Abstract
The N-end rule is one ubiquitin-proteolytic pathway that relates the in vivo half-life of a protein to the identity of its N-terminal residue. NTAN1 deamidates N-terminal asparagine to aspartate, which is conjugated to arginine by ATE1. An N-terminal arginine-bearing substrate protein is recognized, ubiquitylated by UBR1/E3α, and subsequently degraded by 26S proteasomes. Previous research showed that NTAN1-deficient mice exhibited impaired long-term memory in the Lashley III maze. Therefore, a series of studies, designed to assess the role of NTAN1 in short- and intermediate-term memory processes, was undertaken. Two hundred sixty mice (126 −/−; 134 +/ +) received Lashley III maze training with intertrial intervals ranging from 2–180 min. Results indicated that inactivation of NTAN1 amidase differentially affects short-, intermediate-, and long-term memory.
The ubiquitin-proteolytic pathway regulates numerous cellular processes, including signal transduction, cell-cycle progression, DNA repair, apoptosis, antigen processing, and gene expression (Varshavsky 1997). The N-end-rule pathway, one subset of the ubiquitin pathway, relates the in vivo half-life of a protein to the identity of its N-terminal residue (Bachmair et al. 1986). In mice, N-terminal Asn and Gln of the N-end-rule substrates are deamidated to Asp and Glu by NTAN1 and NTAQ1, respectively (Grigoryev et al. 1996; Kwon et al. 2000). Amino-terminal Asp and Glu are conjugated with Arg by ATE1, which exists as two distinct species, ATE1–1 and ATE1–2, the products of alternative splicing (Kwon et al. 1999a). Target proteins bearing Arg (or other primary destabilizing residues) are recognized and ubiquitylated by UBR1/E3α (the E3 component of the N-end-rule pathway) with the help of a ubiquitin-conjugating enzyme (E2) and are processively degraded by 26S proteasomes. UBR1 recognizes N-end-rule substrates through its type 1 and type 2 substrate-binding sites (Varshavsky 1996; Kwon et al. 1999b). Mouse UBR1/E3α cDNA has been recently cloned (Kwon 1998), and the mice lacking UBR1 have been recently constructed (Y.T. Kwon and A. Varshavsky, unpubl.). Recently, mice lacking NTAN1 have been constructed (Kwon et al. 2000). These mice specifically lack the asparagine branch of the N-end-rule pathway and were found to be less active in an open field and impaired on several spatial memory tasks (Kwon et al. 2000).
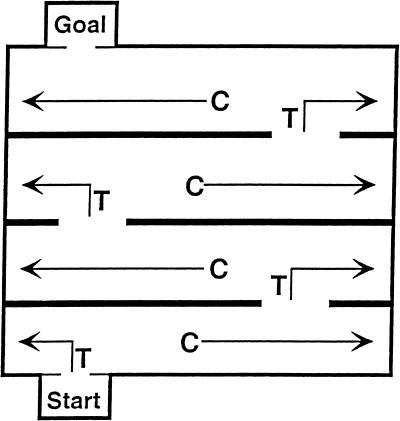
One of these spatial tasks was the Lashley III. This maze contains cul-de-sacs that must be avoided and T-choices at which an animal must learn whether to turn right or left (see Fig. 1). When given one trial per day for 5 d, NTAN1−/− mice were less competent than wild-type mice in learning. Further, when retested ∼8 wk later, NTAN1−/− mice exhibited poorer retention (Kwon et al. 2000). Thus, with an intertrial interval (ITI) of 24 hr, wild-type mice were better learners than NTAN1−/− mice in this maze.
Figure 1.
Overhead view of Lashley III maze. Arrows labelled C indicate a forward-going cul-entry error. Arrows labelled T indicate a forward-going T-choice error.
It is well established that memory is a time-dependent process that requires the consolidation and storage of newly acquired information (McGaugh 1966). In addition, it is well known that the temporal parameters employed during training affect recall (Warden 1923; Travis 1937; Thompson and Pennington 1957; Kamin 1957, 1963; Frieder and Allweis 1982a,b; Yongue and Roy 1985; Bernabeu et al. 1997; Bourtchouladze et al. 1998; Heyser et al. 1999). Research has revealed that memory can be divided into at least two main phases: short-term, which lasts only minutes and requires the modification of preexisting proteins, and long-term, which lasts for days or longer and is accompanied by neural growth and the synthesis of protein (Barondes and Cohen 1966; Cohen and Barondes 1968; Davis and Squire 1984; Bailey et al. 1996). It is important to note, however, that other researchers have further dissected memory into three phases (Ng et al. 1991; Rosenzweig et al. 1991) or four phases (Frieder and Allweis 1982a). Regardless of which system one relies upon, it is generally assumed that an ITI of 24 hr requires the utilization of a long-term presumably protein-dependent memory system (Mark and Watts 1971; Flood et al. 1973; Abel et al. 1997; Bourtchouladze et al. 1998). Inactivation of the NTAN1 gene in mice resulted in impaired long-term memory in the Lashley III maze (Kwon et al. 2000). One interpretation of this result is that a normally short-lived regulatory protein that is targeted for degradation by the N-end-rule pathway through the protein's N-terminal Asn becomes long-lived in NTAN1-deficient mice. The resulting increase in the steady-state concentration of this protein alters the functioning of relevant neural networks in the brain. However, physiological substrates of NTAN1 remain to be identified. The role of NTAN1 in short-term (and intermediate-term) memory systems is still not known.
