Abstract
To gain insight into the mechanisms by which the Myb transcription factor controls normal hematopoiesis and particularly, how it contributes to leukemogenesis, we mapped the genome-wide occupancy of Myb by chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-Seq) in ERMYB myeloid progenitor cells. By integrating the genome occupancy data with whole genome expression profiling data, we identified a Myb-regulated transcriptional program. Gene signatures for leukemia stem cells, normal hematopoietic stem/progenitor cells and myeloid development were overrepresented in 2368 Myb regulated genes. Of these, Myb bound directly near or within 793 genes. Myb directly activates some genes known critical in maintaining hematopoietic stem cells, such as Gfi1 and Cited2. Importantly, we also show that, despite being usually considered as a transactivator, Myb also functions to repress approximately half of its direct targets, including several key regulators of myeloid differentiation, such as Sfpi1 (also known as Pu.1), Runx1, Junb and Cebpb. Furthermore, our results demonstrate that interaction with p300, an established coactivator for Myb, is unexpectedly required for Myb-mediated transcriptional repression. We propose that the repression of the above mentioned key pro-differentiation factors may contribute essentially to Myb’s ability to suppress differentiation and promote self-renewal, thus maintaining progenitor cells in an undifferentiated state and promoting leukemic transformation.
INTRODUCTION
The Myb transcription factor is a master regulator of normal hematopoiesis (1). It is required for the establishment of definitive hematopoiesis as evidenced by the early embryonic death of Myb null mice due to severe anemia with profound defects in the development of multiple other lineages (2). Moreover, Myb has been shown to play an essential role in maintaining adult hematopoietic stem cells (HSC) using a conditional Myb knockout mouse model (3).
Myb also contributes actively to leukemogenesis. Activated Myb induces leukemias in chickens and mice (4,5). Importantly, MYB was found to be activated in human leukemias by genetic lesions, such as translocation, duplication (6–11) and structural alteration (12). Myb is also required for leukemic transformation by other oncogenes, such as MLL-ENL (13) and BCR-ABL (14).
Myb’s oncogenic activities stem primarily from its ability to suppress differentiation and promote self-renewal. Enforced expression of wild-type or activated Myb can block the induced differentiation of established immature, myeloid progenitor-like cell lines (15–17) and can generate such lines from primary myeloid cells (18). Conversely, siRNA-mediated knockdown of MYB induces a phenotype mimicking differentiation induced by phorbol ester in THP-1 myeloid leukemia cell line (19). Similarly to its role in normal HSCs, Myb plays a critical role in maintaining the leukemia stem cell (LSC) population, at least in MLL translocation-induced leukemias (20).
Myb regulates normal hematopoiesis and leukemogenesis by directing orchestrated expression of its transcriptional targets. Although more than 80 genes have been reported to be targeted by Myb [(1); see also Supplementary Table S1 and references therein], many of them have not been thoroughly validated for in vivo DNA occupancy by Myb. Most importantly, the current repository of Myb target genes does not adequately explain the key elements of Myb’s transforming activity, namely suppressing differentiation and promoting self-renewal.
Here we report the transcriptional program instigated by Myb in a myeloid progenitor cell line model. We identified Myb target genes by integrating dynamic genome-wide data from ChIP-Seq with that from global gene expression profiling. Importantly, we uncovered that, despite being usually considered as a transactivator, Myb also functions to directly repress many target genes, including several master regulators of the myeloid lineage, suggesting that this hitherto unappreciated role of Myb may constitute a crucial part of its oncogenic activity. We also showed that the interaction with p300, a known Myb co-activator, is required for both repression and activation by Myb. Thus our results indicate that Myb controls a transcriptional program critical for myelopoiesis and leukemogenesis through both positive and negative transcriptional regulation.
MATERIALS AND METHODS
Cell culture and bone marrow progenitor isolation and transduction
The withdrawal and re-addition of β-E2 from ERMYB cells (21) was performed as described (22). Separation and retroviral transduction of mouse primary bone marrow cells was conducted as described (15). Briefly, c-Kithigh progenitors were enriched from C57BL/6 mice bone marrow cells using EasySep mouse CD117 (c-Kit) positive selection kit (StemCell) and transduced with retroviruses encoding wt-Myb, CT3-Myb or L302A.CT3-Myb. Cells were cultured in Isocove’s modified Dulbecco’s medium supplemented with 10% fetal bovine serum (Hyclone) and 60 FDU/ml GM-CSF for 7 days, and subsequently FACS-sorted for GFP expression (encoded by the retroviral vector) before harvest.
To isolate granulocyte-macrophage progenitors (GMPs) (Lin− c-Kit+ Sca1− CD16/32high CD34high) (23), bone marrow cells of 8 week-old wild-type C57BL/6 or ‘booreana’ (24) mice were enriched for c-Kit expressing cells using CD117 microbeads on an autoMACS cell separator (Miltenyi). Lineage depletion was performed with unconjugated lineage monoclonal antibody cocktail against CD3ε, CD4, CD5, CD8, B220, Gr1, CD11b and Ter119 and PE-conjugated donkey anti-rat IgG. GMPs were identified by a combination of Sca1-PerCP-Cy5.5, cKit-PE-Cy7, CD34-FITC, CD16/32-APC. All antibodies were from eBioscience.
Gene expression profiling analysis
Triplicate total RNA samples from ERMYB cells were collected at 0, 6 and 24 h after β-E2 withdrawal and 6 h after β-E2 re-addition. RNA quality was examined on Agilent 2100 Bioanalyzer using Eukaryote Total RNA Nano chips. Samples were then analyzed by Illumina Mouse Whole Genome arrays (WG6). Probe intensities were corrected and normalized by the Lumi package (25). Differentially expressed genes (B statistic >1 between any two consecutive time points) were identified by the LIMMA package (26) and then grouped into different classes according to their kinetic profiles as follows.
