Abstract
Atlases of the human brain have an important impact on neuroscience. The emergence of ever more sophisticated imaging techniques, brain mapping methods and analytical strategies has the potential to revolutionize the concept of the brain atlas. Atlases can now combine data describing multiple aspects of brain structure or function at different scales from different subjects, yielding a truly integrative and comprehensive description of this organ. These integrative approaches have provided significant impetus for the human brain mapping initiatives, and have important applications in health and disease.
The concept of the brain atlas is not new1. Cartographic approaches have been used for centuries to identify and target specific regions in the brain and to establish spatial relationships between a coordinate and a structure. Comprehensive maps of brain structure have been created, at a variety of spatial scales, from anatomical specimens2-5 and various histological preparations that reveal regional cytoarchitecture6,7, myelination patterns8-10, and protein and mRNA distributions. Most early and some more recent atlases of the human brain were derived from one, or at best a few, individual post-mortem specimens3-5,11-14. Such atlases provide anatomical references or represent a particular feature of the brain15,16, such as a specific neurochemical distribution17 or the cellular architecture of the cerebral cortex6. For example, Brodmann's map (1909) exclusively describes the cytoarchitectonic segregation of the cortex6, Dejerine's map (1901) describes fibre tract anatomy18, and the map by Schaltenbrand and Wahren (1977) describes the thalamus14.
Beyond these traditional, anatomical atlases based on post-mortem tissue, modern brain atlases are being developed that incorporate flexible, computable systems, which accommodate the sometimes considerable variation in a population. The application of magnetic resonance imaging (MRI) to acquire detailed descriptions of anatomy in vivo is a driving force in brain mapping research. Tomographic imaging has the advantage of largely retaining the spatial integrity of the data by maintaining the intrinsic three-axis registration and simple volumetric coordinates19,20. The atlases derived from these images are digital, allowing a wealth of computational algorithms to be applied to automatically align new two- and three-dimensional imaging data into the coordinate systems of these atlases21. Furthermore, there is an increasing ability to image various structural (as well as functional and chemical) features in the brain such as nuclei, cytoarchitectural details and white matter tracts. Technological advances continue to improve spatial and contrast resolution and have led to multispectral characterization (using MRI) of brain anatomy, reflecting features such as lipid content or water diffusivity in different tissues. Recently, chemoarchitectural maps, which describe receptor densities relative to their cytoarchitectonic localization and functional attributes, have been generated21,22 to bridge the gap between anatomical and functional observations.
The transition from a static atlas representation to a computational one has dramatically extended the atlas concept. The brain atlas is now equivalent to a database that incorporates a multitude of data points that are organized, relational, extendable and testable. Initially, the brain atlas was purely neuroanatomical, based on a single representative example. Now it can include population statistics on structure, gene expression, receptor patterns or connectivity over time.
In this review, we describe the features and limitations of traditional methods of creating brain atlases and outline the requirements of a modern brain atlas. So far, most atlasing efforts have consisted of grey matter maps obtained from structural and functional MRI (fMRI) data. Here, we discuss the background and applications of human brain mapping, focusing on a few selected examples of methods that have shown rapid progress in recent years. These newer methods can be used to generate statistical data on cortical cytoarchitecture and receptor distributions, as well as white matter fibre tracts and projections, using techniques such as diffusion tensor imaging (DTI). It is likely that these methods will contribute to the future development of advanced integrative atlases. Brain maps that visualize distributed patterns of task-dependent functional activity are now beginning to be related to anatomical fibre connectivity computed from DTI — each modality contributes to better hypotheses and models regarding neural systems involved in cognitive function. We go on to evaluate several existing integrative, population-based international initiatives. Algorithms that are used to analyse probabilistic and time-varying atlases have led to important findings regarding some of the diseases that affect the brain, and have provided new perspectives on the effects of age, gender and genetic factors. We discuss how these atlases are evolving rapidly as developments in imaging modalities, post-mortem mapping and computational analysis of images are combined to detect new features that were not observable (or measurable) in the past.
The evolution of brain atlases
Lessons from the past
Early research on the cellular composition of the brain culminated in the development of atlases of the human cerebral cortex, which were pioneered by Brodmann6,23, Flechsig8, Vogt and Vogt10, and von Economo and Koskinas7. These studies continued until the 1960s (REFS 24-26). The cortex was segregated into numerous structurally defined areas, based on regional cytoarchitecture (identified mainly by the number of cortical layers, laminar patterns of cell packing density and the shape of neuronal cell bodies) or myeloarchitecture (identified mainly by the degree of myelination and the presence or absence of myelinated fibre bundles in the cerebral cortex). These types of architecture were studied by visual inspection of Nissl- or myelin-stained histological sections in single brains.
However, when maps by different authors of this classical period are considered, a number of problems become apparent. For example, the drawings do not provide the sulcal pattern of a single real brain or a well-defined ‘average’ brain but of an imagined ‘ideal’ brain (for example, Brodmann's schematic drawing of a brain6); so, it is impossible to compare different maps in a common reference space as they are not registered in a stereotaxic system, and brain shape and sulcal contours vary greatly between maps. The number of cortical areas also varies between the different maps, as the delineation procedure for areal boundaries was highly observer-dependent and not assessed quantitatively or statistically. Furthermore, with few exceptions, the drawings or descriptions do not show the positions of areas and their borders in the cortical sectors that are hidden in the sulci. This amounts to nearly two-thirds of the total surface area of the cortex27. Any designation of cortical areas in the sulcal part of the cortex5 is pure supposition, and not supported by original observations. Finally, most classical maps9,10 do not match the high degree of cortical segregation that has more recently been shown by functional imaging — particularly in the multimodal association cortices28-31.
Indeed, architectonic brain mapping based on visual inspection has been severely criticized24,32 because of its lack of clearly stated and objectively verifiable criteria. This led to a decrease in the general interest in architectonic atlases for decades. More recently, axonal tracing techniques have been combined with architectonic observations to produce atlases of selected cortical regions in non-human primates and other mammals28,33. Unfortunately, this experimental approach cannot be used in studies of the human brain. Meanwhile, stereotaxic atlases of the human brain, based on imaging data, began to be published (mostly in book form), primarily in response to the needs of neurosurgeons. These atlases compensate for some of the problems of the classical brain maps — particularly the lack of a spatial reference system. However, stereotaxic atlases tend to focus on subcortical structures and provide only sparse information about the cortex14. Furthermore, they lack quantitative information on the intersubject variability of areal boundaries11, intersubject variability in gross anatomy, left–right asymmetries and the criteria to identify architectonic areas5.
The driving force for modern brain atlases
The current resurgence of interest in brain atlases that provide architectonic maps of the human cerebral cortex has been largely motivated by the introduction of functional imaging techniques such as positron emission tomography (PET) and fMRI. Researchers using these techniques invariably want to define — as far as possible with the limited resolution of PET or fMRI — the location of neural activity and determine whether the focus of activity is associated with a portion of a specific cortical area, an entire cortical area or overlapping areas.
The development of functional imaging techniques has therefore relied on the evolution of structural imaging. These advances have included both in vivo, structural MRI and high-resolution imaging of three-dimensional reconstructed histological sections. An example of the use of these techniques has been the development of the concept of a ‘cortical area’, which has an important role in functional brain mapping. Cortical areas can be defined structurally on the basis of either macroscopic or microscopic (architectonic) criteria, although architectonic borders vary considerably in relation to macroscopic landmarks34-37. The concept of ‘brain mapping’ benefits from the combination of functional imaging of a distinct and well-defined function with a microstructurally defined architectural atlas, because the correlation between function and its underlying cyto- or myeloarchitecture can then be tested by using the architectural map for the definition of regions of interest in functional imaging.
The correlation between the (micro)structure and the function of an area has been established for many cortical regions, in particular for primary sensory and motor areas7,8,38-41. Many functional units in the cortex are consistently found in humans and other primates, such as primary visual area V1, or the motion-sensitive area MT/V5, in the middle temporal gyrus. The spatial layout of these regions in the cortex is also somewhat consistent across individuals38,39,42,43. Visual areas V1–V5 were initially defined electrophysiologically, based on their highly selective responses to visual stimuli with specific intensities, orientations or directions of motion29,30,38,44,45. These classifications allow functional imaging studies to relate their observations to known anatomical units that were initially defined using electrophysiology or direct histological mapping.
Requirements for modern brain atlases
Based on the criticisms of architectonic brain mapping24,32, the requirements of basic and clinical brain research and the current progress in structural and functional brain imaging techniques, the criteria for ideal brain atlases can be established. They should include a multimodal micro-structural approach that combines different independent methods, such as cytoarchitectonic, myeloarchitectonic, chemoarchitectonic and modern fibre-tracking approaches along with macroscopic, in vivo and tomographic imaging methods, and use observer-independent methods46-49 to define cortical boundaries — so-called architectonic parcellation. Brain atlases should contain data on interindividual variations in the geometry and topology of architectonic areas and should allow the use of algorithms that perform adequate linear (such as scaling, rotating, translation and shearing) or nonlinear (based, for example, on an elastic model50,51) alignment of individual brains and their structures to a standard spatial reference system. Nonlinear transformations, which apply local contractions or dilations, are often required (as well as linear transformations such as local rotations and shearing) to equate, or deform, an individual subject's anatomy to match the shape of a brain atlas21. In addition, efforts should be made to establish population brain maps (probability maps), which define the probabilistic position and borders of cortical areas based on studies of a large number of subjects across various population groups. Finally, these atlases should use a database format that provides easy access to the original data on which the atlas is based (for example, anonymized images from individual subjects, or models of specific brain structures). Alternatively, the statistical maps and anatomical delineations in the atlas should be readily importable into software and tool boxes that are commonly used when analysing new brain images. Once imported into other image analysis software, probabilistic maps of cortical regions or fibre pathways can be used to define regions of interest on new images, to provide a priori constraints or search regions for new statistical analyses, or simply to provide visual overlays of data from an alternative modality.
New architectonic atlas approaches
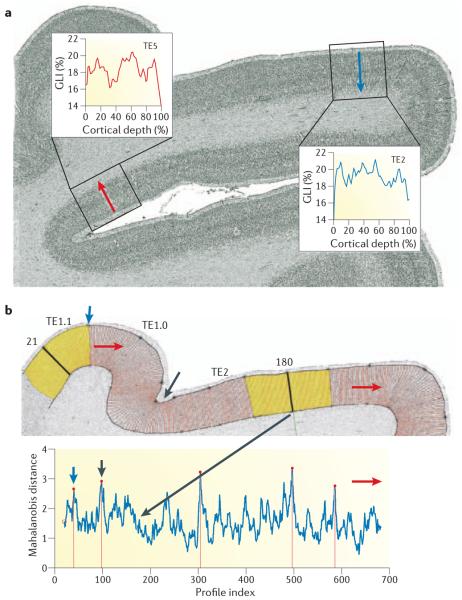
The limitations of classical brain maps led to the evolution of new concepts for generating atlases34,43,52,53. As a result of these conceptual developments, an objective, observer-independent procedure for the parcellation of the cerebral cortex has recently been introduced (FIG. 1). This technique is based on the measurement of the grey-level index (GLI) as an indicator of the volume fraction of cell bodies throughout the cortical layers from the surface to the white matter border in cell body-stained histological sections, and subsequent multivariate analysis of changes in the laminar pattern46. Using this procedure, areal borders can be more reproducibly mapped than in previous studies, which were based on visual inspections34. Three-dimensional reconstructions of post-mortem brains, including cytoarchitectonically defined cortical areas, are then elastically registered or ‘warped’ to the MRI volume of an in vivo brain that is also used as a spatial reference for the registration of functional imaging data50,54,55. This strategy (FIG. 2) allowed the classical maps to be corrected56 and provided a quantitative description of the intersubject variability of cytoarchitectonic areas34-37,40,57,58. It has also facilitated linear or nonlinear registration of individual brains and their structures to a spatial reference system and the discovery of hitherto unknown cytoarchitectonic areas such as intraparietal areas hIp1 and hIp2 (REF. 59), areas of the secondary somatosensory cortex OP1–4 (REF. 60) and area hOc5 of the extrastriate cortex61.