Therefore, in the current set of experiments, wild-type and NTAN1−/− mice were given five trials in the Lashley III maze all on one day with ITIs varying from 2 to 180 min in anticipation that there would be a time-dependent dimension to the NTAN1−/− performance, as a result of the stabilization of proteins. The range of ITI values was selected so as to measure time points preceding, including, and following the ∼84-min half-life of asparagine-bearing proteins.
RESULTS
Preliminary analyses involving genotype, sex, and/or group were done for all ITIs listed in Table 1 (except for the 2-min data) for each behavioral measure. In only two instances did genotype interact with sex or group. This is to be expected by chance alone. Therefore, the genotype main effect was summed over sex and group. A significant main effect for trials was obtained for cul-forward, backward, and learning index at each ITI, showing that learning occurred. Only the 15- and 60-min ITI groups showed learning on the T-forward error measure.
Table 1.
Genotype, Sex, and Age at Start of Lashley III Testing for Groups of NTAN1 Mice Used in Lashley III Intertrial Interval (ITI) Studies
| ITI
|
Genotype
|
Sex
|
Age
|
||
|---|---|---|---|---|---|
| −/−
|
+/+
|
Male
|
Female
|
(weeks)
|
|
| 2 minutes | |||||
| Group 1 | 11 | 11 | 22 | — | 9 |
| 15 minutes | |||||
| Group 1r | 10 | 10 | 10 | 10 | 13 |
| Group 2r | 11 | 12 | 11 | 12 | 10 |
| 30 minutes | |||||
| Group 1r | 14 | 15 | 12 | 17 | 6 |
| 45 minutes | |||||
| Group 1r | 7 | 11 | 10 | 8 | 21 |
| Group 2r | 13 | 10 | 13 | 10 | 12 |
| 60 minutes | |||||
| Group 1r | 10 | 10 | 20 | — | 18 |
| Group 2r | 12 | 12 | 11 | 13 | 10 |
| 90 minutes | |||||
| Group 1r | 7 | 11 | 11 | 7 | 21 |
| 120 minutes | |||||
| Group 1r | 14 | 15 | 14 | 15 | 11 |
| Group 2r | 7 | 7 | 7 | 7 | 13 |
| 180 minutes | |||||
| Group 1r | 10 | 10 | 10 | 10 | 11 |
Received twenty-four-hour retest following trial five.
Learning Across All ITIs
In order to assess the consequences of varying the ITI upon learning, a 2 (genotype) × 8 (ITI) × 5 (trials) ANOVA was performed,with repeated measures on trials for each behavioral measure. We report only main effects and interactions involving the genotype factor.
These analyses revealed significant genotype × ITI linear trends for cul-forward and backward errors [Fs (1,183) = 11.273 and 3.99, respectively, ps < .05], as well as a significant genotype × ITI quadratic trend for learning index [F(1,183) = 4.92, p < .03]. A significant main effect for genotype was observed for cul-forward errors [F(1,244) = 6.04, p < .02]. The genotype × ITI × trials interaction was not significant in any analysis.
