Abstract
C2H2 zinc fingers (C2H2-ZFs) are the most prevalent type of vertebrate DNA-binding domain, and typically appear in tandem arrays (ZFAs), with sequential C2H2-ZFs each contacting three (or more) sequential bases. C2H2-ZFs can be assembled in a modular fashion, providing one explanation for their remarkable evolutionary success. Given a set of modules with defined three-base specificities, modular assembly also presents a way to construct artificial proteins with specific DNA-binding preferences. However, a recent survey of a large number of three-finger ZFAs engineered by modular assembly reported high failure rates (∼70%), casting doubt on the generality of modular assembly. Here, we used protein-binding microarrays to analyze 28 ZFAs that failed in the aforementioned study. Most (17) preferred specific sequences, which in all but one case resembled the intended target sequence. Like natural ZFAs, the engineered ZFAs typically yielded degenerate motifs, binding dozens to hundreds of related individual sequences. Thus, the failure of these proteins in previous assays is not due to lack of sequence-specific DNA-binding activity. Our findings underscore the relevance of individual C2H2-ZF sequence specificities within tandem arrays, and support the general ability of modular assembly to produce ZFAs with sequence-specific DNA-binding activity.
INTRODUCTION
The C2H2 zinc finger (C2H2-ZF) is among the most prevalent DNA-binding domains in eukaryotes, and genes that encode this domain constitute nearly one-half of all known and predicted transcription factors in human and mouse (1–5). C2H2-ZF proteins typically have multiple C2H2-ZFs arranged in tandem, with each C2H2-ZF binding 3 (or more) bases, and with the fingers offset by three bases, so that a multi-fingered protein recognizes a longer DNA sequence that is thought to be largely a concatenation of each finger’s specificity (6). The dramatic expansion of the number of C2H2-ZFs in mammals appears to be a recent evolutionary event, with their loci residing in clusters, indicating that the C2H2-ZF family evolved through tandem duplications (2,3,7). The C2H2-ZF family is known to have remarkably diverse sequence specificity (6), and sequence analyses have suggested that the diversification of C2H2-ZF paralogs may be driven by positive selection on DNA-contacting residues (2,8).
The evolutionary success of C2H2-ZFs may also be explained in part by their capacity for modular assembly: individual C2H2-ZFs (‘modules’) can be recombined to produce proteins (Zinc Finger Arrays, or ZFAs) with new binding specificities, and both natural and artificial C2H2-ZFs have been used successfully in modular assembly of ZFAs with new sequence specificities (9,10) [reviewed in (6,11,12)]. Modular assembly of ZFAs has received much attention because of its utility in engineering artificial transcription factors or zinc-finger nucleases (ZFNs) with desired sequence specificity: for example, ZFNs constructed by modular assembly have been used to successfully make targeted genome modifications in both plants and animals (13). It is also reasonable to posit that modular assembly serves as a mechanism for natural evolutionary diversification of C2H2-ZF proteins (14). In addition, modularity is an assumption that underlies efforts to identify the sequence specificity of the thousands of natural ZFAs—most of which have not been experimentally characterized—by concatenating the known or predicted sequence specificities of their individual C2H2-ZF components (15–17).
Given the conceptual and practical importance of the modularity of C2H2-ZFs, it is important to know the limits and constraints of modular assembly, and in this regard the evidence is mixed. While there are many examples supporting the retention of sequence specificity of individual C2H2-ZFs within ZFAs constructed by modular assembly [e.g. (6,11,12,18)], it is also known that the sequences recognized by a given C2H2-ZF can be influenced by the neighboring C2H2-ZF (19,20). The most straightforward explanation for dependence among neighboring C2H2-ZFs has been referred to as the ‘target site overlap problem’ (21): C2H2-ZFs often contact four-base subsites, such that there is one base of overlap between adjacent C2H2-ZFs (22,23). Alternative docking modes and contacts of up to five bases have also been observed (6,24). Interactions between side-chains also occur between sequential C2H2-ZFs and may be important for both stability of the DNA–protein complex and for sequence specificity (24). Moreover, the spacing between adjacent C2H2-ZFs is not precisely equivalent to three bases [discussed in (25)], raising the possibility that interactions between adjacent C2H2-ZFs may impact the alignment of individual C2H2-ZFs with their subsites.
A recent large-scale examination of modular assembly, hereafter referred to as Ramirez et al. (26), concluded that the modular assembly method of engineering ZFAs has an unexpectedly high failure rate of roughly 70%, in contrast to previous reports claiming 60% or 100% success (9,18). Ramirez et al. constructed a total of 204 ZFAs using three different collections of C2H2-ZF modules (9,27–29). The study tested 27 ZFAs by electrophoretic mobility shift assay (EMSA), among which seven succeeded. A subset of these failed ZFAs was then tested by a plant single-stranded annealing assay; all of these also failed. The study then tested 168 additional ZFAs by a bacterial-2-hybrid (B2H) assay, which tests a ZFA’s ability to activate a reporter gene containing the intended ZFA binding site in the promoter, and obtained only 53 successes. Twenty-two of these ZFAs were tested by an episomal recombination assay, which supported the results of the B2H assays. In total, 144 of 204 ZFAs failed at the assay(s) used to test them.