Figure 1. Observer-independent procedure for cytoarchitectonic parcellations.
a | Two mean cortical profiles (red and blue curves in the insets) sampled from the grey-level index (GLI) image of a cell body-stained histological section (coronal plane) through the superior and the middle temporal gyri of a human brain. Each mean profile is the average of 20 equally spaced, individual profiles (red lines in panel b, top), which were sampled from the rectangular regions of interest (boxes centred around the red or blue arrows). The shape of each profile is described as a feature vector46, and is a measure of the cytoarchitecture of the area. High GLI values indicate high volume densities of cell bodies. b | Top panel shows the sliding window procedure used to establish the distance function (bottom panel). The cortical region of interest is divided into a set of profiles (positions shown in red). The positions of the profiles are consecutively numbered starting from n = 1 (at left margin) to k. A sliding window consists of two cortical segments (yellow, to the left and to the right of a central profile) made up of two neighbouring groups of n individual profiles. As an example, sliding windows are shown at positions n = 21 and n = 180. When a certain profile position has been analysed, the sliding window is moved one step (profile) to the next position. The red arrows indicate the direction of the movement of the sliding window across the cortical ribbon. The blue and black arrows indicate positions where the feature vectors show significant changes. TE1.0, TE1.1 and TE2 are distinct cytoarchitectonic areas of the human auditory cortex64,82. Bottom panel shows the Mahalanobis distance, which indicates the dissimilarity in laminar pattern between two cortical segments and can be calculated from the feature vectors from each profile. The Mahalanobis distance at each position of the sliding window is plotted. Significant maxima at positions n = 40 (blue arrow), n = 97 (black arrow), n = 305, n = 497 and n = 586 indicate the positions of putative areal borders. Performing this procedure in serial histological sections of several (usually ten) brains, three-dimensional reconstructed histological data sets of post-mortem brains and their areas are registered to common standard reference space, and cytoarchitectonic probability maps are calculated for each area.
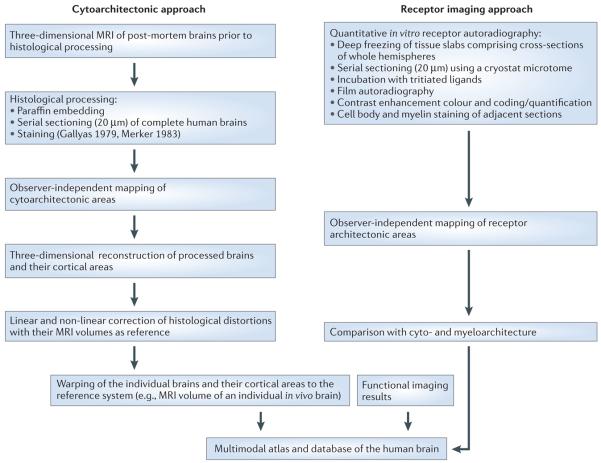
Figure 2. Summary of procedures for generating a multimodal probabilistic atlas.
Probabilistic atlases are generated by entering cytoarchitectonic, receptor architectonic and functional imaging data into a common spatial reference system.
This integrated approach has also been used to create cytoarchitectonic probability maps of the human cortex22,34-36,40,52,53,56,58,62-70. The resulting probability maps have been used for the anatomical interpretation of functional imaging observations40,41,45,52,70-75. For example, it has been shown that left area 45 is more involved in semantic processing than left area 44 (REF. 71). So, schematic drawings of maps and areal borders have been replaced with probability and maximum probability maps55 (FIG. 3). This allows the visualization of the intersubject variability in cytoarchitecture and quantitative definition of the presence and extent of cytoarchitectonic borders and cortical areas in all voxels of the reference brain or space.
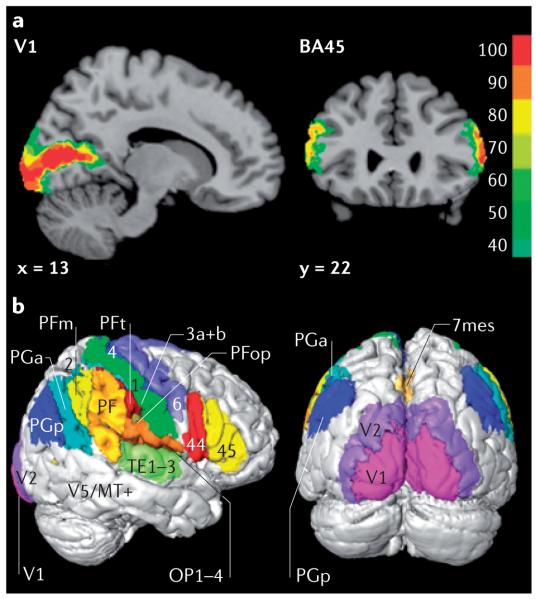
Figure 3. Cytoarchitectonic probability maps of the cortex.
a | Probability maps of the primary visual cortex V1 in a sagittal section (left) and the rostral part (Brodmann's area 45 (BA45); right) of Broca's speech region in a coronal section. x and y refer to the spatial location of the sections in Montreal Neurological Institute (MNI) space54. The colour scale indicates the probabilities that the areas are present in a certain voxel of the reference space. b | Right lateral and occipital views of the maximum probability map in which each voxel of the three-dimensional space has been assigned to the cortical area with the highest probability. Cortical areas shown include areas 44 and 45 (Broca's region), primary motor (4) and premotor (6) areas, the somatosensory cortex (areas 3a, 3b, 1 and 2), inferior posterior parietal association areas (PFt, PFop, PF, PFm, PGa, PGp), parietal opercular areas (OP 1–4), mesial superior parietal area (7mes), auditory areas (TE 1–3) and visual areas (V1, V2 and V5/MT).
Architectonic probability maps from a number of post-mortem brains can be spatially warped onto an individual or ‘average’ brain (represented by a high-resolution MRI volume) as a common spatial reference system. This provides a powerful tool for the anatomical interpretation of functional imaging data, which can also be developed into a topographically organized multimodal database of the human brain.
Molecular architectonics
Histochemical and immunohistochemical methods have greatly improved our knowledge of the regional chemoarchitecture of the cerebral cortex, mainly in animals. However, the often demanding requirements of these methods regarding tissue preservation and fixation have limited their application in human post-mortem brains. Moreover, a large-scale analysis of serial sections of whole human brain hemispheres is hampered by the time-consuming procedures, regionally unequal staining quality, lack of precise reproducibility and high costs. However, analysis of large brain sections is a necessary prerequisite in many mapping projects. Recent developments in the detection of neurotransmitter receptors have overcome this limitation and allowed the molecular mapping of human brains22,34,76-80. Neurotransmitter receptors are important for signal transduction in the brain; many types of receptor are expressed in any given cortical area, and even within a single neuron or glial cell22. So, information on regional differences in receptor density (receptor architecture) is an important feature of a modern brain atlas.
The distribution patterns of neurotransmitter receptors can be assessed using quantitative in vitro receptor autoradiography. This method is based on the binding of radioactively labelled ligands selective for binding sites of a defined receptor type or subtype, and subsequent analysis of binding site densities. The analysis of numerous receptor types in the same brain region is also feasible, and allows insight into the balance of neurotransmitter systems. Receptor autoradiography has numerous advantages. Most importantly, it is possible to study brain tissues without major loss in binding properties, up to a maximum post-mortem delay of approximately 18–28 hours (depending on the post-mortem conditions and receptor types). It also yields strictly quantifiable results, and allows analysis of large serial sections through complete, undissected hemispheres. Finally, it is possible to label numerous different receptors in immediately adjacent cryostat sections (10–20 μm thick), and to match cytoarchitectonic and myeloarchitectonic data derived from alternating sections of the same brains that have been used for receptor labelling.
Receptor autoradiography has been used to study the regional distribution patterns (that is, differences between brain areas) and laminar distribution patterns (that is, differences between cortical layers) of selected neurotransmitter receptor types and subtypes in the human cortex22,34,40,69,70,79-82. It has been shown that localized changes in the laminar distribution patterns and/or mean densities (which are averaged over all cortical areas) of receptor binding sites resemble cytoarchitectonic borders22,34. For example, the border between the primary (V1) and secondary (V2) visual cortices is clearly visible due to the considerably higher muscarinic M2 receptor density in V1 than in V2 (REFS 22,34) (FIG. 4a,b). The V1/ V2 border is also visible in myelin-stained sections owing to the unique presence of a heavily myelinated sublayer (Gennari's stripe) in area V1 (REFS 22,34). Compared with V2, α2-adrenergic, GABAA (γ-aminobutyric acid type A) and serotonergic 5-HT2 receptors are also present at a higher density in V1, whereas glutamatergic kainate and AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid) receptors are present at a lower density in V1. Such receptor architectonic features have led to the discovery of cortical areal borders and intracortical areal subdivisions that had not previously been detected in cytoarchitectonic or myeloarchitectonic studies. These include the subdivision of the primary motor area 4 (REF. 40), as well as the TE1.0 and TE1.1 (temporal cortex82) subdivisions of the primary auditory cortex, which were revealed by the distribution pattern of receptors (FIG. 4c). So far, this receptor autoradiography approach has also been used to examine various neurotransmitter receptors in more than 50 different cyto architectonically defined cortical areas. Receptor mapping not only adds multimodal data about the molecular basis of their areas and nuclei to brain atlases, but it is also a powerful tool for the identification of cyto- or myeloarchitectonically undetectable borders of cortical areas. Finally, as receptors have a key role in neurotransmission, receptor mapping enriches brain atlases with functionally relevant data that are directly related to anatomical criteria such as cyto- or myelo-architecture.
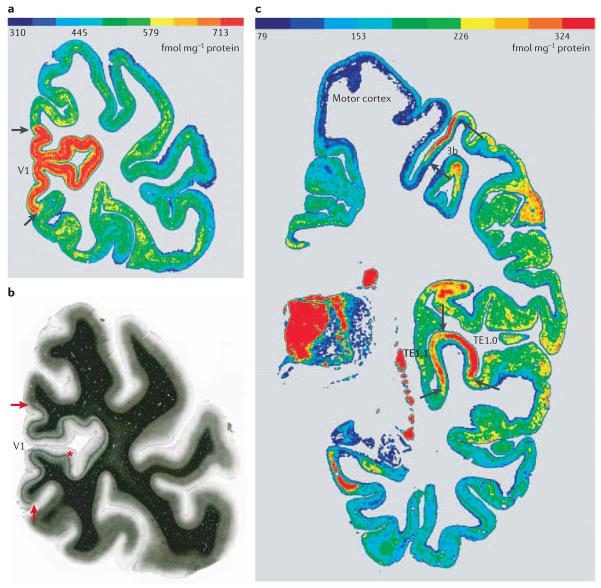
Figure 4. Correlation between receptor autoradiography and myeloarchitectonic borders.
The regional and laminar distribution patterns of muscarinic cholinergic M2 receptors (a) and myelin fibres (b) were visualized in neighbouring coronal sections through the human occipital cortex. The borders (arrows) between the primary visual cortex V1 and the adjacent secondary visual cortex V2 are visible by considerable differences in receptor densities (a) and the abrupt disappearance of Gennari's stripe (b, asterisk) at the border. The regional and laminar distribution patterns of muscarinic cholinergic M2 receptors were visualized in a coronal section through a human hemisphere by agonist ([3H]oxotremorine-M) binding (c). Area 3b is the primary somatosensory cortex63 and areas TE1.0 and TE1.1 (REF. 64) are subdivisions of the primary auditory cortex. Arrows show the borders with adjacent areas. Considerable regional and laminar variations in binding site densities are also visible in the other cortical regions. The colour scales in panels a and c indicate the M2 receptor binding site densities in fmol mg−1 protein.
White matter maps
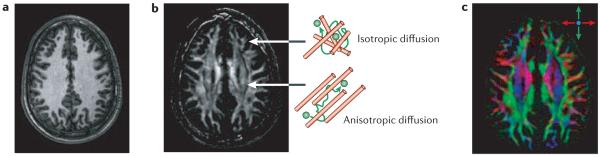
In the past, most atlas efforts have focused on grey matter structure and function. Multimodal atlases, however, are incomplete without information about white matter. The white matter has an important role in connecting different regions of the brain. Perhaps surprisingly, our understanding of human brain connectivity is still limited. One possible reason for this is that it is extremely complex: given a specific number, n, of distinct functional locations in the cortex, there are as many as n2 possibilities for the presence or absence of interconnections (as connections can be intraregional and be in either direction between two regions). Another reason is that the available tools to investigate brain connectivity in vivo are inadequate, as most existing methods are based on invasive techniques such as chemical tracers and lesion-based studies, which are less practical for population-based studies and inappropriate for humans. In addition, conventional MRI has also been powerless on this front. As shown in FIG. 5a, the white matter seems to be homogeneous in T1-weighted images. Even using relaxometry to derive quantitative measures that depend on tissue type, conventional MRI has been unable to provide sufficient contrast to decipher fibre tract organization in the white matter.
Figure 5. Comparison of a conventional T1-weighted image and DTI-based contrasts.
These images show the same slice level from the same subject. a | The T1-weighted image shows the detailed anatomy of the cortex but the white matter appears homogeneous. b | The anisotropy map shows the regions that have high diffusion anisotropy, a hallmark of axonal fibres with a coherent orientation within a pixel. c | For high anisotropy regions, we can map the fibre orientation using colours. In this map, red, green and blue represent fibres running along the right–left, anterior–posterior and superior–inferior axes, respectively. Fibres running along oblique angles are represented by a mixture of the three principal colours.