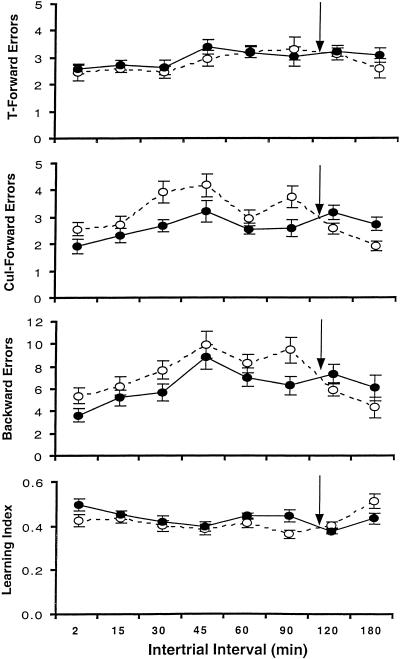
Figure 2 plots mean learning scores for each measure across all ITIs for both genotypes. The significant interactions appear to be due to the crossover of the genotype curves between 90 and 120 min. To determine whether this was so, the data were broken into several subsets and tested for interactions. The first subset consisted of two groupings of the ITIs: 2–60 min and 90–180 min. There were no significant genotype × ITI interactions for any measure in the 2–60 min grouping (Fs < 1), but there were for cul errors, backward errors, and learning index in the 90–180 min grouping [Fs(2,75) = 5.51, 3.78, 4.82, respectively, ps < .05]. The second subset used the ITI groupings of 2–90 min and 120–180 min. This resulted in no significant genotype × ITI interactions for either grouping. The final arrangement was to drop the 180-min ITI and test the remaining groups. This yielded a significant genotype × ITI quadratic trend for cul errors [F(1,226) = 4.07, p < .05]. Because the lack of interactions allows us to generalize within the ITIs, the data were split into two sets: one containing the ITIs of 2–90 min (called “short”); the other containing the ITIs of 120 and180 min (called “intermediate”).
Figure 2.
Lashley III learning data for all intertrial intervals (means ± s.e.m.) for both genotypes. The arrow indicates the point at which performance of the genotypes reverses (closed circles, +/+ mice; open circles, −/− mice)
Short ITIs (2–90 min)
A 2 × 6 × 5 (genotype × ITI × trials) ANOVA was run for all four measures. All interactions involving genotype were non-significant. NTAN1−/− mice made significantly more cul and backward errors [Fs(1,185) = 14.55, 8.92, ps < .01] and had lower learning index scores [F(1,185) = 7.59, p < .01].
Intermediate ITIs (120–180 min)
A 2 × 2 × 5 (genotype × ITI × trials) ANOVA was run for all measures. Again, all interactions involving genotype were non-significant. NTAN1−/− made fewer cul and backward errors [Fs(1,59) = 6.80 and 3.40, ps < .02 and .07] and had higher learning index scores [F(1,59) = 5.99, p < .02].
24-Hr Retest Data
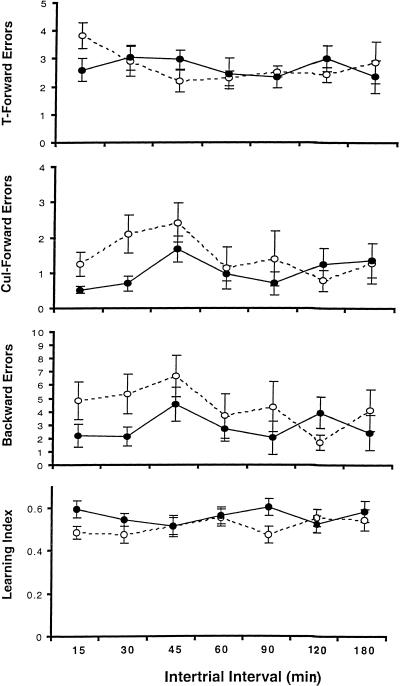
Figure 3 shows the 24-hr retest data for the groups that were retested. These were broken into short ITIs (2–90 min) and intermediate ITIs to parallel the analyses described above. For the short ITIs, 2 × 5 (genotype × ITI) ANOVAs found that NTAN1−/− mice made more cul-forward and backward errors [Fs(1,145) = 7.45 and 6.08, ps < .05] and had lower learning index scores [F(1,145) = 4.69, p < .05]. For the intermediate ITIs, 2 × 5 (genotype × ITI) ANOVAs revealed no differences between the genotypes.
Figure 3.
Twenty-four-hr retest data (means ± s.e.m.) for both genotypes (closed circles, +/+ mice; open circles, −/− mice).
It was of interest to determine if the two genotypes had the same rate of change over the 24-hr interval. Therefore, the analyses described above were redone, this time with trial 5 data included as an additional repeated measures variable. Within the short ITIs, the genotype effect was significant for cul-forward errors and learning index and was near significant for backward errors (p < .08). The only significant trials effect was for cul errors for the short ITI, in which the number of errors on the 24-hr retest was less than on trial 5 of the previous day. We did not find a genotype × trials effect in any of the analyses, indicating that the rate of change was equivalent for the two genotypes.