Ramirez et al. found that much of the discrepancy between their findings and previous reports (9,18) can be accounted for by the fact that the previous reports were biased toward GNN subsites (i.e. the C2H2-ZF modules bound to sequences in which the 5′-base is a guanine). There are at least two reasons to expect a higher success rate with GNN subsites. First, in GNN-binding C2H2-ZFs, the amino acid Arg is typically found at position +6 of the recognition helix (which directly contacts the bases in the major groove), and Arg can make two hydrogen bonds with the 5′-base guanine, creating a particularly strong DNA–protein interaction (22). Second, GNN subsites may be the most compatible with the scaffolds used in current artificial ZFAs because many of the individual C2H2-ZF modules are variants of finger 2 of Zif268 (30–32), which naturally prefers GGG-G or TGG-G (the fourth base is a contact to the next triplet, which would further bias the neighboring triplet toward GNN). Other modules are derived from fingers 1, 2 or 3 of Sp1, which naturally prefer GG(G/T), G(C/A)G and (G/T)GG, respectively (33). Indeed, Ramirez et al. obtained 59% success for ZFAs with three GNN subsites, but only 29, 12 and 0% success for ZFAs with 2, 1 and 0 GNN subsites.
The high failure rates observed by Ramirez et al. call into question the general modularity of the C2H2-ZF motif. However, Ramirez et al. were seeking ZFAs that would function in specific assays, and in most cases did not directly assay DNA-binding: only a minority (27, or 13%) were tested by EMSA. Moreover, the assays tested only the single anticipated 9-mer target. High specificity and/or affinity may be a requirement for ZFNs (and for the B2H assay) (34,35), but is not necessarily a constraint for the evolution of natural transcription factors; most transcription factors display degeneracy at multiple bases of the binding site (36). In fact, if recombination among C2H2-ZFs is used as an evolutionary mechanism for the generation of novel TFs, as has been previously proposed (14), one can imagine that flexibility and degeneracy in the binding preferences of modular C2H2-ZFs could be beneficial for creating new DNA-binding activities. Analysis of useful engineered ZFAs by SELEX has also suggested degeneracy at some base positions (18,37–39). Given these considerations, the blanket declaration that modular assembly generally fails may require qualification, since success and failure are dependent on the assays used and the goals of individual researchers. For example, modular assembly of a new ZFA with sequence-specific DNA-binding activity might be considered a ‘success’ by evolutionary biologists, and indeed many molecular biologists, even if the sequence preference contains degeneracy, or is otherwise not exactly what would have been predicted from the constituent modules. Moreover, to our knowledge, the general concept of modularity does not require invariant behavior of modules in different contexts. Rather, it simply requires that the individual modules can function in different contexts.
Here, we have more closely examined the DNA-binding specificities of 28 of the ‘failed’ ZFAs from Ramirez et al., using protein-binding microarrays (PBMs). PBMs have emerged in the last decade as a rapid and powerful tool for the analysis of sequence specificity of diverse proteins, including C2H2-ZFs (40). The PBM technique can be summarized as follows: a tagged DNA-binding protein is ‘hybridized’ to a microarray that contains a diverse set of approximately 41 000 35-mer probes, and subsequent addition of a fluorescently tagged antibody reveals the DNA sequences that the protein has bound, and to what degree. The DNA probes are designed such that all possible 10-mers are present once and only once; thus, all non-palindromic 8-mers are present 32 times, allowing for a robust and unbiased assessment of sequence preference to all possible 8-mers, and inference of DNA-binding motifs up to 14 bases wide (36,41,42). We and others have used PBMs to determine the binding specificities of hundreds of different transcription factors, from a wide range of species, with very little discrepancy between motifs obtained by PBM and motifs previously defined by more traditional methods, when available (36,41,43–47). In fact, JASPAR (48)—an open-access database for high-quality transcription factor binding site information—currently has more data derived from PBM experiments than it has for all other data in the literature.
In summary, for the failed ZFAs of Ramirez et al., PBM analysis reveals that most have sequence preferences similar to those intended. In addition, most of the individual modules within functional ZFAs bind sequences that are identical or related to their known targets. Our analysis does recapitulate the bias toward GNN subsites. However, we conclude that the high failure rates observed by Ramirez et al. do not reflect a general failure of modular assembly to produce ZFAs with sequence-specific DNA-binding activity.
MATERIALS AND METHODS
Protein-binding microarray experiments
Sequences of the two PBM ‘all-10-mer’ designs are given at http://hugheslab.ccbr.utoronto.ca/supplementary-data/C2H2_modularity/. Details of the design and use of PBMs has been described elsewhere (41,47,49,50). Plasmids are listed in Supplementary Table S1. ZFAs were cloned as SacI–BamHI fragments into pTH5325, a modified T7-driven GST expression vector (see Supplementary Document of the Supplementary Data). Briefly, we used 150 ng of plasmid DNA in a 25 μl in vitro transcription/translation reaction using a PURExpress In Vitro Protein Synthesis Kit (New England BioLabs) supplemented with RNase inhibitor and 50 μM zinc acetate. After a 2-h incubation at 37°C, 12.5 μl of the mix was added to 137.5 μl of protein-binding solution for a final mix of PBS/2% skim milk/0.2 mg per ml BSA/50 μM zinc acetate/0.1% Tween-20. This mixture was added to an array previously blocked with PBS/2% skim milk and washed once with PBS/0.1% Tween-20 and once with PBS/0.01% Triton-X 100. After a 1-h incubation at room temperature, the array was washed once with PBS/0.5% Tween-20/50 μM zinc acetate and once with PBS/0.01% Triton-X 100/50 μM zinc acetate. Cy5-labeled anti-GST antibody was added, diluted in PBS/2% skim milk/50 μM zinc acetate. After a 1-h incubation at room temperature, the array was washed three times with PBS/0.05% Tween-20/50 μM zinc acetate and once with PBS/50 μM zinc acetate. The array was then imaged using an Agilent microarray scanner at 2 μM resolution.