However, this situation has changed since DTI was developed in the mid-1990s (REF. 83). The contrast of this MRI technique is based on directionality of water motion (random Brownian diffusion) in the brain, and it can provide two types of information. First, it can generate a so-called anisotropy map83-86 (FIG. 5b). If the fibre architecture surrounding water molecules has a coherent orientation, as in the case of axonal bundles, water tends to diffuse preferentially along that orientation, in a process called anisotropic diffusion. If the molecular environment is random, water motion is also random, yielding isotropic diffusion. Therefore, the anisotropy map can reveal which brain regions have a more orderly structure (high diffusion anisotropy). Not surprisingly, many regions of the white matter have coherent fibre orientation with respect to the image resolution, and so have high anisotropy, as can be seen in FIG. 5b. The second type of information obtainable from DTI is the orientation of these ordered structures83,87-91; colour is often used to symbolize this information. In FIG. 5c, red, green and blue colours represent fibres running along the right–left, anterior–posterior and superior–inferior axes, respectively. The rich anatomical and spatial information conveyed by this orientation map can be clearly appreciated. Pixel-by-pixel information on fibre orientation can also be extrapolated to reconstruct three-dimensional trajectories of prominent fibre tracts based on a fibre-tracking approach92-98 or a probability-based approach99. FIGURE 6 shows examples of such a three-dimensional fibre-tract reconstruction based on DTI data, which is remarkably similar to known white matter anatomy based on post-mortem samples100.
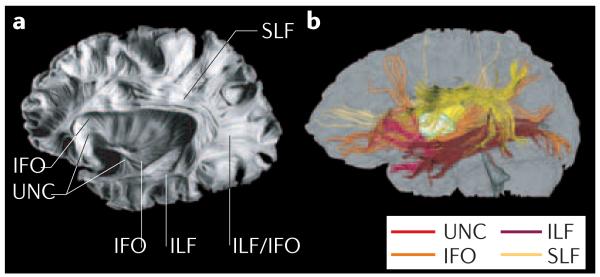
Figure 6. Comparison between a post-mortem brain sample and the results of DTI-based three-dimensional tract reconstruction.
a | Post-mortem sample showing 4 main association fibres: the superior longitudinal fasciculus (SLF), inferior longitudinal fasciculus (ILF), inferior fronto-occipital fasciculus (IFO) and uncinate fasciculus (UNC). b | These tracts can be reconstructed from in vivo human DTI data and presented with different colours. There is excellent agreement between the two. Panel a reproduced, with permission, from REF. 196 © (1997) Univ. of Iowa.
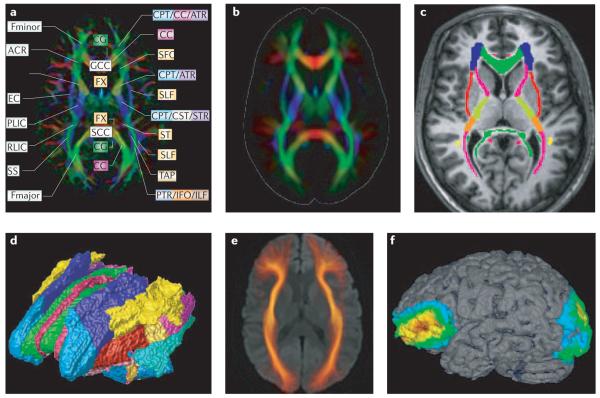
The orientation information from DTI provides essential contrast in the white matter and opens up new opportunities for brain mapping and atlasing. Just as the cortical folding pattern provides clues to identify anatomical locations and map functions, the orientation contrast can be used to identify various functional units in the white matter. Combined with three-dimensional reconstruction, coordinates of specific white matter tracts can also be mapped. FIGURE 7 shows several examples of DTI-based white matter maps. Using a high-quality single-subject dataset, various white matter tracts have been identified and a white matter atlas generated85,90,101-105. By normalizing DTI data from multiple subjects into a common template coordinate system, a probabilistic map of white matter anatomy can be created for normal and diseased patient populations, allowing for quantitative comparisons of white matter anatomy106-108 (FIG. 7b). FIGURE 7c,d show a manually reconstructed map with parcellated white matter tracts based on DTI data. Once the white matter is parcellated into different tracts, these can serve as a valuable framework to map anatomical and functional information. If one is interested in the shape and size of a specific white matter tract, the fibre-tracking approach can be applied to create population-based maps103 (FIG. 7e). Using these mapping techniques, axonal degeneration after stroke or surgery can be followed in a tract-specific manner and these changes can be correlated with functional outcomes109. Therefore, we can investigate such questions as ‘which white matter tracts are affected by stroke?’110,111, or ‘what is the functional outcome of degeneration in a specific white matter tract?’112. DTI can also be used to generate connectivity maps (FIG. 7f) by identifying brain regions associated with a specific white matter tract113. Such connectivity maps could provide important clues to deduce which cortical areas are likely to be affected when white matter injuries occur.
Figure 7. Examples of various methods of mapping the white matter based on DTI.
a | For each individual dataset, many white matter structures can be discretely identified and anatomical labels can be assigned to them. b | Normal or abnormal variability of white matter anatomy can be investigated by using a pixel-based population-averaging approach. This image was created using data from 30 healthy volunteers normalized to a common template using a 12 parameter affine transformation. c | Various structures are parcellated manually or by using three-dimensional tract reconstruction into individual tract families based on orientation (colour) information. d | This parcellation/reconstruction allows a three-dimensional white matter parcellation map to be created. e | The morphology of a specific white matter tract can be investigated, based on a three-dimensional tract reconstruction technique followed by group averaging. Here, the inferior fronto-occipital fasciculus (IFO) was reconstructed from the normal population and mapped onto a common template. f | This approach can also be used to identify cortical regions associated with a specific tract, where the trajectory of the IFO is extrapolated to identify associated cortical regions, and a probabilistic cortical map is created from the 30 subjects. These kinds of map pave the way to investigate white matter anatomy and function in a systematic and quantitative manner. ACR, anterior corona radiata; ALIC, anterior limb of internal capsule; ATR, anterior thalamic radiation; CC, corpus callosum; CG, cingulum; CPT, corticopontine tract; CST, corticospinal tract; EC, external capsule; Fmajor, forceps major; Fminor, forceps minor; FX, fornix; GCC, genu corporis callosi; ILF, inferior longitudinal fasciculus; PLIC, posterior limb of internal capsule; PTR, posterior thalamic radiation; RLIC, retrolentricular part of internal capsule; SCC, splenium corporis callosi; SFO, superior fronto-occipital fasciculus; SLF, superior longitudinal fasciculus; SS, sagittal striatum; ST, stria terminalis; TAP, tapetum. Panel a modified, with permission, from REF. 20 © (2005) Elsevier Science.
Although DTI is a powerful technique for brain mapping, it has several limitations. First, it generates tensor-valued information at each pixel, accumulating large amounts of complicated anatomical information. Data analysis methodology is not yet sufficiently developed to harness this detail, making it challenging to quantify morphology from these datasets. Second, with current imaging resolution (2–3 mm), only the macroscopic anatomy of large white matter tracts can be studied. For example, the notion of ‘fibre orientation at each pixel’ is based on the assumption that there is only one fibre bundle with the same orientation in a given pixel. Currently, many new approaches alternative to the tensor-based method are being proposed, which can extract more anatomical information (such as identifying multiple fibre populations) from each pixel114-117. Although these approaches have successfully demonstrated their effectiveness, with current imaging resolution, the information inherent in diffusion measurement is macroscopic and there is no way to obtain cellular-level or even synaptic connectivity information.
This resolution issue is also related to the validity of DTI-based tract reconstruction results. Although good overall agreement has been reported between the three-dimensional reconstruction results and existing anatomical knowledge, there are frequent false-negative and false-positive results due to noise and partial volume effects. In the future, it will be important to develop approaches to reliably extract anatomically and clinically valuable information from such contaminated data. Although this is a challenging task, several effective approaches have been suggested, such as knowledge-based tract reconstruction (namely, reconstructing only anatomically known fibres)92, probabilistic tracking99 and population-average approaches103.
Nevertheless, DTI can advance our knowledge of the correlations between white matter anatomy and function by providing a refined white matter map to which we can register various experimental findings. Finally, macroscopic DTI information can be compared directly with microscopic post-mortem data on fibre tracts118-120, yielding an integrative atlas system that evaluates both methods with considerable synergistic effects.
Multimodality and population-based atlases
Multimodal mapping
Before the advent of brain mapping methods1, atlasing efforts of various research groups were typically independent of one another. Most atlases had a different spatial scale and resolution, utilized different data structures, described different structural or functional characteristics and were inherently incompatible with the others. Although each of those brain mapping strategies has its unique advantage, the brain atlases that are derived from a single method will be limited, unless certain integrative approaches such as spatial normalization are implemented. An integrated, comprehensive approach requires these diverse mapping methods to be combined and correlated.
Characterizing a single subject with multiple imaging devices clearly combines the strengths of each imaging modality. Data from single subjects, pre-mortem and post-mortem, provide a unique view of the relationship between in vivo imaging and histological assessment. For example, Mega et al.121 scanned patients in the terminal stages of Alzheimer's disease using both MRI and PET. These data were combined with a stain of neurofibrillary tangles and post-mortem three-dimensional histological images that show the gross anatomy122. This multimodal, but single-subject, atlas of Alzheimer's disease relates the anatomical and histopathological underpinnings to in vivo metabolic and perfusion maps of this disease. Recent work on neurodegenerative diseases has also warped histological data into MRI scans to better identify regions (such as the CA fields of the hippocampus and the basal nucleus of Meynert123) that are relevant to understanding the pathology.
Population-based atlases
Modern atlases not only incorporate information from multiple modalities — they also incorporate information from multiple members of a population. Normal anatomical complexity and variability between human brains are so great that group-specific patterns of anatomy and function are often obscured. Reports of structural differences in the brain linked to gender, IQ and handedness are a topic of intense controversy124,125, and it is even less clear how these factors affect disease-specific abnormalities. The importance of these linkages has propelled computational anatomy to the forefront of brain imaging investigations. In particular, population atlases can be compiled into subpopulations to represent specific disease types, and subsequently stratified by age, gender, handedness or genetic factors.
To distinguish abnormalities from normal variants, a realistically complex mathematical framework is required to encode information on anatomical variability in homogeneous populations126. Warping algorithms can be used to equate individual brain datasets with an atlas template, and the applied transformations can be studied to examine patterns of anatomical variation and detect pathology. Cortical anatomy is altered in schizophrenia127, Alzheimer's disease128,129 and in various developmental disorders, including fetal alcohol syndrome130, autism131 and Williams syndrome132. By using specialized strategies for averaging anatomy across individuals, specific features of anatomy emerge that are not observed in individual representations due to their considerable variability; population-specific patterns of cortical organization or asymmetry can then be mapped and visualized124.
Population-based brain atlases54,133 provide an expandable framework to synthesize the results of disparate imaging studies. These atlases use novel analytical tools to combine data across subjects, modalities and time and to detect group-specific features not apparent in individual scans. A notable example is the pattern of anatomical asymmetry in the major cortical gyri and sulci, such as the prominent asymmetries in the perisylvian language-related cortices, which are well known134 but not easily discernible in individual brain scans. The average patterns clearly emerge after averaging computational models of the cortical sulci across a population124. So, a computational framework can be used to answer neuroscientific questions regarding the development of these asymmetries135, and how they are modulated by gender136, aging or in psychiatric illnesses such as schizophrenia37,137,138. Furthermore, population-based atlases can be stratified into subpopulations to reflect a particular (clinical or demographic) subgroup1. Design of appropriate reference systems for brain mapping data presents considerable challenges, as these systems must capture how brain structure and function vary in large populations, across age and gender, in different disease states, across imaging modalities and even across species139.
As imaging data from different studies can now be compared in a common coordinate system, large databases of functional imaging data — and associated meta-data on experimental paradigms and findings — are now beginning to be assembled, along with tools developed to interact with them140. Notable examples of these neuroinformatics efforts include the ICBM53 (see International Consortium for Brain Mapping in Online links box), the ADNI (see Alzheimer's Disease Neuroimaging Initiative in Online links box) consortium project to build a data repository of MRI, PET and other clinical and biomarker data on aging and Alzheimer's disease, the NIMH paediatric imaging study (which is scanning nearly 1000 children every 2 years for 10 years141,142), and the Finnish twin registry, whose scans have been used to identify genetic influences on brain structure125,143,144. Over the next decade, population-based atlases are also likely to gain widespread applicability in genetic studies. In an exciting development, genetic linkage data have been incorporated into brain imaging studies to discover previously unknown effects on the brain of variations at specific genetic loci145 (FIG. 8e). Given their power to store and compute statistics on expected rates of brain development and degeneration for different clinical populations, dynamic brain atlases are also likely to be used in drug trials or studies of factors that influence disease expression and therapeutic response146, and studies on the effects of specific medications such as antipsychotics or mood stabilizers147-149.