Comparative Learning Curves
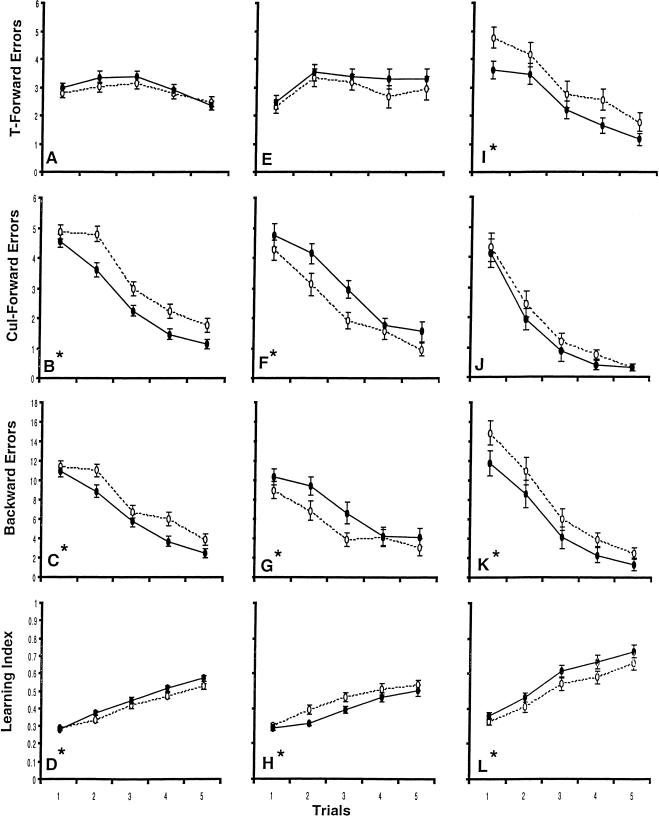
With an ITI of 24 hr, Kwon et al. (2000) found significant genotype effects favoring the wild-type mice on all measures but cul-forward errors. Those learning curves are reproduced in Figure 4I–L. To obtain comparable curves for the animals used in this study, all mice in the short ITIs of 2–90 min were pooled, as were the mice for the intermediate ITIs of 120 and 180 minutes. Their learning curves for the four measures are shown in Figure 4A–H.
Figure 4.
Means (± s.e.m.) per trial for each measure over short intertrial intervals (ITIs) (A–D: +/+ N, 102; −/− N, 95), intermediate ITIs (E–H: +/+ N, 32; −/− N, 31), and long ITIs (I–L: +/+ N, 29; −/− N, 30). The long ITI data are from Kwon et al. (2000). An asterisk indicates a significant main effect for genotype (closed circles, +/+ mice; open circles, −/− mice).
Correlations among the Measures
The learning curves for T- and cul-forward errors in Figure 4 bear no relationship to each other, suggesting that the measures may be independent. To determine this, Pearson correlations among the four Lashley measures were calculated for each genotype for the short, intermediate, and long ITIs. The correlations did not differ between the genotypes, so this classification was dropped and the correlations were recalculated. Within the short and intermediate ITIs, the correlations between T- and cul-forward errors were non-significant (see Table 2), while all the remaining correlations were significant. A similar dissociation between T- and cul-forward has been noted previously (see Jackson and Strong 1969). Within the long ITI, the T- and cul-forward error scores were significantly related, although the correlation was modest. The remaining correlations were numerically higher.
Table 2.
Correlations Between T-Forward and Cul-Forward Errors for Mice in the Short, Intermediate, and Long Intertrial Interval Groups
| Measure | Short | Intermediate | Long |
|---|---|---|---|
| r | .094 | .113 | .4161 |
| N | 197 | 63 | 59 |
p < .01
DISCUSSION
There are three major issues to address: first, why T- and cul-forward errors are independent during short and intermediate memory and modestly correlated in long-term memory; second, justification for a three-part memory system; and third, the relationships between the genotypes, the memory systems, and the behavioral measures.
T- and Cul-Forward Errors
In order to expedite reaching the goal, an animal must learn two things when navigating through the Lashley III maze: (1) where not to go and (2) the correct forward-going path. Figure 1 shows that the mouse must learn the correct sequence of right/left turns at T-choices and to avoid cul-de-sacs (or dead ends).
We have found that both rats and mice eliminate cul-forward errors more quickly than they eliminate T-forward errors (Denenberg et al. 1991; Boehm et al. 1996). Thus, it appears easier to apply a general rule throughout the maze (i.e., never enter a cul), than it is to remember which of two choices is to be made at each of the four T-junction points. Learning not to enter a blind alley may be thought of as a form of inhibitory learning, whereas learning which way to turn at each of the T-choices requires storing specific information in one's memory. We propose that these involve different learning processes and different memory systems and that this leads to a non-significant correlation between T- and cul-forward errors at the short and intermediate ITIs. Although these are different memory systems, the modest correlation between these measures at the long ITI is presumably because of a common requirement for protein synthesis.
