Abstract
Retinoic acid-related orphan receptors (RORs) and the basic helix–loop–helix-PAS transcription factor Npas2 have been implicated in the control of circadian rhythm. In this study, we demonstrate that RORγ directly regulates Npas2 expression in vivo. Although the rhythmicity of Npas2 mRNA expression was maintained in RORγ−/− mice, the peak level of expression was significantly reduced in several tissues, while loss of RORα had little effect. Inversely, overexpression of RORγ in hepatoma Hepa1-6 cells greatly induced the expression of Npas2. RORγ-activated Npas2 transcription directly by binding two ROREs in its proximal promoter. ChIP analysis demonstrated that RORγ was recruited to this promoter in the liver of wild-type mice, but not RORγ-deficient mice. Activation of Npas2 correlated positively with chromatin accessibility and level of H3K9 acetylation. The activation of Npas2 by RORγ was repressed by co-expression with Rev-Erbα or addition of the ROR inverse agonist T0901317. Npas2 expression was also significantly enhanced during brown adipose differentiation and that this induction was greatly suppressed in adipose cells lacking RORγ. Our results indicate that RORγ and Rev-Erbα are part of a feed-back loop that regulates the circadian expression of Npas2 suggesting a regulatory role for these receptors in Npas2-dependent physiological processes.
INTRODUCTION
The retinoic acid-related orphan receptor (ROR) subgroup of nuclear receptors consists of three members, RORα, -β and -γ (NR1F1-3 or RORA-C) (1). As a result of alternative promoter usage and/or exon splicing each ROR gene generates several isoforms that differ only in their amino terminus. These variants exhibit a distinct pattern of tissue-specific expression and are involved in the regulation of different physiological processes and target genes. The RORs bind as a monomer to ROR response elements (ROREs) in the promoter regulatory region of target genes (2,3). Transcriptional regulation by RORs is mediated by the recruitment of co-repressor and co-activator complexes which, through their histone(de)acetylase activities, induce changes in chromatin conformation (1,4). Recent studies have demonstrated that RORs function as ligand-dependent transcription factors. All-trans retinoic acid and several other retinoids act as partial antagonists of RORβ and T0901317 and 7α-hydroxycholesterol as inverse agonists of RORα and RORγ, while cholesterol sulfate has been reported to function as an RORα agonist (5–8).
RORs regulate several important physiological processes and have been implicated in a number of pathologies (1). RORα is critical for cerebellar development and bone formation (9,10), while RORβ regulates functions in the brain and retina (11,12). RORγ plays a key role in lymph node development and thymopoiesis (13,14). Furthermore, both RORα and RORγ are involved in regulating various metabolic pathways, inflammatory responses and immune functions, including Th17 cell differentiation (1,15–19).
In addition to these functions, all three RORs have been implicated in the regulation of various aspects of circadian rhythm (1,11,20–27). Circadian rhythm is fundamental in the regulation of a wide variety of physiological and behavioral activities. In mammals, the central clock in the suprachiasmatic nucleus (SCN) integrates light–dark cycle input and synchronizes the autonomous oscillators in peripheral tissues (28–30). The molecular basis of the circadian clock involves interlocking transcriptional/translational feedback loops that regulate the rhythmic expression and activity of a set of core clock genes, including the basic helix–loop–helix/Per-Arnt-Sim (PAS) domain-type transcriptional activators brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like (Bmal1), circadian locomotor output cycles kaput (Clock), two cryptochrome (Cry) and three period (Per) proteins. The positive loop consists of the basic helix–loop–helix/PAS-type transcription activators Bmal1 and Clock, while Cry and Per are involved in the negative control of the oscillator (31–36). The repression of Bmal1 is mediated by the nuclear orphan receptors Rev-ErbAα and Rev-Erbβ, which function as transcriptional repressors, while RORs have been implicated in the positive regulation of several clock genes, including Bmal1 and Rev-Erbα (1,22–24,26,37,38). The circadian clock subsequently regulates the rhythmic expression of downstream genes involved in timing the rhythmic regulation of numerous functions, including behavior, reproductive and neuroendocrine functions, and metabolism.
In this study, we identify neuronal PAS domain protein 2 (Npas2), a paralogs of Clock, as a new ROR target gene. Npas2 heterodimerizes with Bmal1 and regulates the transcription of circadian genes (39–41). Npas2 and Clock have been reported to exhibit overlapping functions. Single nucleotide polymorphisms (SNPs) in Npas2 have been linked to increased risk of cancer, unipolar major depression, metabolic syndrome and hypertension (42–47). In this study, we demonstrate that the loss of particularly RORγ significantly reduced the peak level of Npas2 mRNA expression in several tissues without affecting the rhythmicity of Npas2 expression. We further show that RORγ regulates Npas2 transcription directly through two ROREs in its proximal promoter. This activation is inhibited by the inverse RORγ agonist T0901317 and by Rev-Erbα. Collectively, these data indicate that both RORγ and Rev-Erbα are part of the mechanism that regulates the oscillatory expression of Npas2 and suggest a regulatory role for these receptors in Npas2-dependent physiological processes and pathologies, including circadian behavior disorders, metabolic syndrome and tumorigenesis.
MATERIALS AND METHODS
Experimental animals
Heterozygous C57BL/6 staggerer (RORα+/sg) mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA). The generation and initial characterization of C57BL/6 RORγ−/− mice were described earlier (13). Mice were supplied ad libitum with NIH-A31 formula and water as described earlier (16) and maintained at 25°C on a constant 12 h light:12 h dark cycle. Littermate wild-type (WT) mice were used as controls for both ROR-deficient models. For cold-induced thermogenesis mice were placed at 4°C for 4 h. All animal protocols followed the guidelines outlined by the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the NIEHS.
RNA isolation
To study gene expression during circadian time, tissues were excised from WT, RORγ−/−, and RORαsg/sg mice every 4 h over a period of 24 h, processed overnight in RNAlater® solution (Ambion, Austin, TX, USA) at 4°C, and then stored at −80°C until use. Tissues were then homogenized in RLT buffer (Qiagen, Valencia, CA, USA) with a Polytron PT-3000 (Brinkmann Instruments, Westbury, NY, USA). RNA was then extracted using a QIAshredder column and RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. Hepa1-6 cells and brown adipocytes were lysed in lysis (RLT) buffer and RNA extracted as described for tissues. The brown adipocytes were treated for 24 h (from differentiation Day 4 to 5) with and without 10 µM of the inverse ROR agonist T0901317 (7) (Sigma-Aldrich, St Louis, MO, USA) before RNA extraction.
Quantitative reverse transcription-polymerase chain reaction (QRT-PCR) analysis
QRT-PCR analysis was performed using SYBR Green I or TaqMan system (Applied Biosystems, Foster City, CA, USA) to quantify gene expression. All primer and probe sequences are listed in Supplementary Table S1. The RNA was reverse-transcribed using a High-Capacity cDNA Archive Kit (Applied Biosystems). The reactions were carried out in triplicate using 20 ng of cDNA and the following conditions: 2 min at 45°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. All the results were normalized by the amount of 18S rRNA or Gapdh mRNA.
Isolation of brown preadipocytes
Brown fat preadipocytes were isolated from postnatal day 2 (PND2) WT and RORγ−/− brown adipose tissue by collagenase digestion as described earlier (48). Preadipocytes were immortalized by infection with pBabe retrovirus encoding SV40-Large T antigen. After selection with puromycin (2 mg/ml), resistant clones were isolated. Several clones of the each genotype, WT, RORγ+/− and RORγ−/− were obtained. Cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS), 1 nM thyroid hormone (T3) and 20 nM insulin (differentiation medium). Differentiation in confluent cultures was induced by the addition of 1 µM dexamethasone, 0.125 mM indomethacin and 0.5 mM isobutylmethylxanthine. After 2 days incubation, the medium was changed to the normal differentiation medium and replaced every other day as described earlier (49).