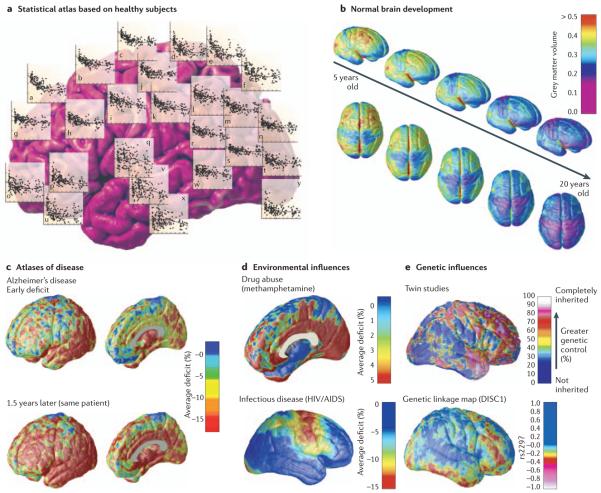
Figure 8. Brain atlases that represent specific subpopulations.
Population-based atlases can be stratified to compare various populations, revealing the impact of development or disease on specific features of the brain. a | Statistical maps show the changing distribution of cortical grey matter (over the human lifespan (n = 176; data from Sowell et al. (2003)130). b | Based on longitudinal MRI scans of children developing normally, dynamic maps can show the rate and anatomical sequence of cortical grey matter maturation by fitting an age-adjusted mean to these trajectories154. c–d | Such maps can be used to reconstruct time-lapse sequences that visualize the spread of cortical atrophy at different stages of Alzheimer's disease (c), changes associated with chronic drug abuse (d, top), or in the course of infectious illnesses such as HIV/AIDS (d, bottom) 161,182,195. Atlases could provide statistical measures of disease burden for drug trials, offering insight into the systems involved and factors that influence them. e | Individual deviations from these average patterns can also be analysed: twin studies have revealed aspects of brain structure (such as frontal grey matter) that are under extremely strong genetic control188. In the new field of imaging genomics, genetic linkage studies have been extended to brain mapping data to identify the statistical linkage between specific genetic variations (single nucleotide polymorphisms) and anatomical variations in the form of a statistical map186. DISC1, disrupted in schizophrenia 1; rs229, denotes a single nucleotide polymorphism marker (rs751229) located in DISC1. The colour bar denotes the correlation between intrapair differences in brain matter density with the number of alleles in common at this genetic marker locus in twins discordant for schizophrenia.
Image processing algorithms are also making digital brain atlases more versatile. For example, probabilistic atlases retain information on cross-subject variations in brain structure and function. These atlases are powerful new tools with broad clinical and research applications52,124,150,151. Deformable brain atlases, which are essentially labelled templates of anatomy that can be locally dilated, stretched and scaled to match scans from new subjects152, are adaptable in that they can be individualized to reflect the anatomy of new subjects. The deformation can transfer all the accumulated data in the atlas onto the new subject's scan, thereby quantifying the degree of deviation in their anatomy, relative to a normative population153-155.
Finally, dynamic brain atlases can also be used to reference and analyse dynamic, time-varying data. Recent studies have analysed longitudinally collected MRI scans to compute probabilistic information on growth rates156-158 (FIG. 8a), lifelong normal changes130 (FIG. 8a) or normative statistics on rates of degenerative tissue loss in drug abuse (FIG. 8d), aging and dementing diseases149,159-161 (FIG. 8c). A complementary development, which has advanced the power of multi-subject atlasing projects, is the widespread use of analysis techniques such as statistical parametric mapping (SPM) (BOX 1).
Box 1. Statistical parametric mapping.
Initially proposed as a method to analyse functional or metabolic images from multiple subjects in a common coordinate space161,192, statistical parametric mapping (SPM) is now widely used by the brain mapping community to detect and identify activated brain regions in functional imaging studies. SPM relates patterns of activation to experimental or subject-specific parameters, and can be used to infer patterns of functional connectivity from time-series data, or to analyse anatomical images193. Atlasing efforts in the early 1990s pointed to the value of registering data from multiple subjects to a common coordinate space. Once aligned, imaging signals could be combined across subjects for statistical analysis, producing a statistic (for example, expressing a group difference) at any given voxel location in the canonical space. Because many millions of voxels are typically tested in a brain mapping study, statistical solutions to the multiple comparisons problem were developed based on the mathematical likelihood that clusters of certain sizes and magnitudes would occur by chance when no signal was present. SPM rapidly became a widely used software package for collating and analysing brain imaging data (see Statistical Parametric Mapping in Online links box), partly because it implemented powerful mathematical formulae — based on new findings in the theory of Gaussian random fields194 — for inferring whether functional activations were present in multisubject studies. The SPM approach shares some affinity with atlasing efforts in that it reports statistical findings in a common coordinate space. In contrast to SPM, which typically analyses registered images on a voxel-by-voxel basis, computational atlasing efforts can also include computational anatomical work that models individual brain structures as geometrical surfaces and curves, such as cortical surface modelling and fibre tract modelling. Ultimately these approaches are complementary.
Disease-specific atlases
Surprisingly few atlases of neuropathology use a standardized three-dimensional coordinate system to integrate data across patients, techniques and acquisitions. Atlases with a well-defined coordinate space54,163, together with algorithms to align data with them21, have enabled the pooling of brain mapping data from multiple individuals and sources, including large patient populations. Automated algorithms can then utilize atlas descriptions of anatomical variance to guide image segmentation164-166, tissue classification167,168, functional image analysis169,170 and pathology detection171. An obvious challenge in integrating pathological data from tissue specimens results from the destructive sectioning and histological staining procedures. Even so, the advent of whole human head cryosectioning techniques (in which the intact blockface is imaged photographically) allows spatially coherent three-dimensional volumetric images to be acquired. Stained pathological material could, in principle, be aligned with these canonical volumes using three-dimensional image deformation strategies that have proven useful in creating cytoarchitectural atlases.
Statistical representations of anatomy resulting from the application of atlasing strategies to specific subgroups of diseased individuals have revealed the profile of structural brain deficits in a number of diseases. These include studies of Alzheimer's disease161, HIV/AIDS132,172, epilepsy172,146, unipolar depression173, childhood and adult-onset schizophrenia174-177, attention-deficit/hyperactivity disorder178, fetal alcohol syndrome179, Tourette's syndrome180, bipolar disorder181,149, autism131,149, Williams syndrome132 and methamphetamine abusers182. Findings from these population atlases have often led to unexpected neuroscientific findings, such as a spreading wave of cortical changes in schizophrenia176,177.
Without methods to overcome the problems of anatomical variability, the statistical power to resolve disease and treatment effects is seriously undermined. First, owing to normal anatomical variation, diseased and healthy individuals overlap on most anatomical measures. Second, these difficulties are exacerbated by disease-related change such as atrophy128,149,159,183,184 or other progressive and dynamic anatomical changes. To fully capitalize on neuroimaging data from the diseased brain, an appropriately complex mathematical framework is needed to address these challenges. Only then can brain maps be compared across patients and across time133,160,182,184,185.
Atlases have also revealed how single genes can affect brain structure186. When they have supported existing models of disease, such as the sequence of spreading pathology in Alzheimer's disease, atlas-based descriptions of variance offer statistics on degenerative rates and can elucidate clinically relevant features at the systems level. Atlases have identified differences in atrophic patterns between Alzheimer's disease and Lewy Body dementia187, and differences in atrophic rates between clinically-defined subtypes of psychosis188-190.
Based on well-characterized patient groups, population-based atlases contain composite maps and visualizations of structural variability, asymmetry and group-specific differences. Pathological change can be tracked over time, and generic features resolved, allowing these atlases to offer biomarkers for a variety of pathological conditions, as well as morphometric measures for genetic studies or drug trials.
Conclusions and perspectives
The evolution of brain atlases has seen tremendous advances; they can now accommodate observations from multiple modalities and from populations of subjects collected at different laboratories. The probabilistic systems described here show promise for identifying patterns of structural, functional and molecular variation in large image databases, for pathology detection in individuals and groups and for determining the effects of age, gender, handedness and other demographic or genetic factors on brain structures in space and time. Integrating these observations to enable statistical comparison has already provided a deeper understanding of the relationship between brain structure and function. Importantly, the utility of an atlas depends on appropriate coordinate systems, registration, and deformation methods to allow the statistical combination of multiple observations in an agreed, but expandable, digital reference framework.
In this review, we highlighted two sources of data that will have an increasingly important role in integrative brain atlases: molecular architectonics and DTI. Once stored in a population-based atlas, information from these techniques can help to interpret more conventional functional and structural brain maps by integrating them with data on molecular content, physiology and fibre connections — a development that can help to formulate and test new types of neuroscientific models. A goal of systems neuroscience is to establish brain systems that underlie cognitive processes and the factors that influence them. DTI data on fibre connectivity, stored in an atlas coordinate system, can offer a rigorous computational basis to test how identifiable anatomical systems (for example, visual, limbic or corticothalamic pathways) interact. This atlas information can be invoked as regions of interest that are incorporated into the statistical design of functional brain mapping studies (for example, with fMRI or electroencephalography), even when underlying fibre connections are not evident in the data being collected for a particular study. Molecular architectonic mapping also provides a complementary perspective in which known neurotransmitter and receptor pathways — the physiology and molecular features of which are now well understood — can be associated with functional subdivisions of the cortex, identified with tomographic imaging. For example, an fMRI study of inhibitory cognitive processes in drug abusers might be informed by other modalities of data on limbic–prefrontal connectivity (from DTI), or on cortical monoamine receptor distributions (from architectonic mapping). In each of these contexts, the coordinate system of the atlas, and the transformations that equate different modality data in the same reference frame, provide the means to build and test systems-level models of cognition or disease, incorporating data from traditionally separate domains of neuroscience.
As brain atlases begin to incorporate data from thousands of subjects, new questions in basic and clinical neuroscience can be addressed that were previously out of reach. For example, quantitative genetic studies are underway to link functional, structural and connectivity information with variations in candidate genetic polymorphisms that could influence them. As polygenic disorders involve the interaction of multiple genetic variations, each with a small effect on the overall phenotype, digital atlases provide the ideal setting to mine large numbers of images computationally with hybrid techniques from computational anatomy and quantitative genetics (such as linkage and association studies in which a statistic is computed at each voxel location in the brain191).
Another area of expansion is in the processing of clinical data from therapeutic trials, to determine factors that combat or modify disease progression. Statistical atlases, containing time-varying data, have revealed unforeseen but characteristic brain changes in several dementias and neuropsychiatric illnesses. The population-based atlases of the future will provide the necessary statistical power to identify demographic, genetic and environmental factors that influence therapeutic response. Most important of all, brain atlases are now being enriched with data from newer technologies, such as DTI, fMRI and modern high-throughput cytoarchitectural methods. These efforts are yielding whole new avenues of research into the functional organization of the brain that will be of interest not just to specialists in neuroimaging, but to all basic and clinical neuroscientists.
Acknowledgements
This work was supported by research grants from the National Institutes of Health (NIH), Roadmap Initiative for Bioinformatics and Computational Biology, National Center for Research Resources, the National Institute of Mental Health (NIMH) and the National Institute of Neurological Disorders and Stroke, and by Human Brain Project grants to the International Consortium for Brain Mapping, funded jointly by NIMH and the National Institute on Drug Abuse and one funded by the National Institute of Aging (NIA). Additional support was provided by the National Institute for Biomedical Imaging and Bioengineering, the National Center for Research Resources and the NIA, the National Library of Medicine and the Biomedical Informatics Research Network (BIRN, http://www.nbirn.net), which is funded by the National Center for Research Resources at the NIH. Other funds came from the German Ministry of Science BMBF, the Helmholtz Association of Research Centres and from various grants from the German Research Foundation DFG and the European Union. We thank the many collaborators and doctoral students in our laboratories. Special thanks to P. Roland from the Karolinska Institute Stockholm for an exciting collaboration of many years, and to N. Palomero-Gallagher for her enthusiasm in the receptor project.
Glossary
- Cartographic approaches
Approaches that place brain images from multiple subjects and devices into an anatomical reference system with standardized three-dimensional coordinates (for volumetric images) or two-dimensional spherical or planar coordinates (for cortical regions).
- Cytoarchitecture
Subdivisions (named or numbered) of the cerebral cortex, called cytoarchitectonic maps, based on cellular features (size and shape of cells, cell packing density in different cortical layers, width of layers) and identified in cell body-stained specimens.
- Magnetic resonance imaging
(MRI). A non-invasive method to obtain images of living tissue. It uses radio-frequency pulses and magnetic field gradients; the principle of nuclear magnetic resonance is used to reconstruct images of tissue characteristics (for example, proton density, water diffusion parameters).
- Multispectral characterization
Multispectral imaging devices measure multiple features of an object at each spatial location, such as optical reflectance at different wavelengths, or different relaxometric decay constants (T1 and T2) in MRI.
- Chemoarchitectural maps
Differences in the molecular composition of cortical and subcortical brain regions can be assessed using enzyme- or immunohistochemistry, in situ hybridization, receptor autoradiography and so on, revealing subdivisions with distinctive distribution patterns, such as expression of transmitter receptors.