Analysis of microarray data
Image spot intensities were quantified using ImaGene software (BioDiscovery). To estimate the relative preference for each 8-mer, two different scores were calculated: the Z-score was calculated from the average signal intensity across the 16 or 32 spots containing each 8-mer; the ‘E-score’ (for enrichment) is a variation on Area Under the ROC curve (41) and is used here as it is highly reproducible and facilitates comparison between separate experiments. Each ZFA was tested on two different universal microarrays (designated ME and HK). E-score data are discussed in the text; however, both Z- and E-score data are provided in the supplementary data online at http://hugheslab.ccbr.utoronto.ca/supplementary-data/C2H2_modularity/. Microarray data have been deposited to GEO (accession number GSE25723).
RESULTS
Analysis of the sequence specificity of ZFAs
Using PBMs, we assayed a total of 31 ZFAs, 28 of which were designated as failures by Ramirez et al. and three that were deemed successes, which we used as positive controls (Supplementary Table S1 contains information about the ZFAs we tested; the Supplementary Document gives the sequence and map of the plasmid we used; Supplementary Table S1 and all of the data can be found online at http://hugheslab.ccbr.utoronto.ca/supplementary-data/C2H2_modularity/). We chose the 28 ZFAs such that (i) 20 modules (of a total of 61 in our study) were tested in more than one context; (ii) the DNA triplets that the encompassed modules specified formed a diverse set, including GNN, CNN, ANN and TNN modules; (iii) the modules included both human C2H2-ZFs [Toolgen modules (9)] and C2H2-ZFs obtained by selection methods [Barbas (28) and Sangamo (27,29) modules] and (iv) 10 ZFAs that failed by EMSA in Ramirez et al. were included. We cloned each of the inserts into a GST expression vector and analyzed each of the proteins on two different PBM arrays, i.e. different designs, such that the 10-mers, and hence 8-mers, are in different contexts between the two arrays (the arrays are designated ‘ME’ and ‘HK’, which are the initials of the designers of the arrays). We obtained essentially identical results from the two array types.
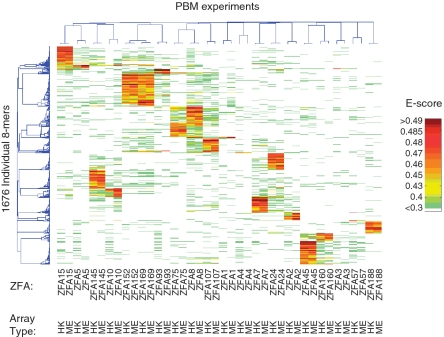
PBM data can be represented in several ways (41,47), including motifs and consensus sequences, as well as a table of relative preferences for individual sequences, most typically all 32 896 possible 8-mers (collapsing reverse complements). A previously established threshold for statistical significance was described by Berger et al. (47) that utilizes 8-mer ‘E-scores’—in essence, a score that reflects the relative ranking of the intensities of the 32 probes that contain each 8-mer, relative to the remaining approximately 41 000 probes. E-scores are similar to the AUC (Area under the ROC curve) statistical metric and range from −0.5 to 0.5. Permutation tests in which the identity of the array probes is scrambled have shown that any score at or above 0.45 would not be observed by chance in a data set much larger than the one used here (47). Using a success criterion that at least one 8-mer must have an E-score of 0.45 or greater, all three of the control proteins were successes, as were 17 of the 28 proteins that failed in Ramirez et al. For the remaining 11, it is possible that these proteins simply lack DNA-binding activity. However, it is also possible that the proteins are misfolded; in our hands, heterologous expression of natural transcription factor DNA-binding domains as GST fusions yields an overall success rate of ∼50% for obtaining a soluble protein with sequence-specific DNA-binding activity (data not shown). Notably, using the E ≥ 0.45 criterion, all six of the ZFAs we assayed that were constructed from natural human C2H2-ZF modules were successful (see below), consistent with a previous claim that naturally occurring human C2H2-ZFs have a high propensity to form functional ZFAs (51), although in our analysis their sequence specificity appears no higher than that of other modules (see below). Figure 1 shows a clustering analysis of all of the 8-mers with E ≥ 0.45 in at least one experiment, illustrating that each ZFA has a distinct and reproducible spectrum of preferences for individual 8-mers.
Figure 1.
Clustering analysis of PBM data. Color scale reflects E-score, as indicated in the legend. All 8-mers that scored at E > 0.45 in at least one experiment are included.