Class A genes (likely Myb target genes) manifested four expression profiles, classified by the changes in expression level between the four time-points, consistent with Myb activity: nfp (negative-flat-positive) and nnp (negative-negative-positive) profiles for Myb activated genes and pfn (positive-flat-negative) and ppn (positive-positive-negative) profiles for Myb repressed genes. Profiles for class B genes (possible Myb target genes) included four for Myb activated genes (fnp, ffp, nff and nnf) and four for Myb repressed genes (fpn, ffn, pff and ppf). These class B profiles deviated from the class A ones but show at least one significant response to β-E2 withdrawal or re-addition. Simplified arrow diagrams representing these dynamic expression profiles are shown in 1C (for Class A) and Supplementary Figure S1 (for Class B). Class C is comprised of genes that were differentially expressed but without assignable profiles, genes with no significant differential expression throughout the time course, and genes not detectably expressed (all samples have a detection P > 0.01) in ERMYB cells at any time point.
Normalized expression levels of Myb regulated genes (classes A and B) were visualized using the Hierarchical Clustering module of GenePattern package (27).
TaqMan low density qPCR arrays and TaqMan assays
Custom TaqMan low density qPCR arrays (TLDAs) (96a format, Applied Biosystems) were made to order containing a total of 89 candidate Myb target genes (Supplementary Tables S3 and S4). Assays were carried out as per manufacturer’s instructions. Expression was normalized to the ‘housekeeping’ gene Hprt1.
Individual inventoried TaqMan assays (Applied Biosystems) were used to measure expression of identified Myb-activated and Myb-repressed genes in mouse GMPs.
ChIP and ChIP-Seq
ChIP was performed as described (28). ERMYB cells were harvested in the presence of β-E2 or 6 h after β-E2 withdrawal. After formaldehyde fixation, chromatin was fragmented to 200–800 bp by sonication. ER Ab-10 (Clone TE111.5D11, NeoMarkers) which recognizes the ERα ligand binding domain of the ER-Myb fusion protein, or mouse IgG1 isotype control (02-6100, Zymed) was used with ERMYB cells. Alternatively, the anti-CT5 (29) and Thelma anti-Myb (30) sera (mixed 1:1) were used. Pooled Myb 1-1 antibody (05-175, Upstate) and Thelma anti-Myb sera (30) (mixed 1:1) were used with ChIP in HL-60 cells. Other antibodies used for ChIP include anti-p300 (sc-585, Santa Cruz), anti-CBP (sc-369, Santa Cruz), anti-acetyl-Histone 3 (06-599, Upstate), anti-H3K4me1 (ab8895, Abcam), anti-H3K4me3 (07-473, Upstate), anti-H3K9me3 (ab8898, Abcam), anti-H3K27me3 (07-449, Upstate) and rabbit IgG (02-6102, Zymed). Primers for ChIP-qPCR are described in Supplementary Table S10.
For ChIP-Seq, biological duplicate ChIP samples were prepared using ER-10 antibody from ERMYB cells cultured in the presence of β-E2 (+β-E2 or ‘activated’-Myb) or deprived of β-E2 for 6 h (−β-E2 or ‘inactivated’-Myb). The employment of the ER antibody rather than Myb antibody raised the potential concern that a subset of identified Myb binding regions (MBRs) might be occupied by ERα rather than the Myb fusion protein. However, our results indicate this is unlikely to be the case. Almost all examined MBRs which were validated using ER antibody have also been validated using mixed anti-Myb sera. Furthermore, almost all of a set of conserved MBRs have been validated in human HL-60 cells using pooled MYB antibodies (Figure 3C). Moreover, the canonical ER binding motif was not detected in the de novo motif discovery analysis (Supplementary Figure S2C). Similar concerns about the Myb regulated genes identified by expression profiling in ERMYB cells are allayed by the fact that very few of these showed significant expression changes in response to β-E2 in a cell line derived by transformation with non-ER-fused, constitutively activate CT3-Myb (A69 cells; Supplementary Table S2). Moreover, our Myb-regulated gene set showed no significant overlap with the well-established estrogen-response signature (data not shown), and furthermore, the majority of a set of identified Myb regulated genes have been validated independently in Myb transduced primary bone marrow cells (Figure 1D and Supplementary Table S4).
Figure 3.