- Diffusion tensor imaging
(DTI). A technique developed in the mid-1990s, based on MRI in which diffusion constants of water molecules are measured along many (>6) orientations and diffusion anisotropy is characterized. It is used to visualize the location, orientation and anisotropy of the brain's white matter tracts, and is sensitive to directional parameters of water diffusion in the brain.
- Myeloarchitecture
Subdivisions (numbered or named) of the cerebral cortex, called myeloarchitectonic maps, based on features (for example, stria of Gennari in the visual cortex) of myelinization (differential density of myelinated fibres and fibre bundles in different cortical layers), and identified in myelin-stained histological specimens.
- Stereotaxic system
A technique or apparatus used in neurosurgery or brain imaging studies to localize a specific anatomical locus using standardized three-dimensional coordinates; used, for example, for directing the tip of a surgical instrument (such as a needle or electrode) to a known location.
- Functional imaging
Distinct from structural imaging techniques such as computerized tomography or MRI that assess anatomical structure, functional imaging techniques (for example, positron emission tomography, functional MRI and electroencephalography) are sensitive to physiological processes such as neuronal activation, electromagnetic properties of living tissue, blood flow or metabolism.
- Multimodal association cortices
The multimodal association cortices of the parietal and frontal lobes integrate somatosensory, auditory and visual information for higher-order cognitive processing.
- Positron emission tomography
A medical imaging technique that uses injected radiolabelled tracer compounds in conjunction with mathematical reconstruction methods to produce a three-dimensional image or map of functional processes in the body, such as glucose metabolism, blood flow or receptor distributions.
- Multimodal microstructural approach
An approach to characterize fine-scale anatomy using multiple histological and neurochemical techniques, revealing different aspects of cellular organization or molecular composition.
- Fibre-tracking approaches
Using this approach, three-dimensional trajectories of white matter tracts can be reconstructed. The algorithm is based on fibre orientation information obtained from diffusion tensor imaging.
- Probability map
A map that depicts the likelihood of a particular feature. It can be used to show how frequently, in percentage, a given anatomical structure is found in a specific location across a population of subjects.
- Cytoarchitectonic probability maps
Based on a sample of brains that have been parcellated using cytoarchitectonic criteria; they display the statistical likelihood, or relative frequency, that a particular voxel in stereotaxic space contains a given cytoarchitectonic unit (for example, a cortical area, or a subcortical nucleus).
- Maximum probability maps
Summary maps in which each voxel of the three-dimensional space has been assigned to the cytoarchitectonically defined unit with the highest probability. They are calculated on the basis of probability maps, such as cytoarchitectonic probability maps, to generate a map with unambiguously defined borders.
- Voxel
The three dimensional equivalent of a pixel. A pixel is a picture element, and a voxel is a volume element.
- T1-weighted images
One of most widely used MRI methods, in which the contrast is based on a selection of MRI acquisition parameters, producing an image that weights signal by the relaxometric parameter, T1, of each tissue (that is, the longitudinal relaxation time). In the brain, T1-weighting causes fibre tracts (nerve connections) to appear white, cortex and basal nuclei to appear grey, and cerebrospinal fluid to appear dark.
- Relaxometry
The measurement of relaxation parameters in nuclear MRI, such as the longitudinal (T1) or transverse (T2) decay constants that characterize signal decay from excited nuclei. By detecting subtle differences in relaxation times, relaxometry is capable of differentiating various tissue types in the brain.
- Anisotropy map
The directional dependency of water diffusion at each point in the brain can be summarized using measures such as fractional anisotropy. High anisotropy values indicate heavily myelinated white matter, whereas decreased anisotropy is often a sign of disease.
- Anisotropic diffusion
Diffusion of a substance (for example, water) that is greater in certain preferred directions, such as along the axons of a fibre tract.
- Isotropic diffusion
Diffusion of a substance (for example, water) that is uniform in all directions.
- Tensor-valued information
Information that can be modelled mathematically as a matrix, or tensor, at each location in an object. Diffusion tensor imaging produces signals with at least six independent parameters at each anatomical point (the diffusion tensor); tensor calculus can then be used to estimate diffusion parameters in any specific direction.
- Partial volume effects
This refers to the blurring of intensity differentiations used to classify contributing tissue types (grey matter, white matter and cerebrospinal fluid). It is the results of pixels placed over a region that contains multiple tissue types. The smaller the pixel size the less frequently this is problematic.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
DATABASES
The following terms in this article are linked online to: OMIM: http://www.ncbi.nlm.nih.gov/entrez/query. fcgi?db=OMIM Alzheimer's disease | autism | schizophrenia | Williams syndrome
FURTHER INFORMATION
Toga's laboratory: http://www.loni.ucla.edu Alzheimer's Disease Neuroimaging Initiative: http://www.loni.ucla.edu/ADNI International Consortium for Brain Mapping: http://www.loni.ucla.edu/ICBM/ Statistical Parametric Mapping: http://www.fil.ion.ucl.ac.uk/spm/ Access to this links box is available online.
References
- 1.Toga AW, Mazziotta JC. In: Brain Mapping: the Methods. Toga AW, Mazziotta JC, editors. Academic; San Diego: 1996. pp. 3–25. [Google Scholar]
- 2.Duvernoy HM. The Human Brain. Springer; New York: 1991. [Google Scholar]
- 3.Ono M, Kubik S, Abernathey CD. Atlas of the Cerebral Sulci. Thieme; Stuttgart: 1990. This widely used atlas reports anatomical descriptions and trends in individual variabilities of cortical sulci. It serves as a reminder of the challenges in developing multisubject reference systems for human brain mapping. [Google Scholar]
- 4.Talairach J, Szikla G. Atlas d'Anatomie Stereotaxique du Telencephale: Etudes Anatomo-Radiologiques. Masson & Cie; Paris: 1967. (in French) [Google Scholar]
- 5.Talairach J, Tournoux P. Co-planar Stereotaxic Atlas of the Human Brain. Thieme; New York: 1988. This atlas influenced the brain mapping field by providing a principled method for spatially transforming anatomical datasets into a coordinate-based reference system based on the anterior commissure–posterior commisure line. The atlas presents post-mortem data from a single subject. [Google Scholar]
- 6.Brodmann K. Some Papers on the Cerebral Cortex, translated as: On the Comparative Localization of the Cortex. Thomas; Springfield, Illinois: 1960. pp. 201–230. [Google Scholar]
- 7.von Economo C, Koskinas GN. Die Cytoarchitektonik der Hirnrinde des Erwachsenen Menschen. Springer; Berlin: 1925. (in German) [Google Scholar]
- 8.Flechsig P. Anatomie des menschlichen Gehirns und Rückenmarks auf myelogenetischer Grundlage. Thieme; Leipzig: 1920. (in German) [Google Scholar]
- 9.Smith GE. A new topographical survey of the human cerebral cortex, being an account of the distribution of the anatomically distinct cortical areas and their relationship to the cerebral sulci. J. Anat. 1907;41:237–254. [PMC free article] [PubMed] [Google Scholar]
- 10.Vogt C, Vogt O. Allgemeinere ergebnisse unserer hirnforschung. J. Psychol. Neurol. 1919;25:292–398. (in German) [Google Scholar]
- 11.Mai J, Assheuer J, Paxinos G. Atlas of the Human Brain. Academic; San Diego: 1997. [Google Scholar]
- 12.Matsui T, Hirano A. An Atlas of the Human Brain for Computerized Tomography. Igako-Shoin; Tokyo: 1978. [Google Scholar]
- 13.Schaltenbrand G, Bailey P. Introduction to Stereotaxis with an Atlas of the Human Brain. Thieme; Stuttgart & New York: 1959. [Google Scholar]
- 14.Schaltenbrand G, Wahren W. Atlas for Stereotaxy of the Human Brain. 2nd edn Thieme; Stuttgart: 1977. [Google Scholar]
- 15.Van Buren JM, Borke RC. Variations and Connections of the Human Thalamus. 1 & 2. Springer; New York: 1972. [Google Scholar]
- 16.Van Buren JM, Maccubin D. An outline atlas of human basal ganglia and estimation of anatomic variants. J. Neurosurg. 1962;19:811–839. doi: 10.3171/jns.1962.19.10.0811. [DOI] [PubMed] [Google Scholar]
- 17.Mansour A, Fox CA, Burke S, Akil H, Watson SJ. Immunohistochemical localization of the cloned mu opioid receptor in the rat CNS. J. Chem. Neuroanat. 1995;8:283–305. doi: 10.1016/0891-0618(95)00055-c. [DOI] [PubMed] [Google Scholar]
- 18.Dejerine J. Anatomie des Centres Nerveux. Rueff; Paris: 1901. [Google Scholar]
- 19.Damasio H. Human Brain Anatomy in Computerized Images. Oxford Univ. Press; Oxford & New York: 1995. [Google Scholar]
- 20.Mori S, Wakana S, Nagae-Poetscher LM, van Zijl PC. MRI Atlas of Human White Matter. Elsevier Science; Amsterdam: 2005. [Google Scholar]
- 21.Toga AW. Brain Warping. Academic; San Diego: 1998. The author surveys the many approaches and computational algorithms for deforming brain imaging data from multiple subjects or modalities to match an atlas or other standardized coordinate space. [Google Scholar]
- 22.Zilles K, et al. Architectonics of the human cerebral cortex and transmitter receptor fingerprints: reconciling functional neuroanatomy and neurochemistry. Europ. Neuropsychopharmacol. 2002;12:587–599. doi: 10.1016/s0924-977x(02)00108-6. [DOI] [PubMed] [Google Scholar]
- 23.Brodmann K. Physiologie des Gehirns. Neue Dtsch Chir. 1914;11:85–426. (in German) [Google Scholar]
- 24.Bailey P, von Bonin G. The Isocortex of Man. Univ. Illinois Press; Urbana: 1951. [Google Scholar]
- 25.Sanides F. Die Architektonik des menschlichen Stirnhirns. Springer; Berlin & New York: 1962. (in German) [Google Scholar]
- 26.Sarkisov SA, Filimonoff IN, Kononowa EP, Preobrachenskaja IS, Kukuew LA. Atlas of the Cytoarchitectonics of the Human Cerebral Cortex. Medgiz; Moscow: 1955. [Google Scholar]
- 27.Zilles K, Armstrong E, Schleicher A, Kretschmann HJ. The human pattern of gyrification in the cerebral cortex. Anat. Embryol. 1988;179:173–179. doi: 10.1007/BF00304699. [DOI] [PubMed] [Google Scholar]
- 28.Nelissen K, Luppino G, Vanduffel W, Rizzolatti G, Orban GA. Observing others: multiple action representation in the frontal lobe. Science. 2005;310:332–336. doi: 10.1126/science.1115593. [DOI] [PubMed] [Google Scholar]
- 29.Sereno MI, et al. Borders of multiple visual areas in humans revealed by functional magnetic resonance imaging. Science. 1995;268:889–893. doi: 10.1126/science.7754376. [DOI] [PubMed] [Google Scholar]
- 30.Bremmer F, et al. Polymodal motion processing in posterior parietal and premotor cortex: a human fMRI study strongly implies equivalencies between humans and monkeys. Neuron. 2001;29:287–296. doi: 10.1016/s0896-6273(01)00198-2. [DOI] [PubMed] [Google Scholar]
- 31.Zeki S, et al. A direct demonstration of functional specialization in human visual cortex. J. Neurosci. 1991;11:641–649. doi: 10.1523/JNEUROSCI.11-03-00641.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lashley KS, Clark G. The cytoarchitecture of the cerebral cortex of atlases: a critical examination of architectonic studies. J. Comp. Neurol. 1946;85:223–306. doi: 10.1002/cne.900850207. [DOI] [PubMed] [Google Scholar]
- 33.Luppino G, Rizzolatti G. The organization of the frontal motor cortex. News Physio. Sci. 2000;15:219–224. doi: 10.1152/physiologyonline.2000.15.5.219. [DOI] [PubMed] [Google Scholar]