ZFA sequence preferences typically resemble intended targets
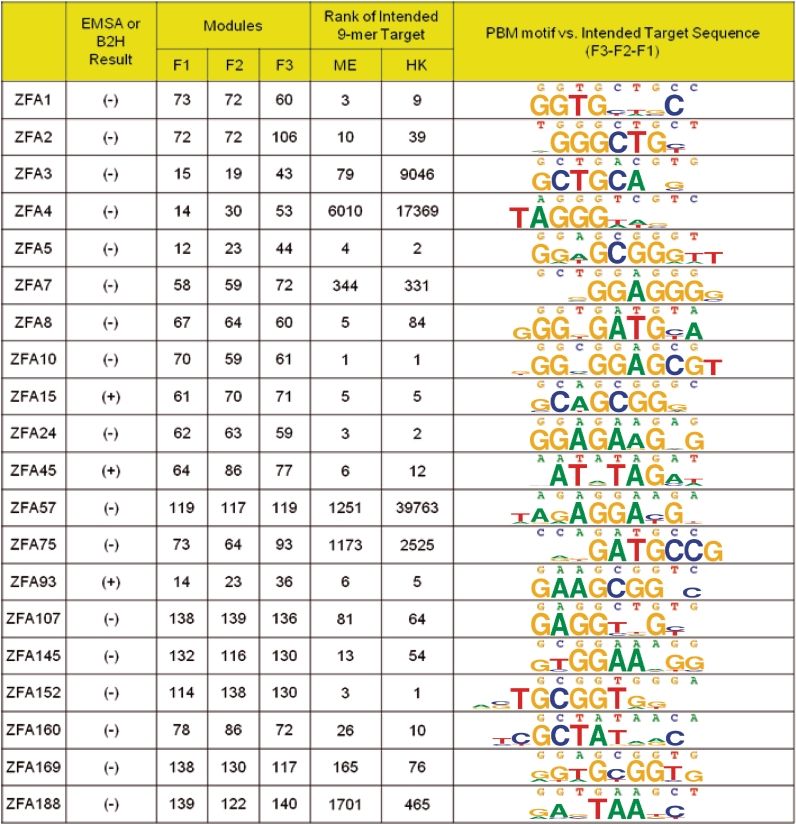
We next asked whether the sequence specificities we obtained corresponded to those intended. Since the ZFAs were designed to recognize 9-base sites, we first examined how the intended target ranked among all 131 072 possible 9-mers, using the same E-score statistic described above. The 9-mer scores are noisier than the 8-mer scores because they are based on a smaller number of probes and the threshold for statistical significance has not been explored as it has been for 8-mers; nonetheless, we observed that the intended 9-mer ranked very highly (above the 99.9th percentile, or top 131, of all 9-mers, on both arrays) in most cases (13/20, including positive controls). For example, for all three of the positive control proteins (ZFA15, ZFA45 and ZFA93), the intended target is within the top 12 most highly ranked 9-mers for both array types (Figure 2). Among the 17 ZFAs that failed for Ramirez et al. but succeeded in the PBM assays, six of them (ZFA1, 5, 8, 10, 24 and 152) recognized the intended sequence with similar precision (within the top 12) (Figure 2), while others appear to prefer many other sequences more highly than the intended 9-mer target. For five ZFAs (4, 7, 57, 75 and 188), the intended 9-mer target did not appear among the top 100 9-mers on either array (Figure 2).
Figure 2.
Sequence specificities of ZFAs constructed by modular assembly, as determined by PBM. ID for ZFA and results of assay for activity follow Ramirez et al. F1, F2 and F3 columns indicate the module numbers used for construction of the ZFA. The rank of the intended 9-mer target (out of all 131 072 possible 9-mers) is determined by E-score; ME and HK refer to the two array designs used. The last column shows the intended target (based on the modules used for assembly) compared to PBM results (the sequence motif shown is generated from the (up to) top 10 8-mers bound by the ZFA, as described in the main text).
We also created motifs by aligning the 10 8-mers with the highest E-scores (or fewer than 10, since we only included 8-mers with E-scores at or above 0.45; we used 8-mers in order to take advantage of the E-score cutoff) (Figure 2; the Document of the Supplementary Data gives the full alignments). Consistent with the results of the 9-mer analysis above, this procedure produced motifs resembling the intended targets for all three of the positive control ZFAs, and also for most of the ZFAs that failed in Ramirez et al. Indeed, the motifs produced could be easily aligned to the intended 9-mer target in all but one case (ZFA188, which we re-sequenced and re-analyzed twice, and obtained essentially identical results). However, it is also evident that there are many cases in which individual C2H2-ZF modules do not behave precisely as intended, including examples of degeneracy or even unanticipated specificity. This is true even for the positive controls, e.g. F1 of ZFA15, F2 and F3 of ZFA45 and F1 of ZFA93 all display nearly complete degeneracy for at least one base position.
