all the essential components of LMWCr for binding chromium and probably binds chromium in an essentially identical fashion to that of LMWCr.
FIGURE 4.
Hill plot of Cr3+ ion binding to synthetic peptide pEEEEGDD. y corresponds to the binding number as defined by the Hill equation.
TABLE 1.
Hill plot constants Kf and n for Cr(III) binding to bovine liver apoLMWCr and synthetic peptides
| Peptide | Kf | n | Cooperativity |
| ApoLMWCr (bovine liver) | 1.10 x 1021 (mol/L)#x22124 | 3.82 | Positively cooperative |
| EDGEECDCGE | 1.48 x 1023 (mol/L)#x22122 | 3.64 | Positively cooperative |
| DGEECDCGEE | 1.01 x 1011 (mol/L)#x22122 | 1.85 | Positively cooperative |
| pEEEEGDD | 1.92 x 1020 (mol/L)#x22124 | 3.82 | Positively cooperative |
Discussion
Attempts to establish which naturally occurring biomolecule binds chromium and then potentially performs an essential function have proved frustrating (1, 32). The first candidate for this biomolecule was coined “glucose tolerance factor”; this species proved to be an artifact generated during its isolation (32–34). In fact, the insulin-stimulating molecule in Brewer’s yeast was ultimately separated from any chromium-containing species (33, 34). Currently, the nutritional status of chromium is in doubt (1, 18). However, chromium taken orally has been shown to have pharmacological effects improving insulin sensitivity and blood lipid parameters in rodent models of insulin resistance and diabetes and possibly recently in healthy rats (33, 35).
Currently, the only biomolecule proposed as a candidate for the pharmacological effects is LMWCr; however, the proposed mechanism of action is based on in vitro studies and needs to be supported by in vivo experiments (1). Additionally, the existence of LMWCr has even recently been questioned because of the failure of its organic component to be characterized (36), even though the inorganic components have been thoroughly investigated (37). Isolating LMWCr has required loading the peptide with chromium so that the peptide could be followed through the isolation procedure. Because the molecule binds Cr3+ so tightly, removing chromium from the oligopeptide is difficult. Because Cr3+ is substitutionally inert and paramagnetic (S = 3/2), characterizing the organic component of the holo-oligopeptide is quite difficult. The 4 paramagnetic centers broaden signals in NMR spectroscopic experiments, so this technique cannot be used to characterize the oligopeptide beyond identifying bridging carboxylate ligands (12). Cr3+ is not released in mass spectral experiments and mass spectra of chromium-acidic peptide complexes have proven difficult to obtain and interpret (38). The presence of a string of terminal glutamate residues prevented successful sequencing by Edman degradation.
Because Cr3+ ions bind so tightly to LMWCr (30 and below) so that only harsh methods were previously successful in removing Cr from LMWCr, gentler methods to remove the Cr3+ ions were investigated. Previously, efforts to generate the apo (or metal-free) form of the oligopeptide involved exposing the LMWCr to an excess of EDTA at low pH (~3.5) and elevated temperatures for substantial periods of time (6, 12). This procedure generated an appreciable amount of denatured, unrecoverable material when performed with milligram quantities of LMWCr; use of catalytic amounts of a reductant, cyanoborohydride (to generate Cr2+ that could be more readily removed from the oligopeptide than substitutionally inert Cr3+), reduced losses but did not completely fix the problem (12). Thus, the current investigation sought to find a new route to remove the chromium rom LMWCr. The reaction
occurs if HO2CR’ is more acidic than HO2CR, where the symbols R and R’ denote attached hydrocarbon side chains, and was previously used to exchange carboxylate ligands in multinuclear chromium assemblies (39). This process was hoped be more gentle and to lead to more recoverable material. TFA was chosen because it is quite acidic and water soluble. The use of TFA to remove chromium from LMWCr from mammalian, avian, and reptilian tissues or body fluids reveals that the LMWCr are all composed of an oligopeptide with an identical molecular weight and apparently identical composition. Unfortunately, the procedure results in the cyclization of the N-terminal glutamate residue. The procedure also results in the separation of a contiguous heptapeptide from additional glycine and cysteine amino acids, although other data suggest these additional amino acids may be connected to the heptapeptide through a nonstandard linkage. The ability of the heptapeptide component of LMWCr to be identified in the urine and/or liver of insulin-utilizing animals of 3 different phyla (mammals, aves, and reptiles) increases the likelihood that LMWCr is not an artifact of its isolation procedure [as proposed in (36)].