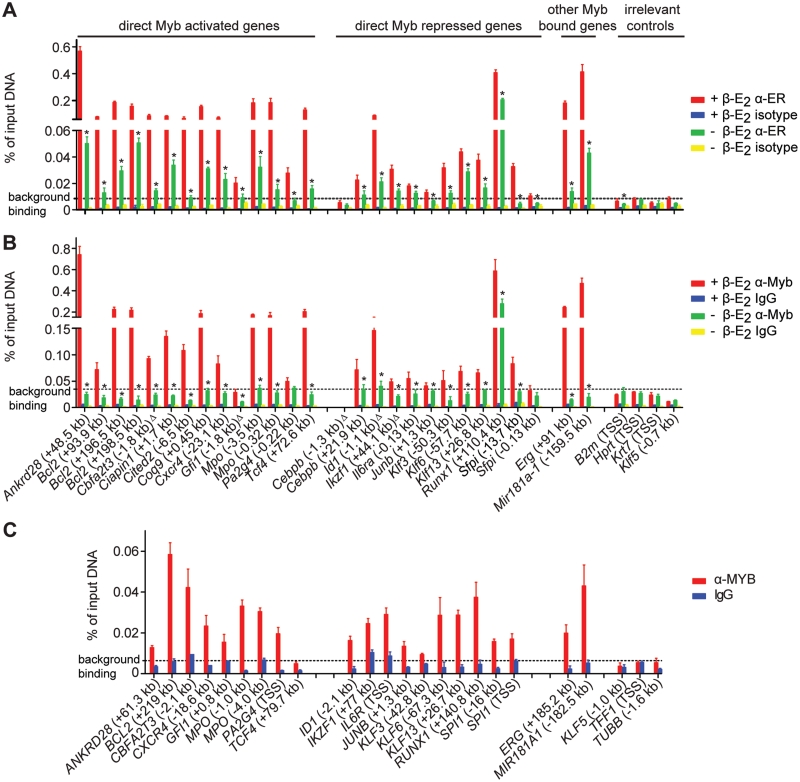
Validation of ChIP-Seq findings. (A) All 23 high-confidence MBRs were validated in ERMYB cells. Four of 5 MBRs (denoted by Δ) which just failed EdgeR P-value filter were also validated. Four irrelevant regions were included as negative controls. (B) Twenty-five of 27 MBRs validated using anti-ER antibody (A) were further validated using anti-Myb sera. Data are presented as mean ± SD. Background binding level represents (mean + 1.64 SD) of α-ER or α-Myb signals of four irrelevant control regions. ‘Asterisk’ denotes significant decreases in Myb occupancy upon β-E2 withdrawal (Student’s t-test, P < 0.05). (C) Twenty of 21 conserved MBRs were further validated in HL-60 cells using pooled MYB antibodies. Background binding level represents (mean + 1.64 SD) of α-MYB signals of three control regions.
Figure 1.
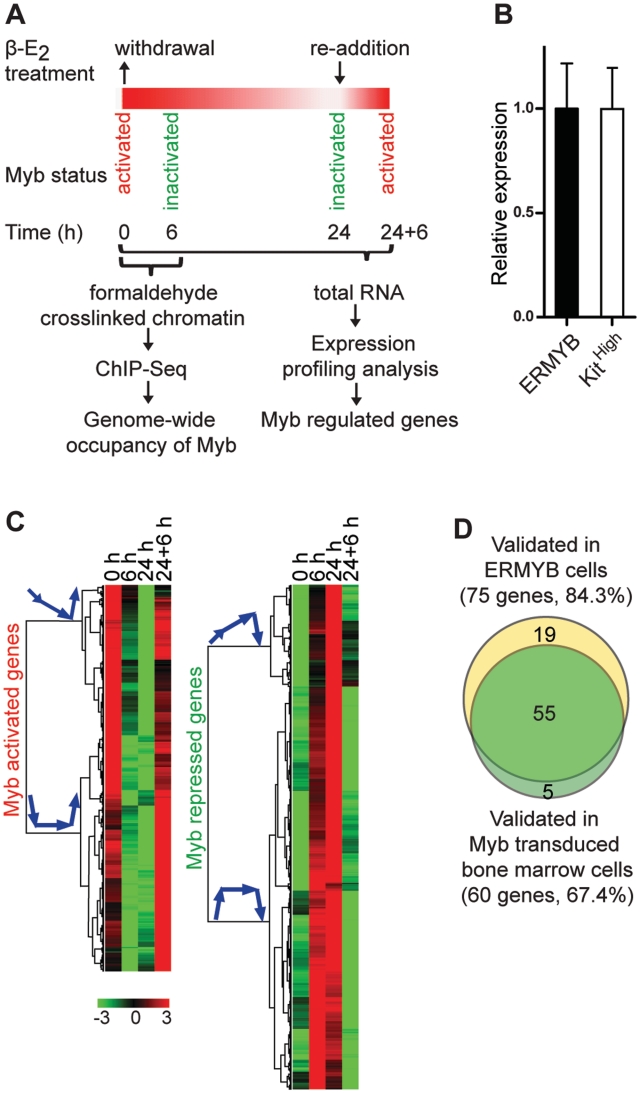
Expression profiling analysis of Myb regulated genes. (A) Schematic diagram of the experimental design. (B) The ER-Myb fusion transcript is expressed at an almost identical level to endogenous Myb mRNA in c-Kithigh bone marrow progenitors. Expression was measured by qRT–PCR and normalized to Hprt. The level of the ER-Myb fusion was set to 1. Data are presented as mean ± SD. (C) Hierarchical clustering of normalized expression levels of class A Myb regulated genes, including 352 Myb-activated and 460 repressed genes. The heat map of relative log2-transformed expression levels during the β-E2 withdrawal and re-addition time course is shown; each row represents a single gene. Simplified arrow diagrams of corresponding kinetic profiles are shown next to each cluster tree. (D) The majority of candidate Myb regulated genes were validated in both ERMYB cells and Myb transduced primary bone marrow cells.
ChIP-Seq DNA libraries were prepared using the Illumina ChIP-Seq Sample Prep Kit, then quality-checked and quantified on Agilent 2100 Bioanalyzer using DNA1000 kit. Sequencing was performed on the Illumina Genome Analyzer II using a standard single read 36-cycle sequencing protocol and Illumina’s sequencing reagents according to the manufacturer’s recommendations. Each library was sequenced individually on a single flow cell lane, except for the +β-E2 replicate 1 (‘activated’-Myb B1), which was sequenced on two lanes (B1T1 and B1T2, respectively). Sequencing yield of −β-E2 isotype control antibody sample was significantly lower than other samples. Thus it was excluded in the subsequent EdgeR analysis (see below).
ChIP-Seq data analyses
The data analysis methodology was summarized in Supplementary Figure S7A. We performed base calling, sequence tags quality assessment, and alignment of sequence tags to the reference mouse genome (mm9, UCSC assembly, July 2007) using Illumina Data Analysis Pipeline software v.1.4. Only high quality sequence tags with unique mapping positions to the reference genome were used for identification of MBRs and further analyses.