Most C2H2-ZF modules display degeneracy
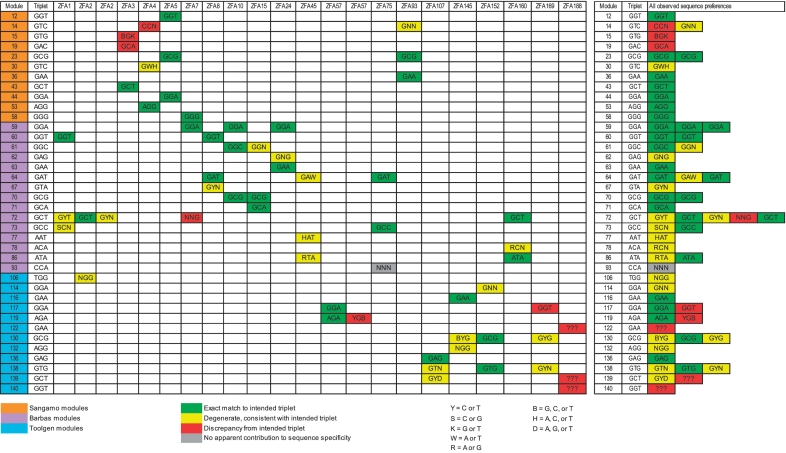
We next asked whether individual modules appeared to bind their intended 3-bp subsite. We manually surmised the apparent specificity of the module in each instance that it was present in a ZFA using the (up to) top 10 DNA 8-mers and 9-mers that the ZFA preferred, aligned to the binding sequence in a way similar to that shown in Figure 2 (full tables of aligned 8-mers and 9-mers and derived motifs are given in Supplementary Document of the Supplementary Data). A summary of this analysis is shown in Figure 3. All 38 C2H2-ZF modules present in at least one successful ZFA are listed, along with their intended target subsite in each of the 20 successful ZFAs. Their apparent specificities are colored according to how closely they resemble the intended target, with green indicating complete agreement, yellow indicating degeneracy (but encompassing the intended target), red indicating disagreement and gray indicating no apparent contribution to sequence specificity despite being present in a successful ZFA.
Figure 3.
C2H2-ZF modules in functional ZFAs typically retain at least degenerate DNA-binding specificity in different contexts. The first row lists ZFAs yielding positive results on PBM, the first column lists the modules used to construct the ZFAs, and the second column indicates their three-base specificity. The remaining cells indicate which modules were used in the array, and what the specificity appeared to be based on the top ten most preferred 8-mers and 9-mers on the PBMs. Colors indicate whether the assayed specificity of the modules in the zinc-finger array was in agreement with its intended target specificity, as described in the main text. To the right is a condensed version of the same data. The source of the modules is also indicated.
This analysis indicates that the majority of the modules do recognize either the intended triplet or a degenerate version, when embedded in a successful ZFA (Figure 3). However, it also underscores the importance of context: of the 15 C2H2-ZF modules that are present in more than one successful ZFA, only four appear to have precisely the same sequence specificity in all contexts. An additional six display different levels of degeneracy in different contexts, while the remaining five appear to specify at least one base differently in different contexts. Nonetheless, degeneracy is most frequently consistent with flexibility of the intended triplet: yellow (degeneracy; 20 instances) is more common than red (disagreement; nine instances) or gray (no contribution; 1 instance) in Figure 3. It is also possible that some of the modules simply have poor intrinsic specificity.
Degeneracy in binding specificities of both artificial ZFAs constructed by modular assembly and natural ZFAs
Degeneracy and context dependence do not seem to be incompatible with success of ZFAs in either our assay or others: as noted above, all three positive controls (i.e. those which Ramirez et al. also scored as successful) displayed some level of degeneracy (Figure 3) (additional examples in the literature are noted in the ‘Introduction’ section). ZFA45 in particular, which is one of the positive controls, displayed degeneracy at all three positions and two of its three constituent modules displayed higher specificity in other contexts (Figure 3). Human C2H2-ZF modules (‘Toolgen’ modules in Figure 3) appear to be particularly prone to degeneracy and context dependence, despite having the highest success rate at producing ZFAs with sequence specificity. These observations are of interest because it is believed that it is desirable that engineered ZFAs are as specific as possible (34).
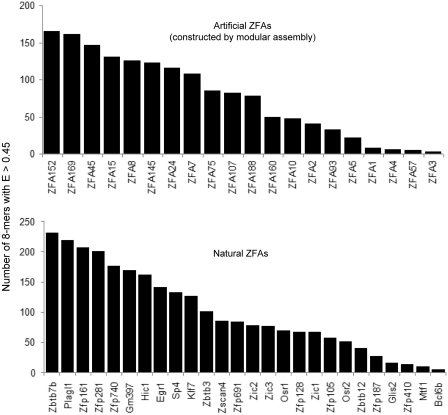
To ask whether degeneracy is a general feature of ZFAs, we again took advantage of the fact that the PBM assay yields the number of 8-mers that are significantly preferred by a given protein, because all 8-mers scoring with E ≥ 0.45 can be considered as significantly preferred (47). Using this criterion, we previously found that human transcription factor DNA-binding domains typically have dozens to hundreds of preferred 8-mers (36). This number is presumably a property of both the width of the binding site, and the tolerance for variation at individual bases. Atf4, for example, has a very specific 8-base binding site, and yields only a single 8-mer with E ≥ 0.45 (TGACGTCA) (I. Mann and T.R. Hughes, unpublished data).