Hepa1-6 and brown preadipocyte stable cell lines
Hepa1-6 cell lines stably expressing Flag-RORα4, Flag-RORγ1 and Flag-Rev-Erbα were generated after transfection with the respective pLXIN plasmid DNA and subsequent selection in G418 (Clontech, Palo Alto, CA, USA). In the remaining of the article, RORα4 and RORγ1 are commonly referred to as RORα and RORγ. Brown adipocyte cell lines BAT(E), BAT(RORγ) and BAT(RORγE502Q) stably expressing empty vector, RORγ or the RORγE502Q mutant containing an inactive activation domain, respectively, were generated in the same way from immortalized preadipocytes. The expression and nuclear localization of RORγ protein were confirmed by western blot analysis and immunochemical staining, respectively. Cell lines established from five individual clones were examined in QRT-PCR and chromatin immunoprecipitation (ChIP) analysis.
ChIP assay
ChIP assay was performed using a ChIP assay kit from Millipore (Billerica, MA, USA) according to the manufacturer’s protocol with a few minor modifications. Briefly, the livers isolated from WT, RORαsg/sg and RORγ−/− mice were homogenized with a polytron PT 3000 (Brinkmann Instruments) and cross-linked by 1% formaldehyde for 20 min at room temperature. After a wash in PBS, an aliquot of the cross-linked chromatin was sonicated and incubated overnight with anti-RORα or anti-RORγ antibody (sc-6062 sc-28559, respectively; Santa Cruz Biotechnology, Santa Cruz, CA, USA). To analyze histone H3 lysine 9 (H3K9) acetylation within the Npas2 promoter region, ChIP assays were performed with an anti-H3K9Ac antibody (ab4441; Abcam, Cambridge, MA, USA). After incubation with protein G agarose beads for 2 h, and several washes, DNA–protein complexes were eluted. The cross-links were reversed by overnight incubation at 65°C in the presence of 25 mM NaCl and subsequent digestion with RNase A and proteinase K, followed by purification of the chiped-DNA. The fold amount of the chiped-DNA relative to each input DNA was determined by QPCR. All QPCR reactions were carried out in triplicate. The procedure for ChIP analysis using Hepa1-6 cells or brown adipocytes was similar as described for tissues except that 2 × 106 cells were cross-linked with 4% formaldehyde for 10 min and the immunoprecipitation was performed with anti-Flag M2 affinity gel (Sigma-Aldrich). The sequences of the primers used for ChIP-QPCR are listed in Supplementary Table S1.
Formaldehyde-assisted isolation of regulatory elements
Formaldehyde-assisted isolation of regulatory elements (FAIRE) was performed as described earlier (50). Briefly, mouse livers were cross-linked and sonicated as described for the ChIP assay. Samples were centrifuged and the DNA in the supernatants isolated by three consecutive extractions with phenol–chloroform–isoamyl alcohol (25:24:1). After the final extraction, the FAIRE samples were reverse-cross-linked as described for the ChIP assay. The enrichment of fragmented genomic DNA in the FAIRE samples relative to each input DNA was measured in triplicate by QPCR.
Reporter gene assay
To generate pGL4.10-Npas2(–1534/+81), the region of −1534/+81 bp of the murine Npas2 promoter was cloned into the promoter-less reporter plasmid pGL4.10 (Promega, Madison, WI, USA). The mutant pGL4.10-Npas2(–1534/+81) plasmids, in which one of the two ROREs or both ROREs were mutated from AGGTCA to AAATCA, were generated using a Quickchange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). The sequences were verified by DNA sequencing. Huh-7 and HEK293T cells were co-transfected with pGL4.10-Npas2(–1534/+81), pGL4.10, or pCMVβ-Gal reporter plasmids and p3xFlag-CMV10-RORγ, p3xFlag-CMV10-RORα or p3xFlag-CMV10-Rev-Erbα expression plasmids as indicated using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After 24 h incubation, the luciferase and β-galactosidase activities were measured by Luciferase Assay Substrate (Promega) and Luminescent β-galactosidase Detection Kit II (Clontech). All transfections were performed in triplicate, and each experiment was repeated at least twice.
RESULTS
Regulation of circadian expression of Npas2 by RORs
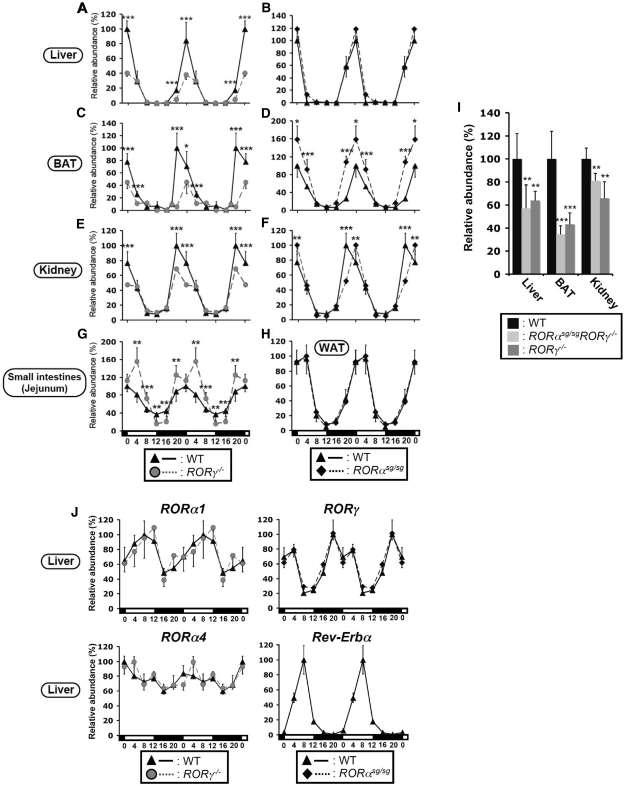
Several studies have provided evidence for a role of RORs in the regulation of several circadian clock genes (1,11,20,23,24,26,27,38). In this study, we focused on the regulation of Npas2 by RORα and RORγ. First, we analyzed the effect of the loss of RORα or RORγ expression on the circadian expression of Npas2 in the liver, brown adipose tissue (BAT), kidney and small intestines using RORαsg/sg and RORγ−/−mice. Consistent with a previous report, Npas2 mRNA showed a strong oscillatory pattern of expression in the liver of WT mice with peak expression between CT20 and CT0 and a low point at CT12 (Figure 1A and B) (51). However, the oscillatory expression of Npas2 was significantly altered in RORγ−/− liver. At CT0, when Npas2 is highly expressed in WT liver, expression of Npas2 mRNA was reduced by about 60% in the liver of RORγ−/− mice (Figure 1A). Npas2 expression exhibited a similar robust oscillatory pattern in BAT, kidney, small intestines and white adipose tissue (WAT) as in WT liver (Figure 1C–H). However, between CT20 and CT4 Npas2 expression was greatly decreased in BAT of RORγ−/− mice (Figure 1C), reduced modestly in RORγ−/− kidney (Figure 1E) and was slightly enhanced in the small intestines (Figure 1G). Little change was observed in the pattern of Npas2 expression in the liver and WAT from RORαsg/sg mice (Figure 1B and H), while a small shift in peak Npas2 expression was seen in RORαsg/sg kidney (Figure 1F) and an increase in RORαsg/sg BAT. RORα is not expressed in small intestines.
Figure 1.
Comparison of the circadian expression of Npas2 in several peripheral tissues from WT, RORγ-/– and RORαsg/sg mice. (A–H) Various tissues from WT, RORγ−/− and RORαsg/sg mice (n = 4) were isolated every 4 h over a period of 24 h and expression of Npas2 was analyzed by QRT-PCR. The 24-h expression pattern was double plotted. (I) Tissues were isolated from RORγ−/−, RORαsg/sgRORγ−/− DKO mice (n = 4) at CT20. Npas2 expression in DKO and RORγ−/− mice was normalized to the expression in littermate WT mice. (J) Circadian pattern of RORγ1, RORα1 and RORα4 mRNA expression in liver from WT, RORγ−/− and RORαsg/sg mice and Rev-Erbα expression in WT mice (n = 4). Data present mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA.
Earlier studies identified a certain degree of redundancy between the regulation of gene expression by RORα and RORγ (16). To examine whether there was any functional redundancy between RORα and RORγ, Npas2 expression was examined in RORαsg/sgRORγ−/− double knockout (DKO) mice. As shown in Figure 1I, at CT20 Npas2 expression was reduced to a very similar degree in the liver, BAT and kidney of DKO mice as observed in RORγ−/− mice. Together, these results indicate that the circadian expression of Npas2 is largely regulated by RORγ and to a minor extent by RORα and that this regulation exhibits a certain tissue specificity.