To identify possible binding regions, we combined replicate ‘activated’-Myb (0 h, +β-E2) and ‘inactivated’-Myb (6 h, −β-E2) samples, respectively, and ran MACS analysis (31) using the optimized parameters -bw110 -d55 -pval1e-3. This very conservative P-value cutoff generated 81 639 unique (MBRs). Of these, only 1417 have adjusted P < 10−40 with the number of binding regions increasing dramatically for P > 10−10 (Supplementary Figure S7B). These regions form the basis of the replicate design whereby the 36 bp sequence tags were extended to 200 bp to reflect the actual sequencing library size and the total number of tag coverage in each region identified by MACS calculated using R-code for each sample. The restriction to regions with bi-modal binding peaks excludes the ‘anomalous’ binding regions (32) that we also observed in our data set.
Individual counts per region were utilized in differential analysis of ‘activated’-Myb, ‘inactivated’- and isotype control samples using the EdgeR package (33). Since at the time of analysis the EdgeR program did not model technical replicates (there were one technical replicate and two biological replicates in the ‘activated’-Myb class), we made the four comparisons comprised of ‘activated’-Myb (B1T1 + B2) and ‘activated’-Myb (B1T2 + B2) sets against the ‘inactivated’-Myb (B1 + B2) set and the isotype control (B1) where B1 or B2 denotes ‘biological replicate’ 1 or 2 and T1 or T2 denotes ‘technical replicate’ 1 or 2. We required that binding regions have P < 10−10 of at least two of these comparisons and a positive fold change (signal of ‘activated’-Myb sample greater than that of ‘inactivated’-Myb sample or isotype control) to pass the P-value and fold change filter. This cutoff identified all but 139 of the regions identified with the MACs program with a P-value threshold <10−40. These additional 139 regions were included in our subsequent analysis.
We then applied additional constraints in the form of the minimum sequence tag number per region, in doing so we did not apply a ‘fixed’ tag count threshold but a position dependent one as follows: for each binding region we calculated the average background coverage per base pair in the isotype control in the same genomic regions ±2.5 kb. This average excludes counts in previously defined binding regions in that range. This average coverage per base pair is normalized by the total number of tags in the isotype control and then used to calculate the expected number of ‘background’ counts in the corresponding Myb-‘activated’ region. We represent the signal above background in the ith Myb-‘activated’ region using a z-score defined as zi = (Ci–pi Li)/sqrt(pi Li(1 − pi)) where Ci is the observed number of counts in the ith Myb-‘activated’ region of length Li and pi is the background coverage per base pair multiplied by the total number of reads [we assume background reads have Poisson distribution: mean = pi Li; SD = sqrt(pi Li (1 − pi)]. As an example, the distribution of zi for chromosome 2 is shown in Supplementary Figure S7C. The regions above the determined threshold for zi are shown in Supplementary Figure S7D. A fixed threshold for zi at the peak of the distribution (Supplementary Figure S7C), is similar to an overall tag cutoff of 8.5 tags/region (denoted by the orange horizontal line in Supplementary Figure S7D), but is biased against long binding regions with few counts.
In total 11 429 candidate binding regions pass the EdgeR P-value and tag number filter. These are enriched for binding regions with small P-values in the MACs analysis but also include a number of regions with large MACs P > 10−4 (Supplementary Figure S7B). Almost all of a selected set of these regions were subsequently validated by independent ChIP-qPCR (Figure 4), providing us with some evidence that the combined replicate and pooled approach has provided additional MBRs without dramatically increasing the false discovery rate.
Figure 4.
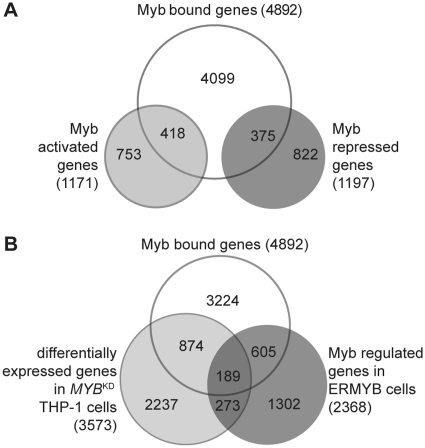
Integration of expression profiling and genome occupancy data. (A) Direct Myb target genes were defined as genes bound and regulated by Myb, including 418 Myb-activated and 375 repressed targets. (B) Significant overlap between Myb bound genes, Myb regulated genes in ERMYB cells and differentially expressed genes in MYBKD THP-1 cells.
We then performed de novo sequence motifs identification and enrichment analyses using the MEME algorithm (34). Two-hundred bp sequences (100 bp on either side of peak summits) were used as MEME input. The following parameters were applied to the input data sets: -text -revcomp -dna -mod oops -nmotifs 4 -minw 6 -maxw 12 -maxsize 5 000 000. Sequence logos were generated using WebLogo 2.8.2 (35).
We subsequently filtered the 11 429 candidate MBRs for presence of Myb binding motif by scanning 160 bp sequences (80 bp on either side of peak summits) of candidate MBRs with the identified position weight matrix for Myb binding (Figure 2C) with a threshold of 0.8 determined empirically. The resultant list comprises 7646 high-confidence MBRs (Supplementary Table S5). We subsequently assigned each MBR to its nearest gene according to its distance to transcription start site (TSS).
Figure 2.