- 34.Zilles K, Schleicher A, Palomero-Gallagher N, Amunts K. In: Brain Mapping: The Methods. 2nd edn Toga AW, Mazziotta JC, editors. Academic; Amsterdam: 2002. pp. 573–602. [Google Scholar]
- 35.Amunts K, et al. Broca's region revisited: cytoarchitecture and intersubject variability. J. Comp. Neurol. 1999;412:319–341. doi: 10.1002/(sici)1096-9861(19990920)412:2<319::aid-cne10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 36.Amunts K, Malikovic A, Mohlberg H, Schormann T, Zilles K. Brodmann's areas 17 and 18 brought into stereotaxic space — where and how variable? NeuroImage. 2000;11:66–84. doi: 10.1006/nimg.1999.0516. [DOI] [PubMed] [Google Scholar]
- 37.Zilles K, et al. Quantitative analysis of sulci in the human cerebral cortex: development, regional heterogeneity, gender difference, asymmetry, intersubject variability and cortical architecture. Hum. Brain Mapp. 1997;5:218–221. doi: 10.1002/(SICI)1097-0193(1997)5:4<218::AID-HBM2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 38.Tootell RBH, et al. Functional analysis of primary visual cortex (V1) in humans. Proc. Natl Acad. Sci. USA. 1998;95:811–817. doi: 10.1073/pnas.95.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Talavage TM, et al. Tonotopic organization in human auditory cortex revealed by progressions of frequency sensitivity. J. Neurophysiol. 2004;91:1282–1296. doi: 10.1152/jn.01125.2002. [DOI] [PubMed] [Google Scholar]
- 40.Geyer S, et al. Two different areas within the primary motor cortex of man. Nature. 1996;382:805–807. doi: 10.1038/382805a0. [DOI] [PubMed] [Google Scholar]
- 41.Young JP, et al. Regional cerebral blood flow correlations of somatosensory areas 3a, 3b, 1, and 2 in humans during rest: a PET and cytoarchitectural study. Hum. Brain Mapp. 2003;19:183–196. doi: 10.1002/hbm.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hagler D, Sereno MI. Spatial maps in frontal and prefrontal cortex. Neuroimage. 2005;29:567–577. doi: 10.1016/j.neuroimage.2005.08.058. [DOI] [PubMed] [Google Scholar]
- 43.Van Essen DC. Surface-based approaches to spatial localization and registration in primate cerebral cortex. NeuroImage. 2004;23:S97–S107. doi: 10.1016/j.neuroimage.2004.07.024. This review article describes many of the neuroanatomical, technical and informatics issues involved in integrating cortically-derived neuroimaging data across subjects and modalities. [DOI] [PubMed] [Google Scholar]
- 44.Tootell H, et al. Functional analysis of primary visual cortex (V1) in humans. Proc. Natl Acad. Sci. 1998;95:811–817. doi: 10.1073/pnas.95.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larsson J, et al. Neuronal correlates of real and illusory contour perception: functional anatomy with PET. Europ. J. Neurosci. 1999;11:4024–4036. doi: 10.1046/j.1460-9568.1999.00805.x. [DOI] [PubMed] [Google Scholar]
- 46.Schleicher A, Amunts K, Geyer S, Morosan P, Zilles K. Observer-independent method for microstructural parcellation of cerebral cortex: a quantitative approach to cytoarchitectonics. Neuroimage. 1999;9:165–177. doi: 10.1006/nimg.1998.0385. [DOI] [PubMed] [Google Scholar]
- 47.Schleicher A, et al. A stereological approach to human cortical architecture: identification and delineation of cortical areas. J. Chem. Neuroanat. 2000;20:31–47. doi: 10.1016/s0891-0618(00)00076-4. [DOI] [PubMed] [Google Scholar]
- 48.Schleicher A, et al. Quantitative architectonic analysis: a new approach to cortical mapping. Anat. Embryol. 2005;210:373–386. doi: 10.1007/s00429-005-0028-2. [DOI] [PubMed] [Google Scholar]
- 49.Annese J, Toga AW. In: Brain Mapping: The Methods. Toga AW, Mazziotta JC, editors. Academic; San Diego: 2002. pp. 537–564. [Google Scholar]
- 50.Hömke L. Numerical Linear Algebra with Applications. Wiley; Copper Mountain: 2006. pp. 215–229. [Google Scholar]
- 51.Thompson P, Toga AW. In: Handbook of Medical Image Processing. Bankman I, editor. Academic; San Diego: 2000. pp. 159–170. [Google Scholar]
- 52.Roland PE, Zilles K. Brain atlases — a new research tool. Trends Neurosci. 1994;17:458–467. doi: 10.1016/0166-2236(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 53.Mazziotta JC, et al. A probabilistic atlas and reference system for the human brain: international consortium for brain mapping (ICBM) Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. A description of an international consortium project that was set up to develop a probabilistic reference system for the human brain, incorporating statistical information on the variations in human brain structure and function in a population of 7000 subjects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans AC, Collins DL, Milner B. An MRI-based stereotactic brain atlas from 300 young normal subjects. Soc. Neurosci. Abstr. 1992;408 [Google Scholar]
- 55.Eickhoff S, et al. A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. NeuroImage. 2005;25:1325–1335. doi: 10.1016/j.neuroimage.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 56.Grefkes C, Geyer S, Schormann T, Roland P, Zilles K. Human somatosensory area 2: observer-independent cytoarchitectonic mapping, interindividual variability, and population map. NeuroImage. 2001;14:617–631. doi: 10.1006/nimg.2001.0858. [DOI] [PubMed] [Google Scholar]
- 57.Eickhoff S, Schleicher A, Zilles K, Amunts K. The human parietal operculum. I. Cytoarchitectonic mapping of subdivisions. Cereb. Cortex. 2005;16:254–267. doi: 10.1093/cercor/bhi105. [DOI] [PubMed] [Google Scholar]
- 58.Geyer S, Schleicher A, Zilles K. Areas 3a, 3b, and 1 of human primary somatosensory cortex: 1. Microstructural organization and interindividual variability. NeuroImage. 1999;10:63–83. doi: 10.1006/nimg.1999.0440. [DOI] [PubMed] [Google Scholar]
- 59.Choi H-J, et al. Cytoarchitectonic identification and probabilistic mapping of two distinct areas within the anterior ventral bank of the human intraparietal sulcus. J. Comp. Neurol. 2006;495:53–69. doi: 10.1002/cne.20849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eickhoff SB, Weiss PH, Amunts K, Fink GR, Zilles K. Identifying human parieto-insular vestibular cortex using fMRI and cytoarchitectonic mapping. Hum. Brain Mapp. 2006;27:611–621. doi: 10.1002/hbm.20205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malikovic A, et al. Cytoarchitectonic analysis of the human extrastriate cortex in the region of V5/MT+: a probabilistic, stereotaxic map of area hOc5. Cereb. Cortex. 2006 April 7; doi: 10.1093/cercor/bhj181. (doi:10.1093/cercor/bhj181) [DOI] [PubMed] [Google Scholar]
- 62.Amunts K, et al. Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: intersubject variability and probability maps. Anat. Embryol. 2005;210:342–352. doi: 10.1007/s00429-005-0025-5. [DOI] [PubMed] [Google Scholar]
- 63.Geyer S, Schormann T, Mohlberg H, Zilles K. Areas 3a, 3b, and 1 of human primary somatosensory cortex. 2. Spatial normalization to standard anatomical space. NeuroImage. 2000;11:684–696. doi: 10.1006/nimg.2000.0548. [DOI] [PubMed] [Google Scholar]
- 64.Morosan P, et al. Human primary auditory cortex: cytoarchitectonic subdivisions and mapping into a spatial reference system. NeuroImage. 2001;13:684–701. doi: 10.1006/nimg.2000.0715. [DOI] [PubMed] [Google Scholar]
- 65.Rademacher J, et al. Probabilistic mapping and volume measurement of human primary auditory cortex. NeuroImage. 2001;13:669–683. doi: 10.1006/nimg.2000.0714. [DOI] [PubMed] [Google Scholar]
- 66.Rademacher J, Bürgel U, Zilles K. Stereotaxic localization, intersubject variability and interhemispheric differences of the human auditory thalamocortical system. NeuroImage. 2002;17:142–160. doi: 10.1006/nimg.2002.1178. [DOI] [PubMed] [Google Scholar]
- 67.Roland PE, Zilles K. The developing european computerized human brain database for all imaging modalities. NeuroImage. 1996;4:39–47. doi: 10.1006/nimg.1996.0050. [DOI] [PubMed] [Google Scholar]
- 68.Roland PE, et al. Cytoarchitectural maps of the human brain in standard anatomical space. Hum. Brain Mapp. 1997;5:222–227. doi: 10.1002/(SICI)1097-0193(1997)5:4<222::AID-HBM3>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 69.Zilles K, Palomero-Gallagher N. Cyto-, myelo- and receptor architectonics of the human parietal cortex. NeuroImage. 2001;14:8–20. doi: 10.1006/nimg.2001.0823. [DOI] [PubMed] [Google Scholar]
- 70.Zilles K. Mapping of human and macaque sensorimotor areas by integrating architectonic, transmitter receptor, MRI and PET data. J. Anatomy. 1995;187:515–537. [PMC free article] [PubMed] [Google Scholar]
- 71.Amunts K, et al. Analysis of the neural mechanisms underlying verbal fluency in cytoarchitectonically defined stereotaxic space — the roles of Brodmann areas 44 and 45. NeuroImage. 2004;22:42–56. doi: 10.1016/j.neuroimage.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 72.Bodegård A, et al. Object shape differences reflected by somatosensory cortical activation. J. Neurosci. 2000;20:1–5. doi: 10.1523/JNEUROSCI.20-01-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bodegård A, et al. Somatosensory areas in man activated by moving stimuli. Cytoarchitectonic mapping and PET. NeuroReport. 2000;11:187–191. doi: 10.1097/00001756-200001170-00037. [DOI] [PubMed] [Google Scholar]
- 74.Eickhoff S, Amunts K, Mohlberg H, Zilles K. The human parietal operculum. II. Stereotaxic maps and correlation with functional imaging results. Cereb. Cortex. 2006;16:268–279. doi: 10.1093/cercor/bhi106. [DOI] [PubMed] [Google Scholar]
- 75.Horwitz B, et al. Activation of Broca's area during the production of spoken and signed language: a combined cytoarchitectonic mapping and PET analysis. Neuropsychologia. 2003;41:1868–1876. doi: 10.1016/s0028-3932(03)00125-8. [DOI] [PubMed] [Google Scholar]
- 76.Pazos A, Probst A, Palacios JM. Serotonin receptors in the human brain — III. Autoradiographic mapping of serotonin-1 receptors. Neuroscience. 1987;21:97–122. doi: 10.1016/0306-4522(87)90326-5. [DOI] [PubMed] [Google Scholar]
- 77.Pazos A, Probst A, Palacios JM. Serotonin receptors in the human brain — IV. Autoradiographic mapping of serotonin-2 receptors. Neuroscience. 1987;21:123–139. doi: 10.1016/0306-4522(87)90327-7. [DOI] [PubMed] [Google Scholar]
- 78.Vogt BA, Plager MD, Crino PB, Bird ED. Laminar distributions of muscarinic acetylcholine, serotonin, GABA and opioid receptors in human posterior cingulate cortex. Neuroscience. 1990;36:165–174. doi: 10.1016/0306-4522(90)90359-c. [DOI] [PubMed] [Google Scholar]
- 79.Zilles K. In: From Monkey Brain to Human Brain. Dehaene S, Duhamel J-R, Hauser M, Rizzolatti G, editors. MIT Press; Cambridge, Massachusetts: 2005. pp. 41–56. [Google Scholar]
- 80.Zilles K, Palomero-Gallagher N, Schleicher A. Transmitter receptors and functional anatomy of the cerebral cortex. J. Anat. 2004;205:417–432. doi: 10.1111/j.0021-8782.2004.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Geyer S, Schleicher A, Zilles K. The somatosensory cortex of human: cytocarchitecture and regional distributions of receptor-binding sites. NeuroImage. 1997;6:27–45. doi: 10.1006/nimg.1997.0271. [DOI] [PubMed] [Google Scholar]
- 82.Morosan P, Rademacher J, Palomero-Gallagher N, Zilles K. In: The Auditory Cortex: Towards a Synthesis of Human and Animal Research. König R, Heil P, Budinger E, Scheich H, editors. Lawrence Erlbaum; Mahwah, New Jersey: 2005. pp. 27–50. [Google Scholar]
- 83.Basser PJ, Mattiello J, LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J. Magn. Reson. B. 1994;103:247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- 84.Conturo TE, McKinstry RC, Akbudak E, Robinson BH. Encoding of anisotropic diffusion with tetrahedral gradients: a general mathematical diffusion formalism and experimental results. Magn. Reson. Med. 1996;35:399–412. doi: 10.1002/mrm.1910350319. [DOI] [PubMed] [Google Scholar]
- 85.Pierpaoli C, Basser PJ. Toward a quantitative assessment of diffusion anisotropy. Magn. Reson. Med. 1996;36:893–906. doi: 10.1002/mrm.1910360612. [DOI] [PubMed] [Google Scholar]
- 86.Ulug AM, Bakht O, Bryan RN, van Zijl PCM. Mapping of Human Brain Fibers Using Diffusion Tensor Imaging. Proc. Int. Soc. Mag. Reson. Med. 1996;41325 [Google Scholar]