To be able to relate the oscillatory pattern of expression of Npas2 in liver to those of RORα and RORγ, we examined their circadian pattern of expression. Figure 1J shows that particularly RORγ1, the only RORγ isoform expressed in liver, showed a robust oscillatory pattern of expression, while RORα1, which is expressed at low levels, has a moderate and RORα4, which in liver is the predominant α isoform, exhibited a weak oscillatory pattern consistent with earlier observations (16,20,52). Loss of RORγ or RORα expression had little effect on the circadian expression of RORα1 or α4 and RORγ1 mRNA, respectively, suggesting that RORα and RORγ are regulated independently from each other (Figure 1J). The phase of the oscillatory expression of RORγ1 in liver (zenith at CT18) was slightly earlier than that of Npas2 (zenith at CT20) and the opposite of that of RORα1 and Rev-Erbα expression (zenith at CT8) (Figure 1J).
Circadian expression of Clock gene in RORαsg/sg and RORγ−/− mice
Npas2 and Clock are paralogues that share several functions (39–41). Each protein can form a heterodimer with Bmal1. Moreover, Npas2 is able to functionally substitute for Clock in the master clock in the SCN and overlapping functions have been reported in the liver. A putative RORE has been identified in the human and mouse promoter region of the Clock gene, while EMSA analysis showed that RORs were able to bind to this RORE (27). To further examine the potential role of RORs in the regulation of Clock, we next determined the effect of the loss of RORα or RORγ on its expression. In the liver, BAT, kidney and intestines Clock exhibited a less-robust oscillatory pattern of expression than Npas2 with maximum expression around CT0 and a similar circadian phase as Npas2 (Supplementary Figure S1). No significant differences in the circadian expression pattern of Clock were observed in the liver, BAT and WAT between WT and RORαsg/sg or RORγ−/− mice (Supplementary Figure S1A–D and H). Interestingly, at CT20 and CT0 Clock expression was greatly downregulated in the kidney of RORαsg/sg mice compared with WT kidneys (Supplementary Figure S1F), while loss of RORγ significantly reduced the amplitude of the oscillatory expression of Clock in kidney (Supplementary Figure S1E). Moreover, Clock expression was significantly reduced (by about 25%) at CT0 and CT4 in the small intestines of RORγ−/− mice compared with WT controls (Supplementary Figure S1G). These results indicate that the regulation of Clock expression by RORs is tissue dependent and less stringent than that of Npas2. Examination of Clock expression in DKO mice showed that in contrast to single KO mice Clock expression was significantly reduced in the liver and BAT of DKO mice (Supplementary Figure S1I).
Activation and in vivo ROR binding to the Npas2 promoter
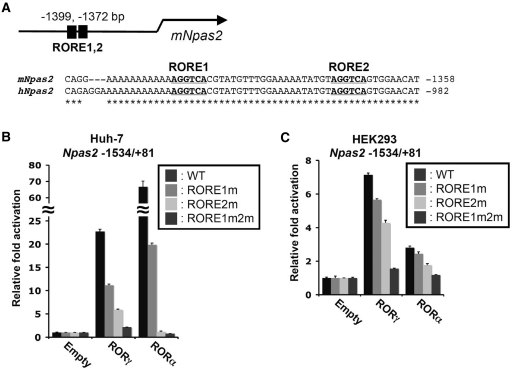
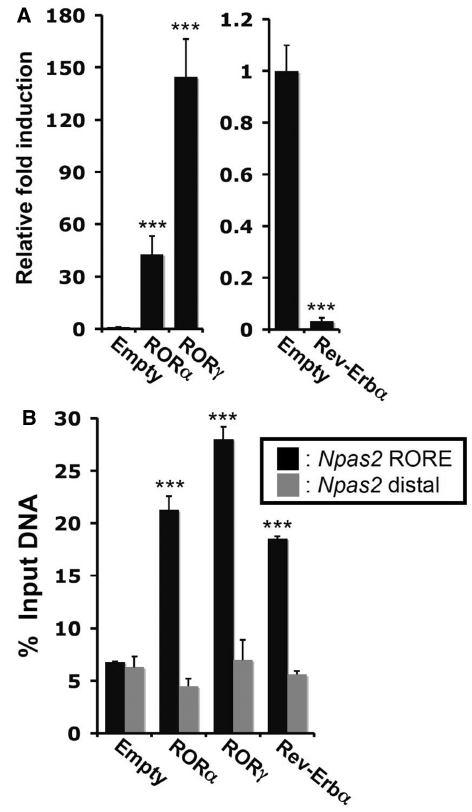
RORs bind as a monomer to ROREs consisting of the consensus sequence AGGTCA preceded by a 6-bp A/T rich region, in the promoter regulatory region of target genes (1–3). The murine Npas2 proximal promoter region contains 2 successive putative ROREs at −1372 and −1399 that are spanned by 21 bp (Figure 2A). These ROREs are conserved between mouse and human Npas2. Although EMSA showed that RORs can bind these ROREs (27), the functionality of these ROREs in the intact promoter and whether RORs are recruited to this Npas2 promoter region in vivo have not been studied. To examine whether RORs were able to activate the Npas2 proximal promoter, reporter gene assays were performed in human hepatoma Huh-7 cells and human kidney epithelial HEK293T cells with a pGL4.10-Npas2(–1534/+81) reporter plasmid in which the Luc reporter was placed under the control of the −1534/+81 proximal promoter region of Npas2. Figure 2B shows that in Huh-7 cells RORγ was able to activate the Npas2 promoter more than 20-fold compared with transfection with the p3xFlag-CMV10 empty vector. Expression of RORα also greatly increased the activation of the Npas2(–1534/+81) promoter. To determine which of the two sites was important in this activation, the effect of AGGTCA to AAATCA mutations in the RORE1 and RORE2 on the activation of the Npas2 promoter was examined. Mutation of RORE1 modestly reduced the activation of the Npas2 promoter by either RORα or RORγ and mutation of the RORE2 decreased the activation more than the RORE1 mutation. Mutation of both ROREs almost completely abolished the activation of the Npas2(–1534/+81) promoter by RORs. Similar results as for Huh-7 were obtained in HEK293T cells (Figure 2C). During submission of this manuscript a different study provided evidence for a role of another RORE-like site (RORE3) at −1207 of the mouse Npas2 promoter in its activation by RORα (53). However, mutation of this RORE within the Npas2(–1534/+81) promoter had little effect on its activation by RORα or RORγ and did not affect its repression by Rev-Erbα, suggesting that this site plays a minor role in the regulation of Npas2 by these receptors (Supplementary Figure S2). Our observations indicate that RORE1 and RORE2, rather than RORE3, contributed to the activation of the Npas2(–1534/+81) promoter by RORα and RORγ, with a more prominent role for RORE2 than RORE1.
Figure 2.
Regulation of Npas2 expression by ROR is mediated through ROREs. (A) Schematic of the location of the RORE1 and RORE2 in the proximal Npas2 promoter region. The bold and underlined sequences of these ROREs are conserved between the human and mouse genomes. The numbers refer to the distance to the transcription start site. (B and C) RORs were able to effectively activate the (–1534/+81) Npas2 proximal promoter in Huh-7 (B) and HEK293T cells (C). Cells were co-transfected with the indicated p3xFlag–CMV10–ROR expression vector, pCMV-β-Gal and a pGL4.10 reporter plasmid driven by either the WT Npas2(–1534/+81) promoter or the promoter in which RORE1, RORE2 or both were mutated (RORE1m, RORE2m and RORE1m2m, respectively). About 24 h later the relative luciferase reporter activities were determined as described in ‘Materials and Methods’ section. Data present mean ± SEM.