The other two measures—backward errors and learning index—were significantly and highly correlated with the cul- and T-forward measures, although for different reasons. Backward errors were positively correlated because the greater the number of backward errors made, the more opportunity there was to make a cul- or T-forward error. The learning index measure was negatively correlated because the denominator of this ratio number is the sum of all errors made. Thus, the cul-and T-forward error measures are the only two that represent different learning processes, and the remainder of the discussion will focus upon them.
Before doing so, however, it is necessary to address an alternative interpretation, one not based upon an effect upon learning and memory. It may be suggested that the larger number of cul errors made by NTAN1−/− mice during the short ITIs is because they swam faster than wild-type mice and, therefore, their momentum carried them past the choice point into a cul-de-sac. We do not measure swimming speed in the Lashley III maze. Nonetheless, we did not find a difference in swimming in the water escape task in this or other studies with these mice, nor have we ever found genotype differences in swimming speed in the Morris water maze (Kwon et al. 2000). Thus, it appears unlikely that speed of swimming was a determining factor in the behavioral results we observed.
Tripartite Memory System
The data from the present studies support a three-part memory system. The short-term and intermediate-term ITI memory systems are distinguished by the genotype reversals on the cul-forward errors, with the wild-type mice exhibiting superior learning from 2 to 90 min but inferior learning from 120 to 180 min. The long-ITI memory system differs from both the short and intermediate systems by having a learning curve for T-forward errors, whereas the short and intermediate ITI groups do not; having a genotype effect favoring wild-type mice on the T-forward errors, whereas no genotype effect was obtained with either the short or intermediate ITIs; not having a genotype effect on the cul-forward measure, whereas significant genotype effects were found for the other two ITI units; and by having a significant correlation between cul- and T-forward errors.
As has been the case with prior research aimed at establishing the temporal boundaries of learning and memory, the short- and intermediate-term memory systems described herein were defined based upon the consistency of genotype differences across the differing ITIs, as indicated by lack of significant interactions at the behavioral level. It is recognized that at the molecular level the various ITIs might each activate a slightly different set of biochemical pathways. However, behavioral homogeneity in the presence of molecular heterogeneity is characteristic of a biological system (Bertalanfy 1969; Weiss 1969).
The present findings are in accord with the conclusions of several researchers that there are at least three memory systems that act either in series or in parallel (Barondes and Cohen 1966; McGaugh 1966; Frieder and Allweis 1978, 1982a). Furthermore, the short- and intermediate-term memory systems found in this study generally fit within the time frames of the short- and intermediate/medium-term memory systems proposed by others (Daniels 1971; Frieder and Allweis 1978, 1982a; Davis and Squire 1984; Rosenzweig et al. 1991; Bailey et al. 1996), despite the often disparate temporal parameters for learning and memory between species and tasks used (Frieder and Allweis 1982b).
Biochemically, the short-term mechanism has been suggested to involve electrical change (i.e., short-term cellular change) (John 1971; Gibbs et al. 1978), whereas the long-term mechanisms are, by and large, considered to be a manifestation of protein synthesis or macromolecular storage (Bailey et al. 1996; Barondes and Cohen 1966; Cohen and Barondes 1968; John 1971; Davis and Squire 1984;). The mechanisms of the intermediate-term processes, or intermediate holding mechanism, however, are unclear. Ionic changes or alterations in RNA synthesis have been proposed (Hyden and Egyhazi 1962; John 1971), although the latter has been shown to be an unlikely candidate (Briggs and Kitto 1962; Smith 1962; Barondes and Jarvik 1964). To date, what exactly occurs at this intermediate time point remains a mystery. Either way, each of these stages has been shown to be differentially affected by pharmacological interventions aimed at their suspected modes of action. This is consistent with the notion that consolidation is a result of parallel rather than serial components (Barondes and Cohen 1966, 1967; Kobiler and Allweiss 1977; Frieder and Allweis 1978). These data suggest that these memory systems are presumably evoked and controlled by different, but perhaps related, biochemical processes.
The Effects of Genotype
Inactivation of the NTAN1 gene in mice revealed a tripartite memory system for Lashley III maze learning as indexed by cul- and T-forward errors. We have suggested that the cul-forward error score is a measure of inhibitory learning and demonstrated that in NTAN1−/− animals this score is depressed during short-term memory but facilitated during intermediate-term memory. We propose that if short-term cellular change (Gibbs et al. 1978; John 1971) is responsible for this type of learning, that perhaps neural overexcitation, brought upon by inactivating the NTAN1 gene and stabilizing asparagine-bearing target proteins, differentially and selectively affects both of these memory processes. We find the correspondence between the ∼84-min half-life of asparagine-bearing proteins and the breakpoint between the short- and intermediate-term memory systems (i.e., between 90 and 120 min) intriguing. At present, however, we have no suggestions to offer as to specific underlying mechanisms of this effect. The failure of the genotypes to differ at the 24-hr ITI suggests that the three systems run in parallel.