The proposed sequence is consistent with the amino acid analysis and the amino terminus being blocked. In fact, a terminal glutamate residue would be expected to cycle under acid treatment. This sequence also explains previous unsuccessful attempts to sequence bovine LMWCr by Edman degradation. The previous efforts generated a low yield of glutamate for the first residue and no appreciable amount of free amino acids in the second cycle (C. M. Davis and J. B. Vincent, unpublished data). Because glutamate residues have a tendency to cyclize under Edman degradation conditions, the string of glutamate residues at the amino terminus of the proposed sequence would make sequencing by Edman degradation impossible. Mass spectra generated by MALDI and ESI both indicate that no Cr exists in the peptide fragment.
Currently, work is continuing to characterize the peptide pEEEEGDD and its complex with 4 bound Cr3+ by a variety of spectroscopic and magnetic techniques and to crystallize the complex. Efforts are underway to examine in vivo the potential of LMWCr to activate components of the insulin signaling cascade. Antibodies raised against the sequence CEEEEGDD conjugated via the cysteine to limpet hemocyanin react with holoLMWCr; unfortunately, although this represents a first step toward developing techniques to examine the intracellular distribution of LMWCr or measure in vivo concentrations of LMWCr, the recognition of holoLMWCr by the antibodies is not sufficient for use in Western blotting and other techniques [(15) and Y. Chen and J. B. Vincent, unpublished data].
In conclusion, the sequence of the contiguous peptide component of LMWCr represents a potentially significant milestone toward understanding the pharmacological role of chromium supplementation at a molecular level nearly 30 y after the initial report of the existence of LMWCr. Hopefully, this work will lead to further studies elucidating or eliminating a potential role for LMWCr in treating the symptoms of type 2 diabetes and other conditions resulting from improper carbohydrate and lipid metabolism.
Supplementary Material
Acknowledgments
J.B.V. and C.J.C. designed the research; Y.C., J.G., and H.M.W. conducted the research with the aid of S.H.S.; J.B.V. wrote the paper with scientific counsel and support from C.J.C. and Y.C.; and J.B.V. had primary responsibility for final content. All authors read and approved the final manuscript.
Footnotes
Supported by funding from the National Center for Complementary and Alternative Medicine of the NIH (R21AT003485) to J.B.V. and C.J.C. and from the National Science Foundation CRIF program for purchase of the ESI/QIT MS (CHE 0639003).
Supplemental Figures 1 and 2 are available from the “Online Supporting Material” link in the online posting of the article and from the same link in the online table of contents at jn.nutrition.org.
Abbreviations used: CID, collision-induced dissociation; ddH2O, doubly deionized water; ESI, electrospray ionization; GP, graphite powder; LC, light chromatography; LMWCr, low-molecular weight chromium-binding substance; MALDI/TOF, matrix-assisted desorption ionization time-of-flight; MS/MS, tandem MS; PE, pyroglutamate; PSD, post-source decay; QIT, quadrapole ion trap; TFA, trifluoroacetic acid.