Inhibition of Npas2(–1534/+81) promoter activation by Rev-Erbα and T0901317
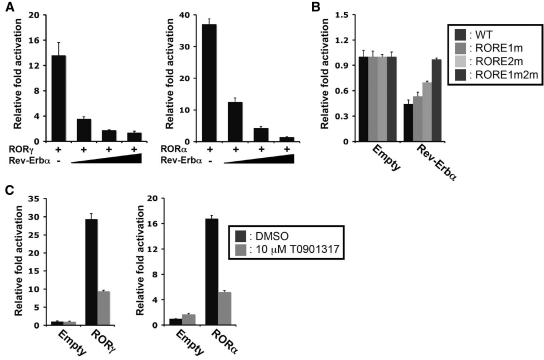
Earlier studies have shown that the nuclear receptor, Rev-Erbα, which also binds ROREs, can compete with RORs for RORE binding (22,23,37,54). Figure 3A shows that co-expression of Rev-Erbα repressed the activation of the Npas2 promoter by either RORα or RORγ in a dose-dependent manner. Moreover, expression of Rev-Erbα in Huh-7 cells repressed the endogenous activation of the Npas2(–1534/+81) promoter (Figure 3B). Mutation of RORE1/2 abolished this repression indicating that this repression was mediated through these ROREs. These observations are consistent with the hypothesis that Rev-Erbα functions as a repressor of Npas2 expression and competes with RORs for RORE binding. Recently, T0901317 was identified as an inverse agonist of RORα and RORγ (7). It was therefore of interest to examine the effect of T0901317 on the expression of Npas2 induction. As shown in Figure 3C, T0901317 was able to repress the activation of the Npas2 promoter by RORγ and RORα in Huh-7 cells.
Figure 3.
Rev-Erbα and the ROR-inverse agonist T0901317 inhibited ROR-induced activation of the Npas2(–1534/+81) promoter. (A) Rev-Erbα expression represses the activation of the Npas2 promoter by RORα and RORγ, Huh-7 cells were transfected with p3xFlag-CMV10-RORγ or p3xFlag-CMV10-RORα, pGL4.10-Npas2(–1534/+81) and increasing concentrations of p3xFlag-CMV10-Rev-Erbα and 24 h later were assayed for reporter activity. (B) The downregulation of basal Npas2 promoter activity by Rev-Erbα was abrogated by mutations in the ROREs. Huh-7 cells were transfected with p3xFlag-CMV10-Rev-Erbα and pGL4.10 driven by the WT Npas2(–1534/+81) promoter or the promoter with the indicated RORE mutations. About 24 h later cells were assayed for reporter activity. (C) The inverse agonist, T0901317, represses the activation of the Npas2 promoter by both RORγ and RORα in Huh-7 cells. Data present mean ± SEM.
Recruitment of RORs to the Npas2 promoter
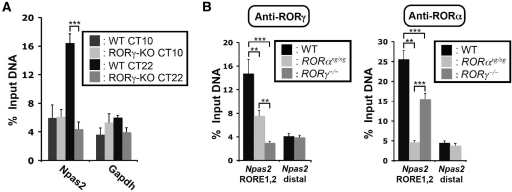
To determine whether RORγ was recruited to the ROREs in the Npas2(–1534/+81) promoter in vivo, ChIP-qPCR analysis was carried out using liver isolated from WT and ROR knockout mice. Tissues were collected at CT22, a time close to the peak expression of RORγ and Npas2 mRNAs, and at CT10, a time at which both RORγ and Npas2 mRNAs are expressed at their lowest level. On this basis, one would expect higher recruitment of RORγ to the Npas2 promoter at CT22 than at CT10. The data in Figure 4A confirmed that RORγ was more efficiently recruited to the −1380 region of the Npas2 promoter in WT liver collected at CT22 compared with the liver collected at CT10. Primers targeting a region within the Gapdh gene were used as negative control and did not show any specific recruitment of RORγ. As expected, at CT22, the level of RORγ associated with the Npas2 promoter in WT liver was significantly higher than in RORγ−/− liver, while RORγ was not recruited to a distal, non RORE-containing region of the Npas2 gene. Although the loss of RORα had only a small effect on Npas2 expression, ChIP analysis indicated that RORα was associated with the Npas2 promoter.
Figure 4.
RORγ and RORα are recruited to the Npas2 promoter. (A) Circadian time-dependent recruitment of RORγ to the Npas2 promoter. ChIP analysis was performed using an anti-RORγ antibody and liver tissues (n = 4) isolated from WT and RORγ−/− mice at CT10 (low expression of RORγ) and CT22 (high expression of RORγ). Amplification of the Gapdh gene was used as a negative control. Data present mean ± SEM, ***P < 0.001 by ANOVA. (B) ChIP analysis was performed with anti-ROR antibodies using liver tissues (n = 4) isolated from WT, RORαsg/sg and RORγ−/− mice at CT22. QPCR amplification of a non RORE-containing distal site of the Npas2 gene was used as a negative control. Data present mean ± SEM, **P < 0.01, ***P < 0.001.
Npas2 activation and repression by RORs and Rev-Erbα, respectively, in Hepa1-6 cells
To obtain further evidence for the involvement of RORα/γ and Rev-Erbα in the regulation of Npas2, we established murine hepatoma Hepa1-6 cell lines stably expressing Flag-tagged RORα, RORγ or Rev-Erbα. Immunocytochemistry using anti-Flag M2 antibody showed that these receptors were expressed in these cells and localized to the nucleus (data not shown). Figure 5A shows that Npas2 expression was dramatically induced in Hepa1-6 cells expressing either RORα or RORγ. In contrast, Rev-Erbα overexpression significantly downregulated the expression of Npas2.
Figure 5.
Overexpression of RORα/γ or Rev-Erbα in Hepa1-6 cells, respectively, induced or repressed Npas2 activation. (A) Npas2 gene expression was examined by QPCR analysis in Hepa1-6 cells (n = 5) stably expressing empty vector, Flag-RORα, Flag-RORγ or Flag-Rev-Erbα. The expression of Npas2 in Hepa1-6(Empty) was normalized to 1. Data present mean ± SEM, ***P < 0.001 by ANOVA. (B) RORs and Rev-Erbα were recruited to the Npas2 promoter in Hepa1-6 cells. ChIP analysis was performed with the Hepa1-6 stable cell lines and anti-Flag M2 antibody. Hepa1-6(Empty) served as a negative control. Data present mean ± SEM, ***P < 0.001.
To analyze the recruitment of RORα, RORγ and Rev-Erbα to the ROREs within the Npas2(–1534/+81) promoter, we performed ChIP assay using the Hepa1-6 stable cell lines and an anti-Flag M2 antibody (Figure 5B). Our data showed that all three receptors were recruited to the −1380 region of the Npas2 promoter, consistent with the conclusion that Npas2 is directly regulated by RORs and Rev-Erbα.
Activation of the Npas2 promoter in relation to changes in DNA accessibility and histone H3K9 acetylation
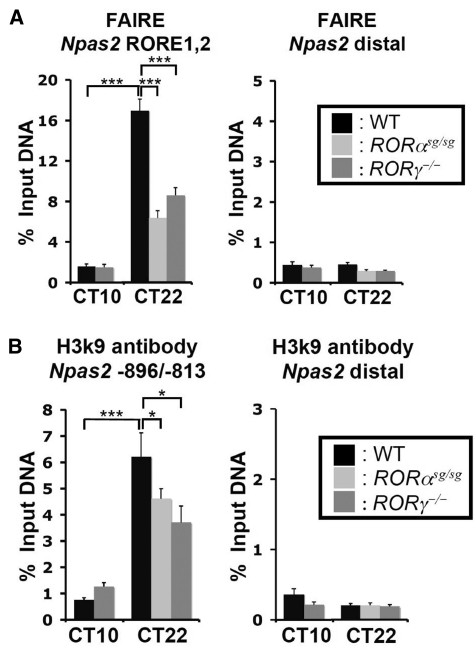
The FAIRE assay has been used to detect active regulatory regions (50) and although the roles of histone acetylation, including H3K9Ac, are far from being understood levels of acetylation correlate positively with actively transcribed genes (55). To study the role of chromatin modulation in Npas2 regulation, we first examined the FAIRE and the H3K9Ac signals within the proximal Npas2 promoter region in DNA samples from liver isolated at CT10 and CT22. As shown in Figure 6, the FAIRE and H3K9Ac signals were markedly increased in DNA samples from liver isolated at CT22, a time at which Npas2 is most highly expressed, compared with that of CT10. These results are consistent with the concept that at CT22 this region of the Npas2 promoter is accessible and associated with functionally active enhancers. Loss of either RORα or RORγ expression led to a reduction in FAIRE signal and H3K9 acetylation at CT22 suggesting that ROR binding promotes open chromatin structure at the site of the Npas2 promoter.
Figure 6.