- 87.Douek P, Turner R, Pekar J, Patronas N, Le Bihan D. MR color mapping of myelin fiber orientation. J. Comput. Assist. Tomogr. 1991;15:923–929. doi: 10.1097/00004728-199111000-00003. [DOI] [PubMed] [Google Scholar]
- 88.Hsu EW, Mori S. Analytical interpretations of NMR diffusion measurements in an anisotropic medium and a simplified method for determining fiber orientation. Magn. Reson. Med. 1995;34:194–200. doi: 10.1002/mrm.1910340210. [DOI] [PubMed] [Google Scholar]
- 89.Nakada T, Matsuzawa H. Three-dimensional anisotropy contrast magnetic resonance imaging of the rat nervous system: MR axonography. Neurosci. Res. 1995;22:389–398. doi: 10.1016/0168-0102(95)00917-i. [DOI] [PubMed] [Google Scholar]
- 90.Makris N, et al. Morphometry of in vivo human white matter association pathways with diffusion weighted magnetic resonance imaging. Ann. Neurol. 1997;42:951–962. doi: 10.1002/ana.410420617. A monumental work in which DTI-based image contrast was correlated with white matter anatomy in a comprehensive manner for the first time. [DOI] [PubMed] [Google Scholar]
- 91.Pajevic S, Pierpaoli C. Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data: application to white matter fiber tract mapping in the human brain. Magn. Reson. Med. 1999;42:526–540. [PubMed] [Google Scholar]
- 92.Conturo TE, et al. Tracking neuronal fiber pathways in the living human brain. Proc. Natl Acad. Sci. USA. 1999;96:10422–10427. doi: 10.1073/pnas.96.18.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jones DK, Simmons A, Williams SC, Horsfield MA. Non-invasive assessment of axonal fiber connectivity in the human brain via diffusion tensor MRI. Magn. Reson. Med. 1999;42:37–41. doi: 10.1002/(sici)1522-2594(199907)42:1<37::aid-mrm7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 94.Mori S, Crain BJ, Chacko VP, van Zijl PCM. Three dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Annal. Neurol. 1999;45:265–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 95.Xue R, van Zijl PCM, Crain BJ, Solaiyappan M, Mori S. In vivo three-dimensional reconstruction of rat brain axonal projections by diffusion tensor imaging. Magn. Reson. Med. 1999;42:1123–1127. doi: 10.1002/(sici)1522-2594(199912)42:6<1123::aid-mrm17>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 96.Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vitro fiber tractography using DT-MRI data. Magn. Reson. Med. 2000;44:625–632. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 97.Poupon C, et al. Regularization of diffusion-based direction maps for the tracking of brain white matter fascicules. NeuroImage. 2000;12:184–195. doi: 10.1006/nimg.2000.0607. [DOI] [PubMed] [Google Scholar]
- 98.Lazar M, et al. White matter tractography using diffusion tensor deflection. Hum. Brain Mapp. 2003;18:306–321. doi: 10.1002/hbm.10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Behrens TE, et al. Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nature Neurosci. 2003;6:750–757. doi: 10.1038/nn1075. [DOI] [PubMed] [Google Scholar]
- 100.Ludwig E, Klingler J. Atlas Cerebrei Human: the Internal Structure of the Brain Demonstrated on the Basis of Macroscopical Preparations. Karger; Basel: 1956. [Google Scholar]
- 101.Stieltjes B, et al. Diffusion tensor imaging and axonal tracking in the human brainstem. NeuroImage. 2001;14:723–735. doi: 10.1006/nimg.2001.0861. [DOI] [PubMed] [Google Scholar]
- 102.Catani M, Howard RJ, Pajevic S, Jones DK. Virtual in vivo interactive dissection of white matter fasciculi in the human brain. NeuroImage. 2002;17:77–94. doi: 10.1006/nimg.2002.1136. [DOI] [PubMed] [Google Scholar]
- 103.Mori S, et al. Imaging cortical association tracts in human brain. Magn. Reson. Med. 2002;47:215–223. doi: 10.1002/mrm.10074. [DOI] [PubMed] [Google Scholar]
- 104.Wakana S, Jiang H, Nagae-Poetscher LM, Van Zijl PC, Mori S. Fiber tract-based atlas of human white matter anatomy. Radiology. 2004;230:77–87. doi: 10.1148/radiol.2301021640. A comprehensive atlas of the human white matter based on two-dimensional and three-dimensional visualization of DTI data. [DOI] [PubMed] [Google Scholar]
- 105.Mamata H, et al. High-resolution line scan diffusion tensor MR imaging of white matter fiber tract anatomy. Am. J. Neuroradiol. 2002;23:67–75. [PMC free article] [PubMed] [Google Scholar]
- 106.Alexander DC, Pierpaoli C, Basser PJ, Gee JC. Spatial transformations of diffusion tensor magnetic resonance images. IEEE Trans. Med. Imaging. 2001;20:1131–1139. doi: 10.1109/42.963816. [DOI] [PubMed] [Google Scholar]
- 107.Jones DK, et al. Spatial normalization and averaging of diffusion tensor MRI data sets. Neuroimage. 2002;17:592–617. [PubMed] [Google Scholar]
- 108.Xu D, Mori S, Shen D, van Zijl PC, Davatzikos C. Spatial normalization of diffusion tensor fields. Magn. Reson. Med. 2003;50:175–182. doi: 10.1002/mrm.10489. [DOI] [PubMed] [Google Scholar]
- 109.Pagani E, Filippi M, Rocca MA, Horsfield MA. A method for obtaining tract-specific diffusion tensor MRI measurements in the presence of disease: application to patients with clinically isolated syndromes suggestive of multiple sclerosis. NeuroImage. 2005;26:258–265. doi: 10.1016/j.neuroimage.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 110.Pierpaoli C, et al. Water diffusion change in Wallerian degeneration and their dependence on white matter architecture. NeuroImage. 2001;13:1174–1185. doi: 10.1006/nimg.2001.0765. [DOI] [PubMed] [Google Scholar]
- 111.Thomalla G, et al. Diffusion tensor imaging detects early Wallerian degeneration of the pyramidal tract after ischemic stroke. NeuroImage. 2004;22:1767–1774. doi: 10.1016/j.neuroimage.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 112.Lee JS, Han MK, Kim SH, Kwon OK, Kim JH. Fiber tracking by diffusion tensor imaging in corticospinal tract stroke: topographical correlation with clinical symptoms. NeuroImage. 2005;26:771–776. doi: 10.1016/j.neuroimage.2005.02.036. [DOI] [PubMed] [Google Scholar]
- 113.Johansen-Berg H, et al. Changes in connectivity profiles define functionally distinct regions in human medial frontal cortex. Proc. Natl Acad. Sci. USA. 2004;101:13335–13340. doi: 10.1073/pnas.0403743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frank LR. Anisotropy in high angular resolution diffusion-weighted MRI. Magn. Reson. Med. 2001;45:935–939. doi: 10.1002/mrm.1125. [DOI] [PubMed] [Google Scholar]
- 115.Tournier JD, Calamante F, Gadian DG, Connelly A. Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using spherical deconvolution. NeuroImage. 2004;23:1176–1185. doi: 10.1016/j.neuroimage.2004.07.037. [DOI] [PubMed] [Google Scholar]
- 116.Tuch DS, Reese TG, Wiegell MR, Wedeen VJ. Diffusion MRI of complex neural architecture. Neuron. 2003;40:885–895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 117.Wedeen VJ, Hagmann P, Tseng WY, Reese TG, Weisskoff RM. Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn. Reson. Med. 2005;54:1377–1386. doi: 10.1002/mrm.20642. [DOI] [PubMed] [Google Scholar]
- 118.Bürgel U, Schormann T, Schleicher A, Zilles K. Mapping of histologically identified long fiber tracts in human cerebral hemispheres to the MRI volume of a reference brain: position and spatial variability of the optic radiation. NeuroImage. 1999;10:489–499. doi: 10.1006/nimg.1999.0497. [DOI] [PubMed] [Google Scholar]
- 119.Rademacher J, Engelbrecht V, Bürgel U, Freund HJ, Zilles K. Measuring in vivo myelination of human white matter fiber tracts with magnetization transfer MR. NeuroImage. 1999;9:393–406. doi: 10.1006/nimg.1998.0416. [DOI] [PubMed] [Google Scholar]
- 120.Rademacher J, et al. Variability and asymmetry in the human precentral motor system. A cytoarchitectonic and myeloarchitectonic brain mapping study. Brain. 2001;124:2232–2258. doi: 10.1093/brain/124.11.2232. [DOI] [PubMed] [Google Scholar]
- 121.Mega MS, et al. Mapping pathology to metabolism: coregistration of stained whole brain sections to PET in alzheimer's disease. NeuroImage. 1997;5:147–153. doi: 10.1006/nimg.1996.0255. [DOI] [PubMed] [Google Scholar]
- 122.Toga AW, Ambach KL, Quinn BC, Hutchin M, Burton JS. Postmortem anatomy from cryosectioned whole human brain. J. Neurosci. Methods. 1994;5:239–252. doi: 10.1016/0165-0270(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 123.Teipel SJ, et al. Measurement of basal forebrain atrophy in Alzheimer's disease using MRI. Brain. 2005;128:2626–2644. doi: 10.1093/brain/awh589. [DOI] [PubMed] [Google Scholar]
- 124.Toga AW, Thompson PM. Mapping brain asymmetry. Nature Rev. Neurosci. 2003;4:37–38. doi: 10.1038/nrn1009. [DOI] [PubMed] [Google Scholar]
- 125.Toga AW, Thompson PM. Genetics of brain structure and intelligence. Ann. Rev. Neurosci. 2005;28:1–23. doi: 10.1146/annurev.neuro.28.061604.135655. [DOI] [PubMed] [Google Scholar]
- 126.Grenander U, Miller MI. Computational Anatomy: an Emerging Discipline. Technical Report, Dept. Mathematics, Brown Univ.; 1998. This highly-cited article was influential in stimulating mathematical developments in the field of computational anatomy. A new framework is proposed that represents anatomical variation by defining statistics and probability measures on three-dimensional elastic or fluid mappings that deform a canonical template of anatomy. [Google Scholar]
- 127.Narr KL, et al. Mapping morphology of the corpus callosum in schizophrenia. Cereb. Cortex. 2000;10:40–49. doi: 10.1093/cercor/10.1.40. [DOI] [PubMed] [Google Scholar]
- 128.Thompson PM, et al. Cortical variability and asymmetry in normal aging and alzheimer's disease. Cereb. Cortex. 1998;8:492–509. doi: 10.1093/cercor/8.6.492. [DOI] [PubMed] [Google Scholar]
- 129.Thompson PM, Mega MS, Toga AW. In: Brain Mapping: the Disorders. Toga AW, Mazziotta JC, editors. Academic; San Diego: 2000. pp. 131–177. [Google Scholar]
- 130.Sowell ER, et al. Mapping cortical change across the human life span. Nature Neurosci. 2003;6:309–315. doi: 10.1038/nn1008. [DOI] [PubMed] [Google Scholar]
- 131.Levitt JG, et al. Proton magnetic resonance spectroscopic imaging of the brain in childhood autism. Biol. Psychiatry. 2003;54:1355–1366. doi: 10.1016/s0006-3223(03)00688-7. [DOI] [PubMed] [Google Scholar]
- 132.Thompson PM, et al. Cortical complexity and thickness are increased in Williams Syndrome. J. Neurosci. 2005;25:4146–4158. doi: 10.1523/JNEUROSCI.0165-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mazziotta JC, Toga AW, Evans AC, Fox P, Lancaster J. A probabilistic atlas of the human brain: theory and rationale for its development. NeuroImage. 1995;2:89–101. doi: 10.1006/nimg.1995.1012. [DOI] [PubMed] [Google Scholar]
- 134.Geschwind N, Galaburda AM. Cerebral lateralization. Biological mechanisms, associations, and pathology: III. A hypothesis and a program for research. Arch. Neurol. 1985;42:634–656. doi: 10.1001/archneur.1985.04060050026008. [DOI] [PubMed] [Google Scholar]
- 135.Sowell ER, et al. Brain abnormalities in early-onset schizophrenia spectrum disorder observed with statistical parametric mapping of structural magnetic resonance images. Am. J. Psychiatry. 2000;157:1475–1484. doi: 10.1176/appi.ajp.157.9.1475. [DOI] [PubMed] [Google Scholar]
- 136.Luders E, et al. Mapping cortical gray matter in the young adult brain: effects of gender. NeuroImage. 2005;26:493–501. doi: 10.1016/j.neuroimage.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 137.Crow TJ. Handedness, language lateralisation and anatomical asymmetry: relevance of protocadherin XY to hominid speciation and the aetiology of psychosis. Point of view. Br. J. Psychiatry. 2002;181:295–297. doi: 10.1192/bjp.181.4.295. [DOI] [PubMed] [Google Scholar]
- 138.Narr KL, et al. Mapping cortical thickness and gray matter concentration in first episode schizophrenia. Cereb. Cortex. 2005;15:708–719. doi: 10.1093/cercor/bhh172. [DOI] [PubMed] [Google Scholar]