The goal of engineered ZFAs is typically to achieve preference to a single 9-base sequence, which we reason would correspond to two or fewer highly preferred 8-base sequences. However, the ZFAs we analyzed typically yielded dozens of 8-mers with E ≥ 0.45 (Figure 4, top). This number is comparable to what we previously observed with natural human ZFAs (Figure 4, bottom). Thus, both natural ZFAs and artificial ZFAs created by modular assembly display a level of degenerate binding that is comparable to other types of eukaryotic transcription factors.
Figure 4.
Comparison of the degeneracy of binding sites for artificial ZFAs constructed by modular assembly and natural ZFAs. Shown are the number of 8-mers with E > 0.45 (average for two array designs) for this study (top) and Badis et al. (36), which examined mouse transcription factors (bottom).
GNN C2H2-ZF modules have the highest success rate
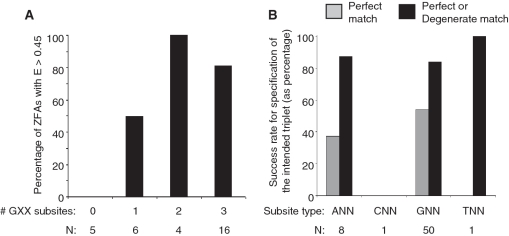
Finally, we re-examined the conclusion of Ramirez et al. that GNN C2H2-ZF modules account for most of the success of engineered ZFAs. Indeed, consistent with the findings of Ramirez et al., we observed that the success of ZFAs in PBMs is lowest for those that lack GNN modules (Figure 5A). Our success rates are notably higher than those of Ramirez et al., particularly for those with two GNN subsites, where we obtained 100% success. The specificity of individual modules within the 20 successful ZFAs is also highest for GNN subsites (Figure 5B), which specified an exact match to the intended triplet (i.e. no degeneracy) in 27 of 50 instances. Most of the eight ANN modules present in successful ZFAs also specified either an exact (three cases) or degenerate (four cases) match to the intended triplet. In contrast, the one CNN module present in a successful ZFA made no apparent contribution to sequence specificity. The one TNN module present in a successful ZFA did contribute to sequence specificity, but specified NGG instead of TGG.
Figure 5.
Success rates of GNN and other C2H2-ZF module subtypes. (A) Proportion and number of ZFAs considered successful by PBM, as a function of the number of GNN modules. (B) Proportion and number of ZFAs of different XNN subtypes with the indicated outcome, considering only ZFAs that were successful by PBM. The number indicated reflects the proportion of times the module satisfies the condition indicated, i.e. gray bars indicate the proportion of instances in which the module gives a perfect match; black bars indicate the proportion of instances that are either a perfect or degenerate match.
DISCUSSION
Our analysis shows that modular assembly of C2H2-ZFs into ZFAs does not result in overwhelming failure with respect to obtaining proteins that bind DNA in a sequence-specific manner. The poor behavior of non-GNN modules (especially CNN and TNN modules), which may be explained by reasons outlined in the Introduction, does appear to account for many if not most of the failures in the PBM assay. Since most of the currently available CNN and TNN modules are derived from C2H2-ZFs that prefer GNN (or GNN-G), it is possible that the low success rates obtained with them is a property of the modules, rather than a property of the modular assembly procedure.
We propose several possible explanations for the apparent discrepancy between our conclusions and those of Ramirez et al. The most obvious is that the PBM assay can detect binding to sequences that are different from the intended targets, whereas all of the assays in Ramirez et al. tested only a single intended target sequence. However, when we specifically asked whether the intended target 9-mer is highly preferred in the PBM assay, we found that it was often very highly ranked. Deviation in the actual versus intended sequence specificity can only explain approximately 1/3 of all cases where we scored a success and Ramirez et al. did not.
A second possible explanation is that the sensitivity of the PBM assay may be higher than that of other assays. B2H fold activation scales roughly with affinity of the ZFA, with a threshold of ∼100 nM (35). In the PBM assay, the protein concentration is typically ∼100 nM before washing, but the microarray probes have a very high local concentration at the surface of the array, which may facilitate re-binding. The PBM assay also does not require high specificity to a single 9-mer sequence; in previous analyses we and others have used PBMs to determine sequence preferences of proteins that bind well to many 8-mers [e.g. (36)]. Cornu et al. (34) found for several ZFAs that sequence specificity is important for ZFN function. However, in our analysis, positive controls selected from Ramirez et al. appeared to possess at least some degeneracy in their binding specificity, indicating that the B2H assay is compatible with some degenerate binding.
A third possibility is that multiple parameters determine success of ZFAs in the assays used by Ramirez et al. (and success as ZFNs), and that there is not a direct linear mapping between any single property of the protein (including its sequence specificity) and its performance in these assays. Properties of proteins that determine success in in vivo assays with heterologous fusion constructs could conceivably include expression level and solubility, as well as unanticipated protein–protein and protein–RNA interactions, both of which C2H2-ZFs can mediate (52). In addition, DNA sequence specificity itself can be defined and described in different ways, including relative preference for target versus random sequence, and tolerance to degeneracy in the target sequence. Consistent with a relatively poor relationship between sequence specificity in vitro and nuclease targeting capacity in vivo, Kim et al. (51) recently reported that 44% of ZFN pairs displayed restriction activity in vitro, but only 7% (23/315) yielded activity in a cell culture assay.