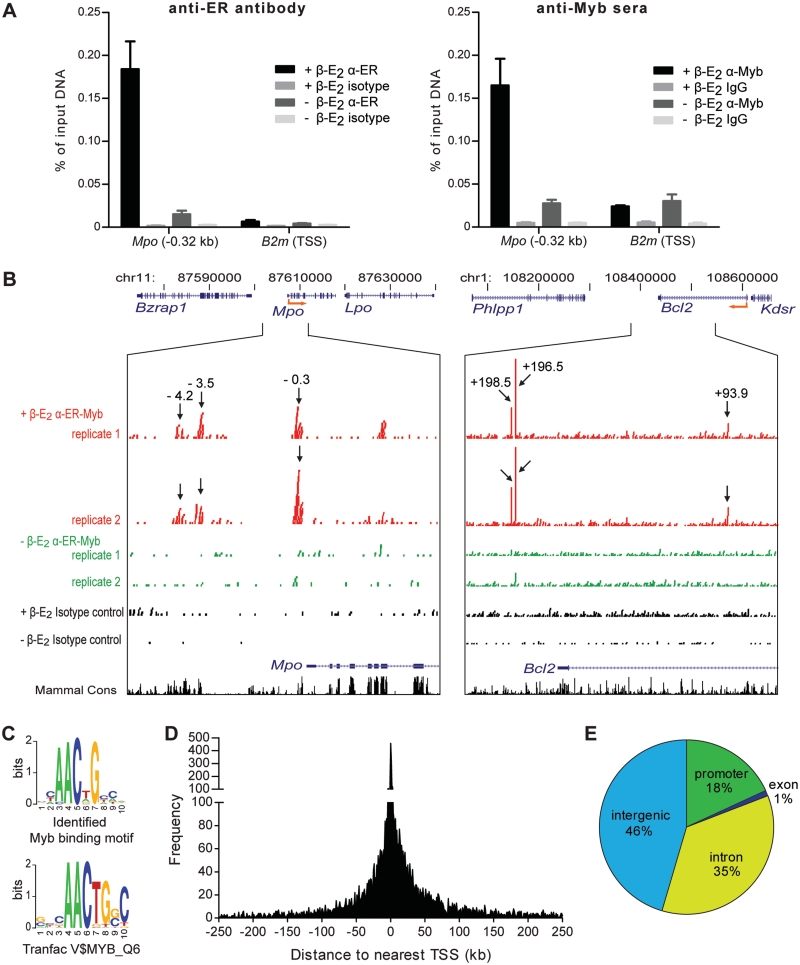
Global identification of MBRs in vivo. (A) Both anti-ER antibody and anti-Myb sera worked in ChIP assays in ERMYB cells: the ER-Myb fusion protein occupied a known Myb binding site in Mpo promoter (Mpo -0.32 kb). B2m TSS region served as a negative control. (B) Representation of ChIP-Seq sequencing reads (raw data) across loci of two established Myb targets—Mpo and Bcl2. (Arrows) ChIP-Seq peak locations relative to TSS of the respective gene (kb). (C) Identified Myb motif shares the AACNG core binding consensus sequence with the previously reported Myb binding consensus in Transfac database. (D) Distribution of MBRs relative to the nearest TSS (kb). Regions 5′ to the TSS are indicated as negative values on the x-axis. (E) Distribution of MBRs relative to Refseq gene features.
Upon closer examination of the 3783 regions which passed all filters except the Myb binding consensus filter, we found that many of these contain Myb binding motif-like sequences, so at least some of them may be true Myb occupied regions. Thus we have also provided a list of these additional regions (Supplementary Table S6).
To identify transcription factors that could potentially bind to other enriched sequence motifs within MBRs, we compared these motifs to known transcription factor binding matrices in the Transfac database (v11.3) using STAMP (36).
Gene set enrichment analysis
We ranked genes detectably expressed in ERMYB cells or in MYBKD THP-1 cells by the additive inverse of fold change values (for ERMYB data, fold change values from 0 to 24 h after β-E2 withdrawal were used). Top 200 Myb activated or repressed genes in either data set were determined based on their fold change values. Curated gene sets were either extracted from the indicated references or downloaded from MSig database (37). Gene set enrichment analysis (GSEA) was performed using GSEA v2.0 software with pre-ranked list and 1000 data permutations.
Pathway analysis and network construction
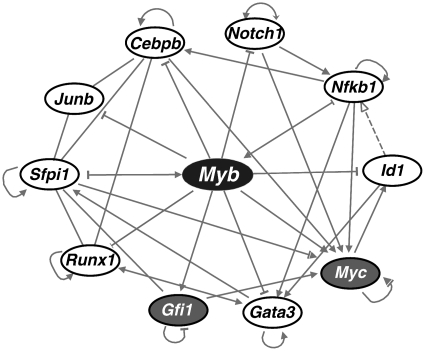
We identified the enriched physiological development and molecular function terms in direct Myb target genes were identified using the core analysis tool of Ingenuity Pathway Analysis program (IPA) program (http://www.ingenuity.com/). We then extracted transcriptional factors from direct Myb targets and constructed a transcriptional regulatory network consisting of Myb and its target transcription factors using IPA (Supplementary Figure S6). The interconnections between the nodes were visualized by adding direct interactions. Subsequently the most interconnected hubs were extracted to reveal the core regulatory network shown in Figure 7.
Figure 7.
Myb controls a core transcriptional regulatory network. Transcriptional factors directly targeted by Myb were used to construct a network using IPA. The most interconnected factors were subsequently extracted to construct a core regulatory network. Ellipses with gray background represent Myb activated transcription factors, whereas open ellipses with white background represent ones repressed by Myb.