Activation of the Npas2 promoter in relation to changes in DNA accessibility and histone H3K9 acetylation. (A) Accessibility to Npas2(RORE1,2), as assessed by FAIRE-QPCR analysis, correlates with Npas2 promoter activation. FAIRE-QPCR at Npas2(RORE1,2) and at distal downstream site of the Npas2 gene was performed using DNA samples isolated from liver of WT, RORαsg/sg and RORγ−/− mice at CT10 and CT22. (B) Activation of the Npas2 promoter correlates with H3K9 acetylation. ChIP-QPCR was performed at the proximal Npas2 promoter (Npas2 –896/–813) and the distal site using anti-H3K9Ac antibody and ChIPed-DNA samples isolated from liver of WT, RORαsg/sg and RORγ−/− mice at CT10 and CT22. Data present mean ± SEM, *P < 0.05, ***P < 0.001.
Regulation of RORγ and Npas2 expression in brown adipocytes
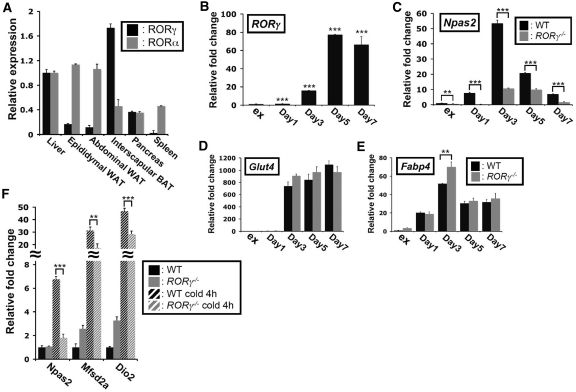
Because of the observations that the expression of Npas2 was most significantly affected in BAT of RORγ−/− mice (Figure 1C) and that RORγ is highly expressed in BAT compared with WAT and liver (Figure 7A), we were interested in exploring the relationship between the expression of RORs and Npas2, and BAT differentiation and function. Analysis of RORγ and Npas2 expression showed that both were induced about 80- and 50-fold, respectively, during differentiation of primary brown preadipocytes (Figure 7B and C). In contrast to RORγ, Npas2 expression was repressed at later stages of differentiation. The induction of Npas2 mRNA expression was greatly reduced during differentiation of brown preadipocytes isolated from RORγ−/− mice. However, the expression of the adipocyte marker genes, Glut4 and Fabp4, was at most time points not significantly different between WT and RORγ−/− adipocytes (Figure 7D and E) indicating that the downregulation of Npas2 in RORγ−/− brown adipocytes was rather specific. BAT plays an important role in heat generation and a number of genes, including Mfsd2a and Dio2, are greatly induced during cold-induced thermogenesis (49,56). Figure 7F shows that the level of Npas2 mRNA in the BAT was increased about 7-fold during cold-induced thermogenesis and that this induction was inhibited by 90% in BAT of RORγ−/− mice. Loss of RORγ also repressed the induction of Dio2 and Mfsd2a during cold-induced thermogenesis. These data suggest that, in addition to its circadian regulation of Npas2, RORγ plays a critical role in the regulation of Npas2 expression during BAT differentiation and cold-induced thermogenesis.
Figure 7.
The induction of Npas2 expression during brown adipocyte differentiation was regulated by RORγ. (A) Comparison of RORα and RORγ expression in the WAT, BAT and several other tissues. RORα and RORγ were highly expressed in the WAT and BAT, respectively. (B–E) RORγ mRNA was highly induced during brown adipocyte differentiation. Primary cultures of BAT preadipocytes obtained from WT and RORγ−/− mice were induced to differentiate as described in ‘Materials and Methods’ section. RNA was isolated at the times indicated and the expression of RORγ, Npas2 and BAT differentiation markers, such as Glut4 and Fabp4, was examined by QPCR analysis. The data are an average of three isolates of brown adipocytes from each WT and RORγ−/− BAT, respectively. e.g. preadipocytes in exponential phase. (F) The induction of Npas2 expression during cold-induced thermogenesis was inhibited in the BAT of RORγ−/−mice. WT and RORγ−/− mice (n = 5) were placed at 4°C for 4 h before BAT was collected for analysis at CT18. Data present mean ± SEM, **P < 0.01, ***P < 0.001 by ANOVA.
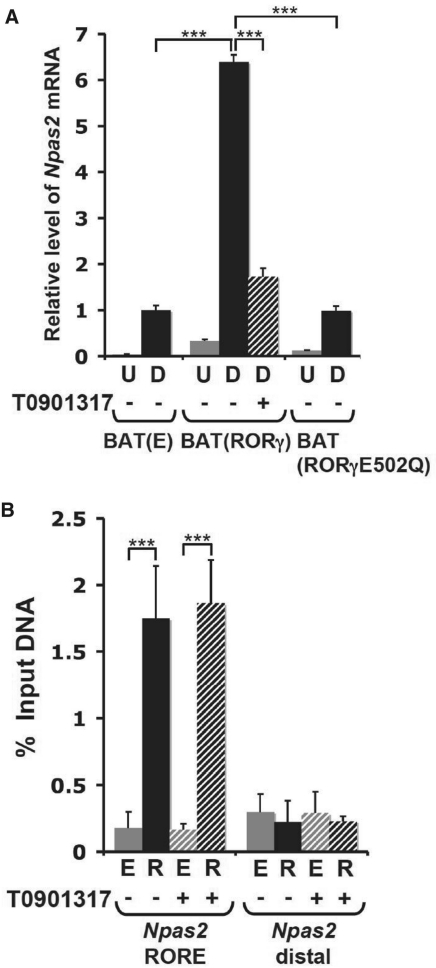
To study the role of RORγ in the regulation of Npas2 in brown adipocytes further, we examined its expression in immortalized brown preadipocyte cell lines stably expressing empty vector, RORγ or the mutant RORγ(E502Q), referred to as BAT(E), BAT(RORγ) and BAT(RORγE502Q). As shown in Figure 8A, Npas2 was induced at 6- to 7-fold higher levels during differentiation of BAT(RORγ) cells compared with BAT(E) cells. In contrast, Npas2 was not elevated in BAT(RORγE502Q) cells expressing an RORγ mutant lacking transactivation function. Treatment of BAT(RORγ) cells with the inverse agonist, T0901317, significantly reduced the induction of Npas2 mRNA (Figure 8A).
Figure 8.
RORγ-activated Npas2 gene expression in brown adipocytes through a direct mechanism. (A) Npas2 expression was examined in brown adipocyte cell lines, BAT(E), BAT(RORγ) and BAT(RORγE502Q) stably expressing empty vector, RORγ or the RORγE502Q mutant containing an inactive activation domain, respectively. Expression of Npas2 was compared between brown preadipocytes at the exponential phase (U) and at differentiation Day 5 (D) as indicated. BAT(RORγ) cells were also treated at Day 4 for 24 h with 10 µM of the inverse agonist T0901317. (B) ChIP analysis was performed using anti-Flag M2 antibody, BAT(E) cells (E) and BAT(RORγ) cells (R) treated with or without T0901317 as described under (A). BAT(E) cells and the distal region were used as a negative controls. The experiment was performed in triplicate. Data present mean ± SEM, ***P < 0.001 by ANOVA.
ChIP analysis with anti-Flag M2 antibody and BAT(RORγ) or BAT(E) cells showed strong recruitment of Flag–RORγ to the RORE-containing region of the Npas2(–1534/+81) promoter (Figure 8B). These data support our conclusion that RORγ is recruited to the Npas2(–1534/+81) promoter. The inverse agonist, T0901317, did not significantly influence the recruitment of RORγ to the Npas2 promoter suggesting that T0901317-bound RORγ was still recruited to ROREs within the Npas2 promoter.
DISCUSSION
In this study, we provide evidence that RORs, RORγ in particular, are important regulators of Npas2 expression. In WT mice, Npas2 displayed a strong oscillatory expression pattern in several tissues, including the kidney, liver and BAT, with peak expression at CT20–CT24/0 (Figure 1) (24,51). Although the rhythmicity of Npas2 expression in these tissues was maintained in RORγ−/− mice, the peak level of Npas2 mRNA expression was significantly reduced. Except for the kidney, loss of RORα had relatively little effect on Npas2 expression in the tissues analyzed despite its recruitment to the Npas2 promoter. That loss of RORα has little effect on Npas2 expression might be related to weak RORα activity and involve low levels of RORα protein bound relatively to RORγ or posttranslational modifications of RORα. Additional evidence that RORs regulate Npas2 expression was provided by data showing that overexpression of either RORα or RORγ in murine hepatoma Hepa1-6 cells greatly induced the expression of Npas2 mRNA (Figure 5A).