- 139.Van Essen DC. Surface-based approaches to spatial localization and registration in primate cerebral cortex. NeuroImage. 2005;23:S97–S107. doi: 10.1016/j.neuroimage.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 140.van Horn JD. Neuroimaging databases as a resource for scientific discovery. Int. Rev. Neurobiol. 2005;66:55–87. doi: 10.1016/S0074-7742(05)66002-3. [DOI] [PubMed] [Google Scholar]
- 141.Giedd JN, et al. Brain development during childhood and adolescence: a longitudinal MRI study. Nature Neurosci. 1999;2:861–863. doi: 10.1038/13158. [DOI] [PubMed] [Google Scholar]
- 142.Rapoport JL, Addington A, Frangou S. The neuro developmental model of schizophrenia: what can very early onset cases tell us? Curr. Psychiatry Rep. 2005;7:81–82. doi: 10.1007/s11920-005-0001-z. [DOI] [PubMed] [Google Scholar]
- 143.Thompson PM, et al. Early cortical change in Alzheimer's disease detected with a disease-specific population-based brain atlas. Cereb. Cortex. 2001;11:1–16. doi: 10.1093/cercor/11.1.1. [DOI] [PubMed] [Google Scholar]
- 144.Cannon TD, et al. Early and late neurodevelopmental influences in the prodrome to schizophrenia: contributions of genes, environment and their interactions. Schizophr. Bull. 2003;29:653–669. doi: 10.1093/oxfordjournals.schbul.a007037. [DOI] [PubMed] [Google Scholar]
- 145.Cannon TD, et al. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter and impaired short- and long-term memory. Arch. Gen. Psychiatry. 2005;62:1205–1213. doi: 10.1001/archpsyc.62.11.1205. [DOI] [PubMed] [Google Scholar]
- 146.Lin JJ. 3D Pre-operative maps of hippocampal atrophy predict surgical outcomes in temporal lobe epilepsy. Neurology. 2005;65:1094–1097. doi: 10.1212/01.wnl.0000179003.95838.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Moore GJ, Bebchuk JM, Wilds IB, Chen G, Manji HK. Lithium-induced increase in human brain grey matter. Lancet. 2000;356:1241–1242. doi: 10.1016/s0140-6736(00)02793-8. [DOI] [PubMed] [Google Scholar]
- 148.Lieberman DZ, Goodwin FK. Use of olanzapine in the treatment of bipolar I disorder. Expert Rev. Neurother. 2004;4:759–767. doi: 10.1586/14737175.4.5.759. [DOI] [PubMed] [Google Scholar]
- 149.Leow A, et al. Brain structural mapping using a novel hybrid implicit/explicit framework based on the level-set method. NeuroImage. 2005;24:910–927. doi: 10.1016/j.neuroimage.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 150.Kikinis R, et al. A digital brain atlas for surgical planning, model-driven segmentation, and teaching. IEEE Trans. Vis. Comput. Graph. 1996;2:232–241. [Google Scholar]
- 151.Mangin JF, et al. Brain morphometry using 3D moment invariants. Med. Image Anal. 2004;8:187–196. doi: 10.1016/j.media.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 152.Collins DL, Peters TM, Evans AC. An automated 3D non-linear image deformation procedure for determination of gross morphometric variability in the human brain. Proc. Visualization. Biomed. Comput. 1994;3:180–190. [Google Scholar]
- 153.Thompson PM, et al. Detection and mapping of abnormal brain structure with a probabilistic atlas of cortical surfaces. J. Comput. Asst. Tomogr. 1997;21:567–581. doi: 10.1097/00004728-199707000-00008. [DOI] [PubMed] [Google Scholar]
- 154.Chung MK. Cortical thickness analysis in autism with heat kernel smoothing. NeuroImage. 2005;25:1256–1265. doi: 10.1016/j.neuroimage.2004.12.052. [DOI] [PubMed] [Google Scholar]
- 155.Carmichael OT, et al. Mapping ventricular changes related to dementia and mild cognitive impairment in a large community-based cohort. IEEE Int. Symp. Biomed. Imaging. 2006:315–318. [Google Scholar]
- 156.Gogtay N, et al. Dynamic mapping of human cortical development during childhood and adolescence. Proc. Natl Acad. Sci. 2004;101:8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sowell ER, Thompson PM, Toga AW. Mapping changes in the human cortex throughout the span of life. Neuroscientist. 2004;10:372–392. doi: 10.1177/1073858404263960. [DOI] [PubMed] [Google Scholar]
- 158.Thompson PM, et al. Growth patterns in the developing brain detected by using continuum-mechanical tensor maps. Nature. 2000;404:190–193. doi: 10.1038/35004593. [DOI] [PubMed] [Google Scholar]
- 159.Lerch JP, et al. Focal decline of cortical thickness in Alzheimer's disease identified by computational neuroanatomy. Cereb. Cortex. 2005;15:995–1001. doi: 10.1093/cercor/bhh200. [DOI] [PubMed] [Google Scholar]
- 160.Janke AL, et al. 4D deformation modeling of cortical disease progression in Alzheimer's dementia. Magn. Reson. Med. 2001;46:661–666. doi: 10.1002/mrm.1243. [DOI] [PubMed] [Google Scholar]
- 161.Thompson PM, et al. Dynamics of gray matter loss in Alzheimer's disease. J. Neurosci. 2003;23:994–1005. doi: 10.1523/JNEUROSCI.23-03-00994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Friston KJ, et al. Spatial registration and normalisation of images. Hum. Brain Mapp. 1995;2:16. [Google Scholar]
- 163.van Essen DC. A population-average, landmark- and surface-based (PALS) atlas of human cerebral cortex. NeuroImage. 2005;28:635–662. doi: 10.1016/j.neuroimage.2005.06.058. [DOI] [PubMed] [Google Scholar]
- 164.Fischl B, et al. Sequence-independent segmentation of magnetic resonance images. NeuroImage. 2004;23:S69–S84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 165.Pitiot A, Delingette H, Thompson PM, Ayache N. Expert knowledge-guided segmentation system for brain MRI. NeuroImage. 2004;23:S85–S96. doi: 10.1016/j.neuroimage.2004.07.040. [DOI] [PubMed] [Google Scholar]
- 166.Zijdenbos AP, Lerch JP, Bedell BJ, Evans AC. Brain imaging in drug R&D. Biomarkers. 2005;10:S58–S68. doi: 10.1080/13547500500214392. The authors describe a recent extension of the atlas concept to include large-scale processing of images from drug trials, analysing images that have been mapped into a stereotaxic coordinate space. [DOI] [PubMed] [Google Scholar]
- 167.Duncan JS, et al. Geometric strategies for neuroanatomic analysis from MRI. NeuroImage. 2004;23:S34–S45. doi: 10.1016/j.neuroimage.2004.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shattuck DW, Sandor-Leahy SR, Schaper KA, Rottenberg DA, Leahy RM. Magnetic resonance image tissue classification using a partial volume model. NeuroImage. 2001;13:856–876. doi: 10.1006/nimg.2000.0730. [DOI] [PubMed] [Google Scholar]
- 169.Dinov ID, et al. Analyzing functional brain images in a probabilistic atlas: a validation of sub-volume thresholding. J. Comput. Asst. Tomog. 2000;24:128–138. doi: 10.1097/00004728-200001000-00024. [DOI] [PubMed] [Google Scholar]
- 170.Rasser PE, et al. Functional MRI BOLD response to Tower of London performance of first-episode schizophrenia patients using cortical pattern matching. NeuroImage. 2005;26:941–951. doi: 10.1016/j.neuroimage.2004.11.054. [DOI] [PubMed] [Google Scholar]
- 171.Thompson P, et al. Mapping hippocampal and ventricular change in Alzheimer's disease. NeuroImage. 2004;22:1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 172.Lin CL, et al. Characterization of perioperative seizures and epilepsy following aneurysmal subarachnoid hemorrhage. J. Neurosurg. 2003;99:978–985. doi: 10.3171/jns.2003.99.6.0978. [DOI] [PubMed] [Google Scholar]
- 173.Ballmaier M, et al. Localizing gray matter deficits in late onset depression using computational cortical pattern matching methods. Am. J. Psychiatry. 2004;161:2091–2099. doi: 10.1176/appi.ajp.161.11.2091. [DOI] [PubMed] [Google Scholar]
- 174.Cannon TD, et al. Cortex mapping reveals regionally specific patterns of genetic and disease-specific gray-matter deficits in twins discordant for schizophrenia. Proc. Natl Acad. Sci. USA. 2002;99:3228–3233. doi: 10.1073/pnas.052023499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Narr KL, et al. Abnormal gyral complexity in first episode schizophrenia. Biol. Psychiatry. 2004;55:859–867. doi: 10.1016/j.biopsych.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 176.Thompson P, et al. Mapping hippocampal and ventricular change in Alzheimer's disease. NeuroImage. 2004;22:1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 177.Vidal C, et al. Dynamically spreading frontal and cingulate deficits mapped in adolescents with Schizophrenia. Arch. Gen. Psychiatry. 2006;63:25–34. doi: 10.1001/archpsyc.63.1.25. [DOI] [PubMed] [Google Scholar]
- 178.Sowell ER, et al. Cortical abnormalities in children and adolescents with attention deficit hyperactivity disorder. Lancet. 2003;362:1699–1707. doi: 10.1016/S0140-6736(03)14842-8. [DOI] [PubMed] [Google Scholar]
- 179.Sowell ER, et al. Regional brain shape abnormalities persist into adolescence after heavy prenatal alcohol exposure. Cereb. Cortex. 2002;12:856–865. doi: 10.1093/cercor/12.8.856. [DOI] [PubMed] [Google Scholar]
- 180.Sowell ER, et al. Gray matter thickness abnormalities mapped in children with Tourette Syndrome. Soc. Neurosci. Abstr. 2004;800:15. [Google Scholar]
- 181.Gogtay N, et al. Dynamic mapping of cortical brain development in pediatric bipolar illness. Int. Conf. Org. Hum. Brain Mapp. 2004:344. [Google Scholar]
- 182.Thompson P, et al. Structural abnormalities in the brains of human subjects who use methamphetamine. J. Neurosci. 2004;24:6028–6036. doi: 10.1523/JNEUROSCI.0713-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mega MS, et al. Sulcal variability in the Alzheimer's brain: correlations with cognition. Neurology. 1998;50:145–151. doi: 10.1212/wnl.50.1.145. [DOI] [PubMed] [Google Scholar]
- 184.Miller M. Computational anatomy: shape, growth and atrophy comparison via diffeomorphisms. NeuroImage. 2004;23:S19–S33. doi: 10.1016/j.neuroimage.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 185.Studholme C, et al. Deformation tensor morphometry of semantic dementia with quantitative validation. NeuroImage. 2004;21:1387–1398. doi: 10.1016/j.neuroimage.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 186.Cannon TD, et al. Mapping heritability and molecular genetic associations with cortical features using probabilistic brain atlases: methods and initial applications to schizophrenia. Neuroinformatics. 2006;4:5–19. doi: 10.1385/NI:4:1:5. [DOI] [PubMed] [Google Scholar]
- 187.Ballmeier M, et al. Comparing gray matter loss profiles between dementia with Lewy bodies and Alzheimer's disease using cortical pattern matching: diagnosis and gender effects. NeuroImage. 2004;23:325–335. doi: 10.1016/j.neuroimage.2004.04.026. [DOI] [PubMed] [Google Scholar]
- 188.Thompson PM, Mega MS, Vidal C, Rapoport JL, Toga AW. Lect. Notes Comput. Sci. 2001;2082:488–501. doi: 10.1007/3-540-45729-1_52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Thompson PM, et al. Mapping adolescent brain change reveals dynamic wave of accelerated gray matter loss in very early-onset schizophrenia. Proc. Natl Acad. Sci. USA. 2001;98:11650–11655. doi: 10.1073/pnas.201243998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Vidal CN, et al. dynamically spreading frontal and cingulate deficits mapped in adolescents with schizophrenia. Arch. Gen. Psychiatry. 2006;63:25–34. doi: 10.1001/archpsyc.63.1.25. [DOI] [PubMed] [Google Scholar]
- 191.Worsley KJ, et al. A unified statistical approach for determining significant signals in images of cerebral activation. Hum. Brain Mapp. 1996;4:58–73. doi: 10.1002/(SICI)1097-0193(1996)4:1<58::AID-HBM4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 192.Friston KJ. Statistical parametric mapping: ontology and current issues. J. Cereb. Blood Flow Metab. 1995;15:361–370. doi: 10.1038/jcbfm.1995.45. [DOI] [PubMed] [Google Scholar]
- 193.Friston KJ, et al. Statistical parametric maps in functional imaging: a general linear approach. Hum. Brain Mapp. 1995;2:189–210. [Google Scholar]
- 194.Ashburner J, Friston KJ. Voxel-based morphometry — the methods. NeuroImage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 195.Thompson PM, et al. Thinning of the cerebral cortex in HIV/AIDS reflects CD4+ T-lymphocyte decline. Proc. Natl Acad. Sci. 2005;102:15647–15652. doi: 10.1073/pnas.0502548102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Williams TH, Gluhbegoric N, Jew JY. The human brain: dissections of the real brain. Virtual Hospital, University of Iowa [online] 1997 < http://www.vh.org/ Providers/Textbooks/BrainAnatomy>.