Runx1 was identified as a Myb bound gene having multiple MBRs associated with (Supplementary Table S5). One of these MBRs was confirmed using independent ChIP-qPCR in both ERMYB and HL-60 cells (Figure 3). Runx1 was likely to be repressed by Myb albeit with an atypical kinetic profile of Myb repressed genes in our ERMYB expression profiling (due to the relatively small and insignificant fold changes from 0 to 6 h and from 6 to 24 h after β-E2 withdrawal, even though the change from 0 to 24 h is significant). Hence it was initially classified as a class C (not Myb-regulated) gene (Supplementary Table S2). Subsequently we validated the repression of Runx1 by Myb using qPCR in both ERMYB cells and Myb transduced bone marrow cells (Supplementary Tables S3 and S4). Based on these results, we conclude that Runx1 is a direct Myb repressed target. Therefore it was included as a direct Myb target gene in Supplementary Tables S7 and S8, and was also included in the pathway analysis and network construction using the IPA program.
RESULTS
Expression analysis of Myb regulated genes
We used the ERMYB cell line (21) as a model system to identify genes regulated by Myb using whole genome microarray expression profiling. ERMYB is a myeloid progenitor cell line derived by transformation of primary fetal liver cells by an activated form of Myb (CT3-Myb) fused to the ligand binding domain of the estrogen receptor α (ERα). This model system offers several advantages for characterizing the Myb transcriptional program. First, these cells strictly require activation of the Myb fusion protein by β-estradiol (β-E2) to maintain a proliferative progenitor-like phenotype and undergo monocytic differentiation when Myb is inactivated by withdrawal of β-E2. Thus this model is ideal for identifying those genes accounting for Myb’s transforming and differentiation-suppressing activities. Second, this model system provided us with a unique opportunity to distinguish between direct and indirect effects of Myb, by integrating expression profiling and genome occupancy data (which currently is technically very difficult in other cell lines or primary cells). Third, re-activation of Myb within 24 h after β-E2 withdrawal can fully reverse the differentiation of these cells, allowing us to investigate the dynamic pattern of target gene expression in response to Myb inactivation and re-activation (depicted in Figure 1A). We reasoned that this strategy is more likely to identify true Myb target genes than conventional approaches in which differential expression is examined only at the endpoint of induction. Finally, the ER-Myb fusion transcript is expressed at levels very similar to physiological levels of endogenous Myb mRNA in primary bone marrow c-Kithigh progenitor cells (Figure 1B), while endogenous Myb protein is undetectable in ERMYB cells (21). Findings from this model system should thus enable us to more accurately define Myb’s actions than would be the case with a simple overexpression system.
We performed whole genome expression profiling with cells from the β-E2 withdrawal and re-addition time course (Figure 1A) and grouped genes according to their kinetic profiles (Supplementary Table S2). We reasoned that genuine Myb target genes (class A) should manifest kinetic profiles consistent with Myb activity: Myb activated genes should be down-regulated when Myb activity is switched off and up-regulated when Myb is turned on again, while Myb repressed genes should show the opposite profiles (Figure 1C). The second group of genes (class B) did not conform to class A profiles but responded partially to inactivation and re-activation of Myb (Supplementary Figure S1). That is, they showed at least one significant response to either β-E2 withdrawal or re-addition. In fact some established Myb target genes, such as Mpo (38) and Elane (39), were found within this class (Supplementary Table S2). The third group (class C) included genes with profiles incompatible with direct regulation by Myb, genes with unperturbed expression and genes not detectably expressed.
In total, we identified 2368 Myb regulated genes (2667 probe IDs) of classes A and B, of which 1171 genes were activated and 1197 genes were repressed by Myb (Supplementary Table S2). Of these we selected 89 candidates and re-examined their expression using quantitative PCR (qPCR) with TaqMan low density arrays (TLDAs, see Supplementary Table S3). qPCR results for 75 genes (84.3%) in ERMYB cells were consistent with the microarray results (Figure 1D and Supplementary Table S3).
We carried out further validation in mouse primary bone marrow cells which were transduced with retroviruses encoding either wild-type Myb (wt-Myb) or an activated form, CT3-Myb, and then cultured in the presence of granulocyte-macrophage colony stimulating factor (GM-CSF) for 7 days. Under these conditions, vector transduced control cells strongly down-regulated Myb (Supplementary Table S4) and underwent myeloid differentiation. In contrast, both wt-Myb and CT3-Myb virus transduced (and transformed) cells overexpressed Myb and did not differentiate (15). Note that Myb overexpression was relatively modest in this system, with wt- and CT3 forms being expressed at ∼3 and ∼5 times, respectively, the level found in Kithigh progenitors (i.e. at Day 0). We again measured expression of the 89 candidate Myb regulated genes using TLDAs. The majority of candidate genes examined (60 genes; 67.4%) showed expression patterns consistent with Myb regulation in that they maintained activation or repression in the presence of ectopic wt-Myb or CT3-Myb expression (Figure 1D and Supplementary Table S4). Notably, the impacts of wt-Myb and CT3-Myb on their target genes largely overlap. That is, 88.3% of the validated genes were consistently regulated by both wt-Myb and CT3-Myb (Supplementary Table S4). However, expression changes caused by CT3-Myb were generally greater than those by wt-Myb, especially of those commonly repressed genes (38 of 39; Supplementary Table S2).
Global analysis of MBRs in vivo
We next examined genome-wide Myb occupancy pattern in ERMYB cells using ChIP-Seq. We first tested the ChIP performance of antibodies recognizing either the ER moiety (ER-10 antibody) or the CT3-Myb portion [anti-CT5 (29) and Thelma (30) sera] of the ER-Myb fusion protein. We detected strong occupancy of Myb at a known Myb binding site in Mpo proximal promoter [−0.32 kb; (38)] using either anti-ER antibody or anti-Myb sera (Figure 2A). Due to limited supply of the anti-CT5 sera and lack of commercially available Myb antibodies recognizing the CT3-Myb portion of the ER-Myb fusion protein (most of these recognize the C-terminus of wt-Myb protein which is absent in the ER-Myb fusion protein), we carried out ChIP-Seq using the ER antibody (see ‘Materials and Methods’ section for more information).