We previously reported a certain degree of functional redundancy between the regulation of gene expression by RORα and RORγ (16). For example, the hepatic expression of several Phases I and II metabolic genes was considerably more affected in DKO than in single knockout mice. Analysis of Npas2 expression in DKO mice indicated no significant functional redundancy between the RORα and RORγ in regulating Npas2. These data are consistent with our conclusion that in these tissues Npas2 expression is regulated selectively by RORγ with a minor role for RORα. In contrast to Npas2, the expression of Clock was significantly reduced in the liver and BAT of DKO mice (Supplementary Figure S1I), while the expression was not changed in single knockout mice suggesting a redundant function of RORα and RORγ in regulating Clock.
RORs can regulate gene transcription directly by binding ROREs in the regulatory region of target genes or through indirect mechanisms (1). The 1534-nt proximal promoter region of the Npas2 gene contains three putative ROREs (27). In this study, we demonstrate that Npas2 is a direct ROR target gene and that its transcriptional activation involves the recruitment of RORs to RORE1 and RORE2. This is supported by reporter gene analysis showing that expression of either RORα and RORγ activated the Npas2(–1534/+81) proximal promoter. Mutation of either RORE1 or RORE2 partially reduced the Npas2 promoter activity, whereas mutation of both ROREs almost totally abolished activation of the Luc reporter suggesting that optimal Npas2 induction by RORγ requires both ROREs. In contrast to an earlier report, mutations in RORE3 had little effect on ROR-induced activation of the Npas2(–1534/+81) promoter suggesting that it plays a minor role in the regulation of Npas2 by RORs. We further demonstrated by ChIP analysis that RORα and RORγ were recruited to these ROREs in the Npas2 promoter in WT liver, Hepa1-6 and BAT cells. As expected no significant recruitment of RORα or RORγ was observed in RORαsg/sg or RORγ−/− liver, respectively. Our data further showed that the recruitment of RORγ was CT dependent. In contrast to CT22, when RORγ is expressed at a high level, RORγ was not found to be associated with the Npas2 promoter at CT10, when RORγ expression is low. Collectively, these data suggest that the regulation of Npas2 transcription by RORγ occurs by a direct mechanism. Transcriptional activation is associated with increased chromatin accessibility, as determined by FAIRE, and histone acetylation, including H3K9 acetylation (50,55). Our data showed that the FAIRE signal and H3K9 acetylation are significantly increased at the Npas2 promoter in liver at CT22 compared with CT10 consistent with increased accessibility and the high level of transcriptional activity of the Npas2 promoter at CT22. Loss of RORγ significantly reduced the FAIRE signal and H3K9 acetylation at the Npas2 proximal promoter consistent with a relationship between RORγ binding, open chromatin structure and transcriptional activation of Npas2. Despite the fact that loss of RORα has little effect on Npas2 expression in liver, it also caused a reduction in the FAIRE signal and H3K9 acetylation suggesting a role for RORα plays a role in chromatin remodeling in Npas2. This is consistent with the ChIP analysis showing binding of RORα to the Npas2 ROREs (Figure 4) (53). Further studies are required to understand the role of RORα in Npas2 gene regulation. Chromatin remodeling might occur prior to and/or following ROR binding. The observation that loss of RORs did not totally inhibit Npas2 expression and reduced FAIRE signal and H3K9 acetylation by about 50% suggests that other transcription factors are involved in the positive regulation of Npas2 and chromatin remodeling of the Npas2 promoter. Thus, at CT22 RORs may bind to this region when it is already quite accessible through the activity of other transcription factors. RORs through their recruitment of histone acetyltransferases, such as CBP/p300, likely contribute to the increased histone acetylation and chromatin remodeling.
Recent studies have demonstrated that RORs function as ligand-dependent transcription factors (5–8,58). T0901317 functions as an inverse agonist for both RORα and RORγ and has been reported to effectively inhibit RORα- and RORγ-mediated induction of Th17 differentiation and IL-17 expression. In this study, we demonstrate that T0901317 inhibited the activation of the Npas2 promoter by RORα and RORγ in hepatoma Huh-7 cells as well as in brown adipocytes. ChIP analysis showed that T0901317 did not significantly affect the recruitment of RORγ to the ROREs in the Npas2 proximal promoter. The recruitment of RORγ to the Npas2 promoter in the presence of T0901317 indicates that T0901317 does not affect the interaction of RORs with these ROREs. The inhibition of transcription is likely due to a T0901317-induced change in the conformation of the ROR receptors that affects their interaction with co-activators and subsequently their transcriptional activity. This hypothesis is supported by a recent study showing that T0901317 inhibits the interaction of RORs with the co-activator SRC-1 (7).
The nuclear receptors, Rev-Erbα and Rev-Erbβ, have been reported to act as transcriptional repressors (59). Moreover, it is well-established that Rev-Erb receptors bind response elements similar to RORE and can compete with RORs for RORE binding (22,23,37,54). In this study, we demonstrated that Rev-Erbα can also function as a repressor of Npas2 expression. In contrast to the effects of RORα or RORγ, overexpression of Rev-Erbα in Hepa1-6 cells caused a reduction in Npas2 mRNA level (Figure 5A). In addition, overexpression of Rev-Erbα inhibited the basal Npas2(–1534/+81) promoter activity (Figure 3B). Mutation of either RORE1 or RORE2, but not RORE3, reduced this inhibition, while the double mutation totally abrogated the repression by Rev-Erbα, suggesting that the inhibitory effect of Rev-Erbα is mediated through these two ROREs. This was supported by ChIP analysis showing recruitment of Rev-Erbα to the RORE-containing region of the Npas2 promoter (Figure 5B). Thus, as RORs, Rev-Erbs play a critical role in the regulation of Npas2 expression through recognition of the same ROREs in the Npas2 promoter. It is likely that competition between RORs and Rev-Erbα for RORE1 and RORE2 is part of the mechanism that regulates the oscillatory expression of Npas2. This is corroborated by data showing that Rev-Erbα effectively antagonized the activation of the Npas2 promoter by RORs (Figure 3A).
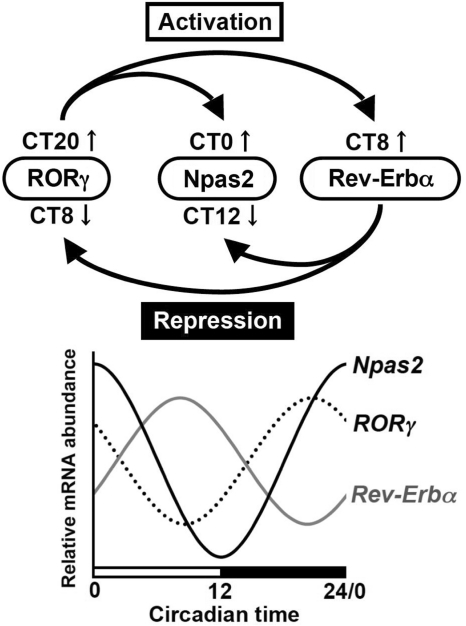
We were interested in relating the regulation of Npas2 by RORγ and Rev-Erbα to their rhythmic pattern of expression. The oscillatory expression pattern of Npas2 and RORγ are in almost opposite phase compared with that of Rev-Erbα, while RORγ reaches a peak slightly earlier than Npas2 (Figures 1 and 9) (16,20,24,51). The lowest level of Rev-Erbα mRNA expression occurred at CT16–CT24 when Npas2 and RORγ are expressed at peak levels, whereas at CT8–CT10 the peak level of Rev-Erbα expression coincides with the lowest level of RORγ and Npas2 expression. These patterns of expression are consistent with the hypothesis that at CT8-10 downregulation of RORγ and induction of Rev-Erbα may act cooperatively in repressing Npas2 expression, while induction of RORγ and repression of Rev-Erbα jointly promote Npas2 expression at CT16–24 (Figure 9). A recent study demonstrated that loss of Bmal1 abrogates Rev-Erbα expression and as a result alleviates the repression of RORγ expression by Rev-Erbα (24), while RORα has been demonstrated to positively regulate Rev-Erbα (22,60,61). Thus, RORγ and Rev-Erbα are part of a regulatory loop that regulates the circadian pattern of expression of Npas2 (Figure 9). The opposite phases of the circadian expression of these receptors and their competition for RORE binding collectively participate in the regulation of the oscillatory pattern of Npas2 expression.