In contrast to cul-forward errors, T-forward errors, which measure how effectively the animal has learned the correct path to the goal, were not reduced for any ITI between 2 and 180 min. Learning did occur at the 24-hr ITI, but NTAN1−/− mice were inferior relative to wild-type mice. Thus, we conclude that protein synthesis is necessary for T-choice learning to occur, and that prolonging the presence of asparagine-bearing proteins interferes with this and/or related processes. Because the two error measures are independent of each other, it well may be that two different biochemical mechanisms are involved in the establishment of memories within these memory systems.
Our findings suggest that proteolytic processes are used to establish different phases of memory, each of which requires presumably related yet unique proteolytic components. Because no specific baseline physiological substrates of NTAN1 involved in learning and memory have been identified, the specific biochemical underpinnings of these three memory systems are yet to be elucidated. We view this as a working hypothesis since it is likely that eliminating this one gene has affected many processes, in addition to learning and memory. Because the genome organizes itself in a systems fashion (Wahlsten 1999; Denenberg 2000), we recognize that the behavioral profile of NTAN1-deficient mice represents the interactive consequences of several of these processes, rather than being the exclusive province of purely cognitive measures.
MATERIALS AND METHODS
Subjects
Table 1 shows the genotype, sex, and age at the start of Lashley III testing for the 260 NTAN1−/− mice that made up the 12 independent groups tested in the current studies. The construction and breeding of these mutant mice have been described previously (Kwon et al. 2000). All of the mice described herein were offspring of homozygous breeding pairs (−/− × −/− and +/ + × +/ +) bred at the University of Connecticut's Developmental Psychobiology Laboratory. All mice were maintained singly housed on a 12-hr light-dark cycle (lights on at 0600 hr) with food and water ad libitum. Experimenters remained blind to the genotype of the mice until behavioral testing was complete.
All mice were given water escape training prior to the Lashley III maze testing. Both genotypes learned to escape and there was no genotype effect. This measure will not be discussed further.
Lashley III Maze Training
Figure 1 is a schematic of the Lashley III maze, showing cul-forward errors and T-choice forward errors. A water version of the maze was used, with the temperature maintained at 19–21°C (Denenberg et al. 1991). Each mouse received a total of five trials given at one of the following ITIs: 2, 15, 30, 45, 60, 90, 120, or 180 min.
The measures of learning included a learning index defined as number of correct entries divided by total number of entries, cul-forward errors, forward T-choice errors, and backward errors (any backward entry was considered an error).
The majority of groups received one additional trial 24 hr after their last training trial (see Table 1).
Acknowledgments
This work was supported in part by NIH Grant HD20806 to V.H.D.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dberg@uconnvm.uconn.edu; FAX (860) 486-3827.
Article and publication are at www.learnmem.org/cgi/doi/10.1101/lm.33500.
REFERENCES
- Abel T, Nguyen PV, Barad M, Deuel TAS, Kandel ER, Bourtchouladze R. Genetic demonstration of a role of PKA in the late phase of LTP and in hippocampus based long-term memory. Cell. 1997;88:615–626. doi: 10.1016/s0092-8674(00)81904-2. [DOI] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Bailey C, Bartsch D, Kandel ER. Toward a molecular definition of long-term memory storage. Proc Nat Acad Sci. 1996;93:13445–13452. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barondes SH, Jarvik ME. The influence of actinomycin-D on brain RNA synthesis and on memory. J Neurochem. 1964;11:187–195. doi: 10.1111/j.1471-4159.1964.tb06128.x. [DOI] [PubMed] [Google Scholar]
- Barondes SH, Cohen HD. Puromycin effect on successive phases of memory storage. Science. 1966;151:594–595. doi: 10.1126/science.151.3710.594. [DOI] [PubMed] [Google Scholar]