Literature Cited
- 1.Vincent JB, Stearns DM. The bioinorganic chemistry of chromium: essentiality, therapeutic agent, toxin, carcinogen? Chichester: Wiley-Blackwell; In press; 2011 [Google Scholar]
- 2.Yamamoto A, Wada O, Ono T. A low-molecular-weight, chromium-binding substance in mammals. Toxicol Appl Pharmacol. 1981;59:515–23 [DOI] [PubMed] [Google Scholar]
- 3.Wu GY, Wada O. Studies on a specific chromium binding substance (a low-molecular-weight chromium binding substance) in urine. Sangyo Igaku. 1981;23:505–12 [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto A, Wada O, Ono T. Distribution and chromium-binding capacity of a low-molecular-weight, chromium-binding substance in mice. J Inorg Biochem. 1984;22:91–102 [DOI] [PubMed] [Google Scholar]
- 5.Wada O, Manabe S, Yamaguchi N, Ishikawa S, Yanagisawa H. Low-molecular-weight, chromium-binding substance in rat lungs and its possible role in chromium movement. Ind Health. 1983;21:35–41 [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto A, Wada O, Manabe S. Evidence that chromium is an essential factor for biological activity of low-molecular-weight Cr-binding substance. Biochem Biophys Res Commun. 1989;163:189–93 [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto A, Wada O, Suzuki H. Purification and properties of biologically active chromium complex from bovine colostrum. J Nutr. 1988;118:39–45 [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto A, Wada O, Suzuki H. Separation of biologically active chromium complex from cow colostrum. Tohoku J Exp Med. 1987;152:211–9 [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto A, Wada O, Ono T. Isolation of a biologically active low-molecular-mass chromium compound from rabbit liver. Eur J Biochem. 1987;165:627–31 [DOI] [PubMed] [Google Scholar]
- 10.Vincent JB. Relationship between glucose tolerance factor and low-molecular-weight chromium-binding substance. J Nutr. 1994;124:117–8 [DOI] [PubMed] [Google Scholar]
- 11.Vincent JB. The biochemistry of chromium. J Nutr. 2000;130:715–8 [DOI] [PubMed] [Google Scholar]
- 12.Davis CM, Vincent JB. Isolation and characterization of a biologically active form of chromium oligopeptide from bovine liver. Arch Biochem Biophys. 1997;339:335–43 [DOI] [PubMed] [Google Scholar]
- 13.Sumrall KH, Vincent JB. Is glucose tolerance factor an artifact produced by acid hydrolysis of low-molecular-weight chromium-binding substance? Polyhedron. 1997;16:4171–7 [Google Scholar]
- 14.Hatfield MJ, Gillespie S, Chen Y, Li Z, Cassady C, Vincent JB. Low-molecular-weight chromium-binding substance from chicken liver and American alligator liver. Comp Biochem Physiol B Biochem Mol Biol. 2006;144:423–31 [DOI] [PubMed] [Google Scholar]
- 15.Chen Y. Low-molecular-weight chromium-binding substance: advanced studies from aves to human [dissertation]. Tuscaloosa (AL): The University of Alabama; 2009 [Google Scholar]
- 16.Chan WC, White PD. Fmoc Solid Phase Peptide Synthesis: a practical approach. New York: Oxford University Press; 2000. p. 345 [Google Scholar]
- 17.Clipston NL, Jai-nhuknan J, Cassady CJ. A comparison of negative and positive ion time-of-flight post-source decay mass spectrometry for peptides containing basic residues. Int J Mass Spectrom. 2003;222:363–81 [Google Scholar]
- 18.Larsen MR, Cordwell SJ, Roepstorff P. Graphite powder as an alternate or supplement to reverse-phase material for desalting and concentration of peptide mixtures prior to matrix-assisted laser desorption/ionization-mass spectrometry. Proteomics. 2002;2:1277–87 [DOI] [PubMed] [Google Scholar]
- 19.Roepstorff P, Fohlman J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed Mass Spectrom. 1984;11:601. [DOI] [PubMed] [Google Scholar]