Figure 9.
Schematic presentation of the molecular mechanism for the antagonistic regulation of Npas2 circadian expression by RORγ and Rev-Erbα. (Lower panel) Oscillatory pattern of expression of Npas2, RORγ and Rev-Erbα in liver. (Upper panel) RORγ functions as a positive regulator of Npas2 transcription in vivo and in cultured cells by binding to two ROREs in its proximal promoter region. RORγ is efficiently recruited to these ROREs at CT18–CT24/0 when RORγ and Npas2 are highly expressed. Rev-Erbα, which peak expression occurs around CT8 (37), represses Npas2 expression through competitive binding to the same ROREs. Rev-Erbα has also been reported to repress RORγ expression, while RORγ can activate Rev-Erbα expression. The reduced RORγ expression and increased Rev-Erbα expression at CT8–10 might act synergistically to reduce Npas2 circadian expression, while the inverse occurs at CT16–24. Thus, RORγ and Rev-Erbα are parts of a regulatory loop regulating each other and consequently the circadian expression of Npas2. Because loss of RORs does not totally block Npas2 expression, other transcription factors likely regulate its transcription as well.
As its paralog Clock, Npas2 forms a heterodimeric complex with Bmal1 and regulates gene transcription by interacting with E-boxes in the regulatory region of target genes (39). Clock and Npas2 have been reported to exhibit overlapping roles in the regulation of the circadian clock genes in the SCN and circadian-controlled genes in liver (39–41). In addition to its role in circadian regulation, epidemiologic studies have provided evidence for a role of Npas2 in several other biological processes. SNPs in Npas2 have been linked to increased risk of cancer, unipolar major depression, metabolic syndrome and hypertension (42–47). Npas2 has been implicated in the regulation of the DNA damage response pathway and several cell cycle and DNA repair genes suggesting a role for Npas2 as a tumor suppressor (45,46). In this study, we demonstrate that both RORγ and Npas2 are induced during differentiation of preadipocytes into brown fat adipocytes. In addition, we show that Npas2 expression is enhanced in BAT during cold-induced thermogenesis, suggesting a regulatory role for Npas2 in this physiological process. However, Npas2 expression was strongly downregulated in RORγ−/− brown adipocytes during differentiation and in BAT of RORγ−/− mice during cold-induced thermogenesis. In addition, we showed that T0901317 was able to inhibit the induction of Npas2 by RORγ in the brown adipocytes. Collectively, these data support the conclusion that RORγ functions as a transcriptional activator of Npas2 in BAT. Although our study suggests a role for Npas2 in thermogenesis, future studies are required to determine the precise function of Npas2 and RORγ in BAT cells. Taken the role of Rev-Erbs, RORs and BAT in metabolic syndrome, these data are of interest in the light of reports showing that Npas2 gene variants have been linked to increased risk of metabolic syndrome (16,17,22,43,62).
Our study shows that RORγ regulates Npas2 expression in vivo. Loss of RORγ had little effect on the rhythmicity, but reduced the peak level of Npas2 expression. RORγ regulates the transcription of Npas2 by binding to two ROREs in its proximal promoter region and enhances the accessibility to the proximal Npas2 promoter enhancer region. The activation of Npas2 promoter by RORs is repressed by co-expression with Rev-Erbα or addition of the inverse agonist T0901317. These observations suggest that RORs and Rev-Erbα may be implicated in the modulation of Npas2-controlled physiological processes and Npas2-linked pathologies, including circadian behavior disorders, metabolic syndrome and tumorigenesis. Thus, changes in the level of the physiological ligands of RORs and Rev-Erbα may play an important role in the development of these pathologies. In addition, because RORs function as ligand-dependent transcription factors, increased Npas2 expression by ROR agonists may provide a novel therapeutic approach for disease in which Npas2 has been implicated, including metabolic syndrome, hypertension and depression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institute of Environmental Health Sciences, the National Institutes of Health (Z01-ES-101586). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
The authors would like to thank Drs. Christina T. Teng and Kristin Lichti-Kaiser (NIEHS) for their comments on the manuscript and Laura Miller (NIEHS) for her assistance.
REFERENCES
- 1.Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl. Recept. Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giguere V, Tini M, Flock G, Ong E, Evans RM, Otulakowski G. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8:538–553. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- 3.Medvedev A, Yan ZH, Hirose T, Giguere V, Jetten AM. Cloning of a cDNA encoding the murine orphan receptor RZR/ROR gamma and characterization of its response element. Gene. 1996;181:199–206. doi: 10.1016/s0378-1119(96)00504-5. [DOI] [PubMed] [Google Scholar]
- 4.McKenna NJ, O’Malley BW. Minireview: nuclear receptor coactivators–an update. Endocrinology. 2002;143:2461–2465. doi: 10.1210/endo.143.7.8892. [DOI] [PubMed] [Google Scholar]
- 5.Kallen JA, Schlaeppi J, Bitsch F, Geisse S, Geiser M, Delhon I, Fournier B. X-ray structure of the RORα LBD at 1.63A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORα. Structure. 2002;10:1697–1707. doi: 10.1016/s0969-2126(02)00912-7. [DOI] [PubMed] [Google Scholar]
- 6.Stehlin C, Wurtz JM, Steinmetz A, Greiner E, Schule R, Moras D, Renaud JP. X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation. EMBO J. 2001;20:5822–5831. doi: 10.1093/emboj/20.21.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar N, Solt LA, Conkright JJ, Wang Y, Istrate MA, Busby SA, Garcia-Ordonez RD, Burris TP, Griffin PR. The benzenesulfoamide T0901317 [N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluorometh yl)ethyl]phenyl]-benzenesulfonamide] is a novel retinoic acid receptor-related orphan receptor-alpha/gamma inverse agonist. Mol. Pharmacol. 2010;77:228–236. doi: 10.1124/mol.109.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Kumar N, Solt LA, Richardson TI, Helvering LM, Crumbley C, Garcia-Ordonez RD, Stayrook KR, Zhang X, Novick S, et al. Modulation of retinoic acid receptor-related orphan receptor alpha and gamma activity by 7-oxygenated sterol ligands. J. Biol. Chem. 2010;285:5013–5025. doi: 10.1074/jbc.M109.080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gold DA, Baek SH, Schork NJ, Rose DW, Larsen DD, Sachs BD, Rosenfeld MG, Hamilton BA. RORalpha coordinates reciprocal signaling in cerebellar development through sonic hedgehog and calcium-dependent pathways. Neuron. 2003;40:1119–1131. doi: 10.1016/s0896-6273(03)00769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinmayr M, Andre E, Conquet F, Rondi-Reig L, Delhaye-Bouchaud N, Auclair N, Daniel H, Crepel F, Mariani J, Sotelo C, et al. Staggerer phenotype in retinoid-related orphan receptor alpha-deficient mice. Proc. Natl Acad. Sci. USA. 1998;95:3960–3965. doi: 10.1073/pnas.95.7.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaeren-Wiemers N, Andre E, Kapfhammer JP, Becker-Andre M. The expression pattern of the orphan nuclear receptor RORbeta in the developing and adult rat nervous system suggests a role in the processing of sensory information and in circadian rhythm. Eur. J. Neurosci. 1997;9:2687–2701. doi: 10.1111/j.1460-9568.1997.tb01698.x. [DOI] [PubMed] [Google Scholar]
- 12.Srinivas M, Ng L, Liu H, Jia L, Forrest D. Activation of the blue opsin gene in cone photoreceptor development by retinoid-related orphan receptor beta. Mol. Endocrinol. 2006;20:1728–1741. doi: 10.1210/me.2005-0505. [DOI] [PubMed] [Google Scholar]
- 13.Kurebayashi S, Ueda E, Sakaue M, Patel DD, Medvedev A, Zhang F, Jetten AM. Retinoid-related orphan receptor gamma (RORgamma) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc. Natl Acad. Sci. USA. 2000;97:10132–10137. doi: 10.1073/pnas.97.18.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, Mebius RE, Littman DR. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 15.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The Orphan Nuclear Receptor RORgammat Directs the Differentiation Program of Proinflammatory IL-17(+) T Helper Cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 16.Kang HS, Angers M, Beak JY, Wu X, Gimble JM, Wada T, Xie W, Collins JB, Grissom SF, Jetten AM. Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol. Genomics. 2007;31:281–294. doi: 10.1152/physiolgenomics.00098.2007. [DOI] [PubMed] [Google Scholar]