An additional consideration underscored by our study is that the expectation that an artificial ZFA created by modular assembly will generally have exclusive specificity for a single 9-mer may be unrealistic. High specificity of ZFNs is believed to be desirable (34), but it is in fact typical for C2H2-ZFs found in nature to prefer a set of variants of a sequence motif [e.g. (36)]. This property (degeneracy) is apparently shared by artificial ZFAs created by modular assembly. To our knowledge, the individual C2H2-ZF modules used here have not been previously characterized for their relative preference to all possible 3-mers in multiple contexts, and rules dictating the effects of interactions among adjacent C2H2-ZF modules are poorly understood at best. Therefore, it is difficult to say what should have been anticipated from our experiments. On the basis of our results, however, it appears that extremely high specificity may not be a general property of the C2H2-ZF domain. Indeed, such strong sequence specificity is not a feature of most eukaryotic TFs (36,48), and the regulatory and evolutionary strategies of metazoan genomes may even rely on flexible assemblies of relatively promiscuous binding factors (53,54).
The fact that modular assembly of ZFAs is successful in the majority of cases in our analysis, and using our success criteria—notwithstanding CNN and TNN modules, which for reasons already outlined deserve further examination—also supports the potential for C2H2-ZF modular assembly as an evolutionary mechanism (14). We further propose that the typically degenerate sequence specificity of individual C2H2-ZFs, and their frequent context dependency within ZFAs, may represent a beneficial evolutionary property. We note that this feature of ZFAs is not inconsistent with the general concept of modularity, as discussed in the Introduction. In any case, in 19 of the 20 successful ZFAs in our analysis, it is easy to manually align the high-scoring 8-mers and 9-mers (and the resulting motifs) to the intended 9-mer target, and most of the modules do behave approximately as intended (i.e. most are colored green or yellow in Figure 3).
Our findings also highlight the importance of characterizing or predicting the sequence preferences of individual C2H2-ZFs, and using them to infer the binding sites of artificial and natural ZFAs (15–17), which would be less relevant (or at least more complicated) if the assumption of modularity were generally untrue. Ultimately, efforts to understand and predict the sequence specificities of ZFAs with high accuracy will require a more complete characterization of individual C2H2-ZFs, including their sequence preferences outside the canonical triplet, as well as a better grasp of the influence of inter-finger interactions. Nonetheless, despite the degeneracy of most C2H2-ZF DNA-binding activities, and the influence of context, the intended 9-mer target typically ranks very highly in the PBM data, and other high-scoring sequences usually bear an obvious relationship to the intended 9-mer. A simple table of the most preferred triplet for all individual natural ZFs would thus be extremely useful even if degeneracy and context were ignored.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Canadian Institutes of Health Research Operating Grant (MOP-77721 to T.R.H.); National Science and Engineering Research Council CGS-M award (to K.N.L.); Canadian Institutes of Health Research post-doctoral fellowship (to H.v.B.). Funding for open access charge: Canadian Institutes of Health Research Operating Grant (MOP-77721 to T.R.H.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Hilal Kazan, Quaid Morris, Mike Eisen and Julian Mintseris for assistance with microarray designs.
REFERENCES
- 1.Messina DN, Glasscock J, Gish W, Lovett M. An ORFeome-based analysis of human transcription factor genes and the construction of a microarray to interrogate their expression. Genome Res. 2004;14:2041–2047. doi: 10.1101/gr.2584104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:e1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huntley S, Baggott DM, Hamilton AT, Tran-Gyamfi M, Yang S, Kim J, Gordon L, Branscomb E, Stubbs L. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–677. doi: 10.1101/gr.4842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fulton DL, Sundararajan S, Badis G, Hughes TR, Wasserman WW, Roach JC, Sladek R. TFCat: the curated catalog of mouse and human transcription factors. Genome Biol. 2009;10:R29. doi: 10.1186/gb-2009-10-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tupler R, Perini G, Green MR. Expressing the human genome. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- 6.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 7.Shannon M, Hamilton AT, Gordon L, Branscomb E, Stubbs L. Differential expansion of zinc-finger transcription factor loci in homologous human and mouse gene clusters. Genome Res. 2003;13:1097–1110. doi: 10.1101/gr.963903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamilton AT, Huntley S, Tran-Gyamfi M, Baggott DM, Gordon L, Stubbs L. Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome Res. 2006;16:584–594. doi: 10.1101/gr.4843906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, et al. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat. Biotechnol. 2003;21:275–280. doi: 10.1038/nbt796. [DOI] [PubMed] [Google Scholar]
- 10.Choo Y, Sanchez-Garcia I, Klug A. In vivo repression by a site-specific DNA-binding protein designed against an oncogenic sequence. Nature. 1994;372:642–645. doi: 10.1038/372642a0. [DOI] [PubMed] [Google Scholar]
- 11.Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 12.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 13.Remy S, Tesson L, Menoret S, Usal C, Scharenberg AM, Anegon I. Zinc-finger nucleases: a powerful tool for genetic engineering of animals. Transgenic Res. 2010;19:363–371. doi: 10.1007/s11248-009-9323-7. [DOI] [PubMed] [Google Scholar]