- 20.Sekiya S, Yamaguchi Y, Kato K, Tanaka K. Mechanistic elucidation of the formation of reduced 2-aminopyridine-derivatized oligosaccharides and their application in matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:3607–11 [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi M, Oka M, Nishida K, Ishida M, Hamazaki A, Kuyama H, Ando E, Okamura T-a, Ueyama N, et al. Enhancement of MALDI-MS spectra of C-terminal peptides by modification of proteins via an active ester generated in situ from an oxazolone. Anal Chem. 2006;78:7861–9 [DOI] [PubMed] [Google Scholar]
- 22.Bonfiglio R, King RC, Olah TV, Merkle K. Effects of sample preparation methods on the variability of electrospray ionization response for model drug compounds. Rapid Commun Mass Spectrom. 1999;13:1175–85 [DOI] [PubMed] [Google Scholar]
- 23.Rink C, Roy S, Khanna S, Rink T, Bagchi D, Sen CK. Transcriptome of the subcutaneous adipose tissue in response to oral supplementation of type 2 Leprdb obese diabetic mice with niacin-bound chromium. Physiol Genomics. 2006;27:370–9 [DOI] [PubMed] [Google Scholar]
- 24.Cheng RYS, Alvord WG, Powell D, Kasprzak KS, Anderson LM. Microarray analysis of altered gene expression in the TM4 Sertoli-like cell line exposed to chromium(III) chloride. Reprod Toxicol. 2002;16:223–36 [DOI] [PubMed] [Google Scholar]
- 25.Dinakarpandian D, Morrissette V, Chaudhary S, Amini K, Bennett B, Van Horn JD. An informatics search for the low-molecular weight chromium-binding peptide. BMC Chem Biol. 2004;4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aksoyoglu S. Cesium sorption on mylonite. J Radioanal Nucl Chem. 1990;140:301–13 [Google Scholar]
- 27.Davis CM, Vincent JB. Chromium oligopeptide activates insulin receptor tyrosine kinase activity. Biochemistry. 1997;36:4382–5 [DOI] [PubMed] [Google Scholar]
- 28.Davis CM, Sumrall KH, Vincent JB. The biologically active form of chromium may activate a membrane phosphotyrosine phopshatase (PTP). Biochemistry. 1996;35:12963–9 [DOI] [PubMed] [Google Scholar]
- 29.Cornish-Bowden A. Principles of enzyme kinetics. London: Buterworths; 1976 [Google Scholar]
- 30.Sun Y, Ramirez J, Woski SA, Vincent JB. The binding of trivalent chromium to low-molecular-weight chromium-binding substance (LMWCr) and the transfer of chromium from transferrin and Cr(pic)3 to LMWCr. J Biol Inorg Chem. 2000;5:129–36 [DOI] [PubMed] [Google Scholar]
- 31.Shiner JS, Solaro RJ. The Hill coefficient for the Ca2+-activation of striated muscle contraction. Biophys J. 1984;46:541–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent JB. Chromium: celebrating 50 years as an essential element? Dalton Trans. 2010;39:3878–94 [DOI] [PubMed] [Google Scholar]
- 33.Vincent JB, Stallings D. Introduction: a history of chromium studies (1955–1995). : Vincent J, editor The nutritional biochemistry of chromium(III). Amsterdam: Elsevier; 2007. p. 1–40 [Google Scholar]
- 34.Vincent JB. The bioinorganic chemistry of chromium(III). Polyhedron. 2001;20:1–26 [Google Scholar]
- 35.Di Bona KR, Love S, Rhodes NR, McAdory D, Sinha SH, Kern N, Kent J, Strickland J, Wilson A, et al. Vincent. Chromium is not an essential trace element for mammals: effects of a “low-chromium” diet. J Biol Inorg Chem. 2011;16:381–90 [DOI] [PubMed] [Google Scholar]
- 36.Levina A, Codd R, Dillion CT, Lay PA. Chromium in biology: toxicology and nutritional aspects. Prog Inorg Chem. 2003;51:145–644 [Google Scholar]
- 37.Jacquamet L, Sun Y, Hatfield J, Gu W, Cramer SP, Crowder MW, Lorigan GA, Vincent JB, Latour J-M. Characterization of chromodulin by X-ray absorption and electron paramagnetic spectroscopies and magnetic susceptibility measurements. J Am Chem Soc. 2003;125:774–80 [DOI] [PubMed] [Google Scholar]
- 38.Pu D, Vincent J, Cassady C. The effects of chromium(III) coordination on the dissociation of peptides. J Mass Spectrom. 2008;43:773–81 [DOI] [PubMed] [Google Scholar]
- 39.Royer AC, Rogers RD, Arrington DL, Street SC, Vincent JB. Spectroscopic studies of he dodecanuclear chromium complex Cr12O9(OH)3(pivalate)15: conformation of the presence of twelve Cr(III) centers and the crystal structure of Cr12O9(OH)3(pivalate)15⋅2PrOH⋅9H2O. Polyhedron. 2002;21:155–65 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.