- ————— Delayed and sustained effect of acetoxycycloheximide on memory in mice. Proc Nat Acad Sci. 1967;58:157–164. doi: 10.1073/pnas.58.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernabeu R, Bevilaqua L, Ardenghi P, Bromberg E, Schmitz P, Bianchin M, Izquierdo I, Medina JH. Involvement of hippocampal cAMP/cAMP-dependent protein kinase signaling pathways in a late memory consolidation phase of aversively motivated learning in rats. Proc Nat Acad Sci. 1997;94:7041–7046. doi: 10.1073/pnas.94.13.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertalanffy LV. Chance or law. In: Koestler A, et al., editors. Beyond Reductionism. Boston, Massachusetts: Beacon Press; 1969. pp. 56–76. [Google Scholar]
- Boehm G, Sherman GF, Rosen GD, Galaburda AM, Denenberg VH. Neocortical ectopias in BXSB mice: Effects upon reference and working memory systems. Cerebral Cortex. 1996;6:696–700. doi: 10.1093/cercor/6.5.696. [DOI] [PubMed] [Google Scholar]
- Bourtchouladze R, Abel T, Berman N, Gordon R, Lapidus K, Kandel ER. Different training procedures recruit either one or two critical periods of contextual memory consolidation, each of which requires protein synthesis and PKA. Learn & Mem. 1998;5:365–374. [PMC free article] [PubMed] [Google Scholar]
- Briggs MH, Kitto GB. The molecular basis of memory and learning. Psych Rev. 1962;69:537–541. doi: 10.1037/h0045270. [DOI] [PubMed] [Google Scholar]
- Cohen HD, Barondes SH. Effect of acetoxycycloheximide on learning and memory of a light-dark discrimination. Nature. 1968;218:271–273. doi: 10.1038/218271a0. [DOI] [PubMed] [Google Scholar]
- Daniels D. Acquisition, storage, and recall of memory for brightness discrimination by rats following intracerebral infusion of acetoxycycloheximide. J Comp Phys Psych. 1971;76:110–118. doi: 10.1037/h0031044. [DOI] [PubMed] [Google Scholar]
- Davis H, Squire LR. Protein synthesis and memory: A review. Psych Bull. 1984;96:518–559. [PubMed] [Google Scholar]
- Denenberg VH. Evolution proposes and ontogeny disposes. Brain & Lang. 2000;73:274–296. doi: 10.1006/brln.2000.2307. [DOI] [PubMed] [Google Scholar]
- Denenberg VH, Talgo N, Carroll DA, Freter S, Deni R. A computer-aided procedure for measuring Lashley III maze performance. Phys & Beh. 1991;50:857–861. doi: 10.1016/0031-9384(91)90031-i. [DOI] [PubMed] [Google Scholar]
- Flood JF, Rosenzweig MR, Bennett EL, Orme AE. The influence of duration of protein synthesis inhibition on memory. Phys & Beh. 1973;10:555–562. doi: 10.1016/0031-9384(73)90221-7. [DOI] [PubMed] [Google Scholar]
- ————— Transient hypoxic-amnesia: Evidence for a triphasic memory-consolidating mechanism with parallel processing. Behav Bio. 1978;22:178–189. doi: 10.1016/s0091-6773(78)92200-9. [DOI] [PubMed] [Google Scholar]
- ————— Memory consolidation: Further evidence for the four-phase model from the time-courses of diethyldithiocarbamate and ethacrynic acid amnesias. Phys & Beh. 1982a;29:1071–1075. doi: 10.1016/0031-9384(82)90300-6. [DOI] [PubMed] [Google Scholar]
- ————— Prevention of hypoxia-induced transient amnesia by post-hypoxic hyperoxia. Phys & Beh. 1982b;29:1065–1069. doi: 10.1016/0031-9384(82)90299-2. [DOI] [PubMed] [Google Scholar]
- Gibbs M, Gibbs CL, Ng KT. A possible physiological mechanism for short-term memory. Phys & Beh. 1978;2:619–627. doi: 10.1016/0031-9384(78)90255-x. [DOI] [PubMed] [Google Scholar]
- Grigoryev S, Stewart AE, Kwon YT, Arfin SM, Bradshaw RA, Jenkins NA, Copeland NG, Varshavsky A. A mouse amidase specific for N-terminal asparagine. The gene, the enzyme, and their function in the N-end rule pathway. J Bio Chem. 1996;271:28521–28532. doi: 10.1074/jbc.271.45.28521. [DOI] [PubMed] [Google Scholar]
- Heyser CJ, McDonald JS, Polis IY, Gold LH. Strain distribution of mice in discriminated Y-maze avoidance learning: Genetic and procedural differences. Behav Neurosci. 1999;113:91–102. doi: 10.1037//0735-7044.113.1.91. [DOI] [PubMed] [Google Scholar]