Genome-wide maps of in vivo Myb chromatin occupancy were generated using ChIP-Seq with two biological replicate samples from ERMYB cells with activated and inactivated Myb (i.e. 0 and 6 h; Figure 1A). We first examined raw ChIP-Seq data across loci corresponding to two known Myb target genes (Figure 2B), Mpo (38) and Bcl2 (40,41). In addition to the previously reported binding site within the proximal Mpo promoter (−0.3 kb relative to the TSS), we identified two peaks further upstream (−3.5 and −4.2 kb). These peaks were absent or greatly diminished when Myb was inactivated by β-E2 withdrawal. Although it was suggested previously that Myb activates Bcl2 transcription by binding to its proximal promoter in T cells (40,41), we were unable to detect any significant Myb occupancy of the Bcl2 promoter. Instead, we found one intronic peak (+93.9 kb) and two downstream peaks (+196.5 and +198.5 kb, respectively). This indicates that Myb may bind to different locations to regulate the same target in different cell types. In fact we have detected Myb binding to both the proximal promoter region and two conserved downstream binding sites (+93.9 and +198.5 kb) of Bcl2 in human breast cancer cells (42).
We identified candidate MBRs using the MACS algorithm (31), and removed those inconsistent between replicate samples (Supplementary Figure S7A) using the EdgeR package (33). These candidates were further filtered for sequence tag number and fold change between samples with activated and inactivated Myb, resulting in the identification of 11 429 candidate MBRs (see also ‘Materials and Methods’ section).
To determine the preferred sequences bound by Myb in vivo, we interrogated the sequences of these candidate MBRs for overrepresented DNA motifs using the MEME program (34). Notably, we identified the Myb binding motif itself to be the most prevalent sequence motif, which shares the same AACNG core with the known Myb binding consensus (Transfac V$MYB_Q6) but differed at flanking positions (Figure 2C). These Myb binding sequences were centered on peak summits as expected (Supplementary Figure S2A). We then applied this consensus sequence filter to further refine the candidate MBRs. The resultant set of 7646 high-confidence MBRs corresponds to 4892 unique genes (hereafter defined as Myb bound genes; Supplementary Table S5), and includes many Myb target genes reported previously in other systems (Supplementary Table S1).
The majority of identified MBRs is distal from the nearest TSS although there is enrichment near TSS (Figure 2D), with only 18% of MBRs occurred within gene promoters [defined here as ±2.5 kb to the TSS; (43)]. Further analysis of MBR distribution relative to annotated Refseq gene features (Figure 2E) revealed that occupancy was predominantly intergenic (46%) and within introns (35%).
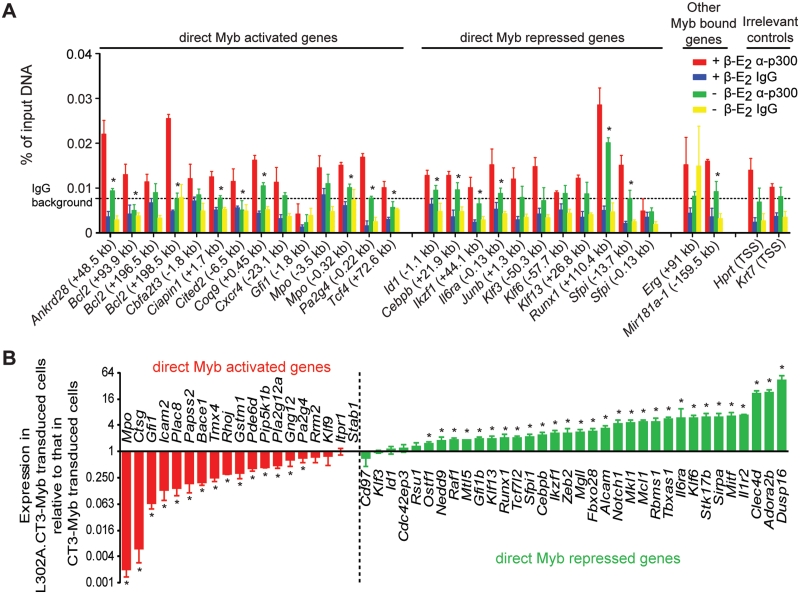
To validate our ChIP-Seq findings, we first performed qPCR on samples from an independent ChIP using ER antibody with primers for 23 high-confidence MBRs, representing those associated with Myb activated, repressed or other bound genes. As shown in Figure 3A, Myb occupancy at all 23 MBRs was significantly higher than that of four irrelevant control regions, and decreased significantly upon Myb inactivation by β-E2 withdrawal. Additionally, we selected five MBRs which had EdgeR P-values just above the empirical cutoff (10−10; see ‘Materials and Methods’ section) but had low MACS P-values, of which, four were also validated (Figure 3A). This suggests that while our list of identified MBRs is of high-confidence, there are probably additional MBRs that remain to be validated.
We further validated these MBRs by ChIP-qPCR using anti-Myb sera (anti-CT5 and Thelma sera). Twenty-five of 27 MBRs validated in ChIP using ER antibody (Figure 3A) were also validated in ChIP using anti-Myb sera (Figure 3B), further confirming that the MBRs we identified in ChIP-Seq were indeed occupied by Myb in vivo. The two MBRs that were validated using anti-ER antibody but not validated using anti-Myb sera (Gfi1 −1.8 kb and Sfpi1 −0.13 kb) were probably due to the high background of the anti-Myb sera (Figure 3A and B). In fact, both of these MBRs were validated in HL-60 cells (Figure 3C; see below).