- 17.Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J. Biol. Chem. 2008;283:18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- 18.Jaradat M, Stapleton C, Tilley SL, Dixon D, Erikson CJ, McCaskill JG, Kang HS, Angers M, Liao G, Collins J, et al. Modulatory role for retinoid-related orphan receptor alpha in allergen-induced lung inflammation. Am. J. Resp. Crit. Care Med. 2006;174:1299–1309. doi: 10.1164/rccm.200510-1672OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP, Shah B, Chang SH, Schluns KS, Watowich SS, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- 21.Andre E, Conquet F, Steinmayr M, Stratton SC, Porciatti V, Becker-Andre M. Disruption of retinoid-related orphan receptor beta changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J. 1998;17:3867–3877. doi: 10.1093/emboj/17.14.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duez H, Staels B. The nuclear receptors Rev–erbs and RORs integrate circadian rhythms and metabolism. Diabetes Vasc. Dis. Res. 2008;5:82–88. doi: 10.3132/dvdr.2008.0014. [DOI] [PubMed] [Google Scholar]
- 23.Guillaumond F, Dardente H, Giguere V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 24.Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masana MI, Sumaya IC, Becker-Andre M, Dubocovich ML. Behavioral characterization and modulation of circadian rhythms by light and melatonin in C3H/HeN mice homozygous for the RORbeta knockout. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007;292:R2357–2367. doi: 10.1152/ajpregu.00687.2006. [DOI] [PubMed] [Google Scholar]
- 26.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 27.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian clocks. Nat. Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 28.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 29.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 2006;15(Spec No 2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 30.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat. Rev. Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 31.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 33.Albrecht U. Invited review: regulation of mammalian circadian clock genes. J. Appl. Physiol. 2002;92:1348–1355. doi: 10.1152/japplphysiol.00759.2001. [DOI] [PubMed] [Google Scholar]
- 34.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 35.Isojima Y, Okumura N, Nagai K. Molecular mechanism of mammalian circadian clock. J. Biochem. 2003;134:777–784. doi: 10.1093/jb/mvg219. [DOI] [PubMed] [Google Scholar]
- 36.Schibler U, Naef F. Cellular oscillators: rhythmic gene expression and metabolism. Curr. Opin. Cell Biol. 2005;17:223–229. doi: 10.1016/j.ceb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 37.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 38.Duez H, Staels B. Rev-erb-alpha: an integrator of circadian rhythms and metabolism. J. Appl. Physiol. 2009;107:1972–1980. doi: 10.1152/japplphysiol.00570.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science. 2001;293:506–509. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- 40.Bertolucci C, Cavallari N, Colognesi I, Aguzzi J, Chen Z, Caruso P, Foa A, Tosini G, Bernardi F, Pinotti M. Evidence for an overlapping role of CLOCK and NPAS2 transcription factors in liver circadian oscillators. Mol. Cell. Biol. 2008;28:3070–3075. doi: 10.1128/MCB.01931-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat. Neurosci. 2007;10:543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soria V, Martinez-Amoros E, Escaramis G, Valero J, Perez-Egea R, Garcia C, Gutierrez-Zotes A, Puigdemont D, Bayes M, Crespo JM, et al. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology. 2010;35:1279–1289. doi: 10.1038/npp.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Englund A, Kovanen L, Saarikoski ST, Haukka J, Reunanen A, Aromaa A, Lonnqvist J, Partonen T. NPAS2 and PER2 are linked to risk factors of the metabolic syndrome. J. Circad. Rhythms. 2009;7:5. doi: 10.1186/1740-3391-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi CH, Zheng T, Leaderer D, Hoffman A, Zhu Y. Cancer-related transcriptional targets of the circadian gene NPAS2 identified by genome-wide ChIP-on-chip analysis. Cancer Lett. 2009;284:149–156. doi: 10.1016/j.canlet.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffman AE, Zheng T, Ba Y, Zhu Y. The circadian gene NPAS2, a putative tumor suppressor, is involved in DNA damage response. Mol. Cancer Res. 2008;6:1461–1468. doi: 10.1158/1541-7786.MCR-07-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Y, Stevens RG, Leaderer D, Hoffman A, Holford T, Zhang Y, Brown HN, Zheng T. Non-synonymous polymorphisms in the circadian gene NPAS2 and breast cancer risk. Breast Cancer Res. Treat. 2008;107:421–425. doi: 10.1007/s10549-007-9565-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicholas B, Rudrasingham V, Nash S, Kirov G, Owen MJ, Wimpory DC. Association of Per1 and Npas2 with autistic disorder: support for the clock genes/social timing hypothesis. Mol. Psychiatry. 2007;12:581–592. doi: 10.1038/sj.mp.4001953. [DOI] [PubMed] [Google Scholar]
- 48.Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 49.Angers M, Uldry M, Kong D, Gimble JM, Jetten AM. Mfsd2a encodes a novel major facilitator superfamily domain-containing protein highly induced in brown adipose tissue during fasting and adaptive thermogenesis. Biochem. J. 2008;416:347–355. doi: 10.1042/BJ20080165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL, Gimble JM. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 52.Mongrain V, Ruan X, Dardente H, Fortier EE, Cermakian N. Clock-dependent and independent transcriptional control of the two isoforms from the mouse Rorgamma gene. Genes Cells. 2008;13:1197–1210. doi: 10.1111/j.1365-2443.2008.01237.x. [DOI] [PubMed] [Google Scholar]
- 53.Crumbley C, Wang Y, Kojetin DJ, Burris TP. Characterization of the Core Mammalian Clock Component, NPAS2, as a REV-ERB{alpha}/ROR{alpha} Target Gene. J. Biol. Chem. 2010;285:35386–35392. doi: 10.1074/jbc.M110.129288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Austin S, Medvedev A, Yan ZH, Adachi H, Hirose T, Jetten AM. Induction of the nuclear orphan receptor RORgamma during adipocyte differentiation of D1 and 3T3-L1 cells. Cell Growth Differ. 1998;9:267–276. [PubMed] [Google Scholar]
- 55.MacDonald VE, Howe LJ. Histone acetylation: where to go and how to get there. Epigenetics. 2009;4:139–143. doi: 10.4161/epi.4.3.8484. [DOI] [PubMed] [Google Scholar]
- 56.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 57.Yin L, Wu N, Lazar MA. Nuclear receptor Rev-erbalpha: a heme receptor that coordinates circadian rhythm and metabolism. Nucl. Recept. Signal. 2010;8:e001. doi: 10.1621/nrs.08001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin L, Martynowski D, Zheng S, Wada T, Xie W, Li Y. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma. Mol. Endocrinol. 24:923–929. doi: 10.1210/me.2009-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramakrishnan SN, Muscat GE. The orphan Rev-erb nuclear receptors: a link between metabolism, circadian rhythm and inflammation? Nucl. Recept. Signal. 2006;4:e009. doi: 10.1621/nrs.04009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raspe E, Mautino G, Duval C, Fontaine C, Duez H, Barbier O, Monte D, Fruchart J, Fruchart JC, Staels B. Transcriptional regulation of human Rev-erbalpha gene expression by the orphan nuclear receptor RORalpha. J. Biol. Chem. 2002;107:49275–49281. doi: 10.1074/jbc.M206215200. [DOI] [PubMed] [Google Scholar]
- 61.Delerive P, Chin WW, Suen CS. Identification of Reverb(alpha) as a novel ROR(alpha) target gene. J. Biol. Chem. 2002;277:35013–35018. doi: 10.1074/jbc.M202979200. [DOI] [PubMed] [Google Scholar]
- 62.Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Sasso GL, Moschetta A, Schibler U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.