- 14.Meng X, Thibodeau-Beganny S, Jiang T, Joung JK, Wolfe SA. Profiling the DNA-binding specificities of engineered Cys2His2 zinc finger domains using a rapid cell-based method. Nucleic Acids Res. 2007;35:e81. doi: 10.1093/nar/gkm385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Stormo GD. Context-dependent DNA recognition code for C2H2 zinc-finger transcription factors. Bioinformatics. 2008;24:1850–1857. doi: 10.1093/bioinformatics/btn331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan T, Friedman N, Margalit H. Ab initio prediction of transcription factor targets using structural knowledge. PLoS Comput. Biol. 2005;1:e1. doi: 10.1371/journal.pcbi.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Persikov AV, Osada R, Singh M. Predicting DNA recognition by Cys2His2 zinc finger proteins. Bioinformatics. 2009;25:22–29. doi: 10.1093/bioinformatics/btn580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segal DJ, Beerli RR, Blancafort P, Dreier B, Effertz K, Huber A, Koksch B, Lund CV, Magnenat L, Valente D, et al. Evaluation of a modular strategy for the construction of novel polydactyl zinc finger DNA-binding proteins. Biochemistry. 2003;42:2137–2148. doi: 10.1021/bi026806o. [DOI] [PubMed] [Google Scholar]
- 19.Choo Y, Klug A. Selection of DNA binding sites for zinc fingers using rationally randomized DNA reveals coded interactions. Proc. Natl Acad. Sci. USA. 1994;91:11168–11172. doi: 10.1073/pnas.91.23.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isalan M, Choo Y, Klug A. Synergy between adjacent zinc fingers in sequence-specific DNA recognition. Proc. Natl Acad. Sci. USA. 1997;94:5617–5621. doi: 10.1073/pnas.94.11.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl Acad. Sci. USA. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 23.Fairall L, Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition. Nature. 1993;366:483–487. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]
- 24.Wolfe SA, Grant RA, Elrod-Erickson M, Pabo CO. Beyond the "recognition code": structures of two Cys2His2 zinc finger/TATA box complexes. Structure. 2001;9:717–723. doi: 10.1016/s0969-2126(01)00632-3. [DOI] [PubMed] [Google Scholar]
- 25.Kim JS, Pabo CO. Getting a handhold on DNA: design of poly-zinc finger proteins with femtomolar dissociation constants. Proc. Natl Acad. Sci. USA. 1998;95:2812–2817. doi: 10.1073/pnas.95.6.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright DA, Thibodeau-Beganny S, Sander JD, Winfrey RJ, Hirsh AS, Eichtinger M, Fu F, Porteus MH, Dobbs D, Voytas DF, et al. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat. Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- 28.Mandell JG, Barbas CF., 3rd Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Q, Xia Z, Zhong X, Case CC. Validated zinc finger protein designs for all 16 GNN DNA triplet targets. J. Biol. Chem. 2002;277:3850–3856. doi: 10.1074/jbc.M110669200. [DOI] [PubMed] [Google Scholar]
- 30.Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF., 3rd Development of zinc finger domains for recognition of the 5'-ANN-3' family of DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 31.Dreier B, Fuller RP, Segal DJ, Lund CV, Blancafort P, Huber A, Koksch B, Barbas CF., 3rd Development of zinc finger domains for recognition of the 5'-CNN-3' family DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 32.Segal DJ, Dreier B, Beerli RR, Barbas CF., III Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5'-GNN-3' DNA target sequences. Proc. Natl Acad. Sci. USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi Y, Berg JM. A direct comparison of the properties of natural and designed zinc-finger proteins. Chem. Biol. 1995;2:83–89. doi: 10.1016/1074-5521(95)90280-5. [DOI] [PubMed] [Google Scholar]
- 34.Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung JK, Cathomen T. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol. Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 35.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. An affinity-based scoring scheme for predicting DNA-binding activities of modularly assembled zinc-finger proteins. Nucleic Acids Res. 2009;37:506–515. doi: 10.1093/nar/gkn962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459:437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- 40.Bulyk ML, Huang X, Choo Y, Church GM. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc. Natl Acad. Sci. USA. 2001;98:7158–7163. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mintseris J, Eisen MB. Design of a combinatorial DNA microarray for protein-DNA interaction studies. BMC Bioinformatics. 2006;7:429. doi: 10.1186/1471-2105-7-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJ. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Philippakis AA, Qureshi AM, Berger MF, Bulyk ML. Design of compact, universal DNA microarrays for protein binding microarray experiments. J. Comput. Biol. 2008;15:655–665. doi: 10.1089/cmb.2007.0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berger MF, Bulyk ML. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat. Protoc. 2009;4:393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iuchi S. Three classes of C2H2 zinc finger proteins. Cell. Mol. Life Sci. 2001;58:625–635. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wunderlich Z, Mirny LA. Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet. 2009;25:434–440. doi: 10.1016/j.tig.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weirauch MT, Hughes TR. Conserved expression without conserved regulatory sequence: the more things change, the more they stay the same. Trends Genet. 2010;26:66–74. doi: 10.1016/j.tig.2009.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.