- Hyden H, Egyhazi E. Nuclear RNA changes of nerve cells during a learning experiment in rats. Proc Nat Acad Sci. 1962;48:1366–1373. doi: 10.1073/pnas.48.8.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson WJ, Strong PN. Differential effects of hippocampal lesions upon sequential tasks and maze learning by the rat. J Comp Phys Psych. 1969;68:442–450. doi: 10.1037/h0027502. [DOI] [PubMed] [Google Scholar]
- John ER. Brain Mechanisms of Memory. In: McGaugh JL, editor. Psychobiology. New York, New York: Academic Press; 1971. pp. 199–283. [Google Scholar]
- Kamin L. The retention of an incompletely learned avoidance response. J Comp Phys Psych. 1957;50:457–460. doi: 10.1037/h0044226. [DOI] [PubMed] [Google Scholar]
- ————— Retention of an incompletely learned avoidance response: Some further analyses. J Comp Phys Psych. 1963;56:713–718. doi: 10.1037/h0043941. [DOI] [PubMed] [Google Scholar]
- Kobiler D, Allweis C. Retrograde amnesia production by the intracisternal injection of 20 micronl of saline in rats. Pharm Biochem Beh. 1977;6:255–258. doi: 10.1016/0091-3057(77)90022-3. [DOI] [PubMed] [Google Scholar]
- Kwon Y, Reiss Y, Fried VA, Hershko A, Yoon JK, Gonda DK, Sangan P, Copeland NG, Jenkins NA, Varshavsky A. The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc Nat Acad Sci. 1998;95:7898–7903. doi: 10.1073/pnas.95.14.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YT, Kashina AS, Varshavsky A. Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway. Mol Cell Bio. 1999a;19:182–193. doi: 10.1128/mcb.19.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YT, Levy F, Varshavsky A. Bivalent inhibitor of the N-end rule pathway. J Bio Chem. 1999b;274:18135–18139. doi: 10.1074/jbc.274.25.18135. [DOI] [PubMed] [Google Scholar]
- Kwon YT, Balogh SA, Davydov IV, Kashina AS, Yoon JK, Xie Y, Gaur A, Hyde LA, Denenberg VH, Varshavsky A. Altered activity, social behavior, and spatial memory in mice lacking the NTAN1p amidase and the asparagine branch of the N-end rule pathway. Mol Cell Bio. 2000;20:4135–4148. doi: 10.1128/mcb.20.11.4135-4148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark RF, Watts ME. Drug inhibition of memory formation in chickens. I. Long-term memory. Proc Royal Soc London. 1971;178:439–454. doi: 10.1098/rspb.1971.0074. [DOI] [PubMed] [Google Scholar]
- McGaugh JL. Time-dependent processes in memory storage. Science. 1966;153:1351–1358. doi: 10.1126/science.153.3742.1351. [DOI] [PubMed] [Google Scholar]
- Ng KT, Gibbs ME, Crowe SF, Sedman GL, Hua F, Zhao W, O'Dowd B, Rickard N, Gibbs CL, Sykova E, et al. Molecular mechanisms of memory formation. Mol Neurobio. 1991;5:333–350. doi: 10.1007/BF02935556. [DOI] [PubMed] [Google Scholar]
- Rosenzweig M, Bennett EL, Martinez JL, Beniston D, Colombo PJ, Lee DW, Patterson TA, Schulteis G, Serrano PA. Stages of memory formation in the chick: Findings and problems. In: Andrew RJ, editor. Neural and Behavioural Plasticity in the Domestic Chick. Oxford, England: Oxford University Press; 1991. pp. 394–418. [Google Scholar]
- Smith CE. Is memory a matter of enzyme induction? Science. 1962;138:889–890. doi: 10.1126/science.138.3543.889. [DOI] [PubMed] [Google Scholar]
- Thompson R, Pennington DF. Memory decrement produced by ECS as a function of distribution in original learning. J Comp Phys Psych. 1957;50:401–404. doi: 10.1037/h0042901. [DOI] [PubMed] [Google Scholar]
- Travis R. The effect of the length of the rest period on motor learning. J Psych. 1937;3:189–194. [Google Scholar]
- Varshavsky A. The N-end rule: Functions, mysteries, uses. Proc Nat Acad Sci. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- Wahlsten D. Single-gene influences on brain and behavior. Ann Rev Psych. 1999;50:599–624. doi: 10.1146/annurev.psych.50.1.599. [DOI] [PubMed] [Google Scholar]
- Warden C. The distribution of practice in animal learning. Comp Psych Mono. 1923;1:1–64. [Google Scholar]
- Weiss P. The living system: Determinism stratified. In: Koestler A, et al., editors. Beyond Reductionism. Boston, Massachusetts: Beacon Press; 1969. pp. 3–42. [Google Scholar]
- Yongue BG, Roy EJ. Adrenalectomy fails to alter radial maze performance of rats at retention intervals of 24 hours or less. Phys & Beh. 1985;35:651–654. doi: 10.1016/0031-9384(85)90157-x. [DOI] [PubMed] [Google Scholar]