To provide further confidence, we carried out ChIP using pooled MYB antibodies (Myb 1-1 antibody and Thelma sera) in HL-60 cells, human leukemic pro-myeloblasts that represent a similar stage to ERMYB cells in myeloid development. We demonstrated that endogenous MYB occupied 20 of 21 conserved MBRs identified in mouse ChIP-Seq (Figure 3C), albeit with significantly lower signals than the ER-Myb fusion protein which was probably due to the lower ChIP efficiency of the MYB antibodies used. These results confirm that our approach was highly effective in identifying bona fide Myb-occupied sites in vivo.
Identification of direct Myb target genes
We next assessed whether the identified MBRs pinpoint Myb regulated genes by integrating the expression profiling data with the Myb chromatin occupancy data. As illustrated in Figure 4A, of 4892 high-confidence Myb-bound genes, 793 are, as members of classes A or B, transcriptionally regulated by Myb and thus are likely to represent direct Myb target genes. Of those, 418 were activated and 375 genes were repressed by Myb (Supplementary Table S7). The fact that we identified similar numbers of direct Myb activated and repressed targets indicates that Myb can genuinely function as a transcriptional repressor in addition to its established role as a transactivator.
To cross validate our findings, we examined a published set of differentially expressed genes upon siRNA mediated MYB knockdown (MYBKD) in human THP-1 leukemia cells (19). This revealed a large overlap between this set and our sets of Myb bound or Myb regulated genes (Figure 4B and Supplementary Table S8): 1063 genes differentially expressed upon MYBKD were bound by Myb in ERMYB cells; and 462 genes differentially expressed in MYBKD THP-1 cells were also regulated by Myb in ERMYB cells. Moreover, GSEA (37) showed that the top 200 Myb-activated or -repressed genes (ranked by fold change) identified in ERMYB cells were significantly enriched in the THP-1 MYBKD set, and vice versa (Supplementary Table S9).
Myb has been shown to be a critical downstream mediator of Hoxa9 and Meis1signaling, and is essential for transformation by these two oncogenes (13). We examined the up- or down-regulated gene sets by Hoxa9/Meis1, reasoning that at least part of these gene signatures should be imposed by Myb, if Myb is a essential downstream target of this signaling pathway. Using GSEA, we found this is indeed the case: the two gene sets were very significantly overrepresented in either Myb activated genes or repressed genes identified in ERMYB cells (Supplementary Table S9), thereby validating our set of Myb regulated genes.
Characterization of MBRs associated with direct Myb target genes
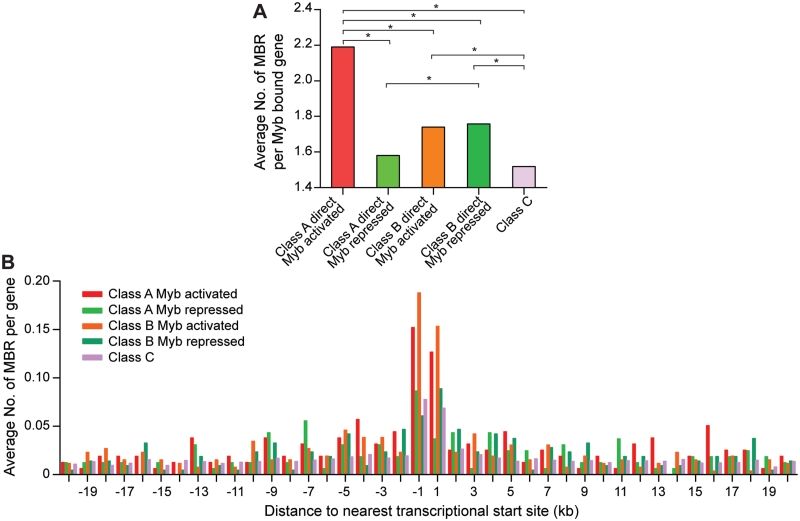
The majority of Myb bound genes (80.7%; Figure 4A) were not clearly regulated by Myb. In order to gain insights into why some Myb bound genes were regulated by Myb whereas others not, we compared the average number of associated MBRs per gene for Myb bound genes in different microarray classes (Figure 5A). Direct Myb targets (bound genes in classes A and B) appeared to have significantly more MBRs than those bound but not Myb-regulated or not expressed (class C), with the highest enrichment seen for Myb bound and activated genes in class A. Consistently, distribution analysis of MBRs relative to genes in each microarray class revealed higher enrichment of MBRs near TSS of Myb activated genes (classes A and B) compared with genes not regulated or expressed (class C) whereas Myb repressed genes (classes A and B) appear to have more MBRs at locations further to TSS (Figure 5B).
Figure 5.
MBRs were enriched near Myb target genes. (A) Direct Myb targets (classes A and B) generally have significantly more MBRs than those Myb bound but not regulated or not expressed genes (class C). *P < 0.05 (Wilcoxon rank sum test). (B) MBRs are enriched in the proximity of TSS of Myb activated genes (classes A and B). Myb repressed genes appeared to have more MBRs at locations relatively distal to TSS. Average numbers of MBR per gene of each microarray class were plotted against their distances relative to the nearest TSS in 1 kb bins up to 20 kb away from TSS.
To identify other transcription factors that might contribute to Myb regulation of its target genes, we analyzed overrepresented sequence motifs within MBRs (100 bp window from either side of peak summit) associated with Myb activated, repressed and not Myb-regulated/not expressed genes. We then compared these motifs to the Transfac database to identify putative corresponding factors (Supplementary Figure S2C). We found that the binding motifs for Runx1 and for Ets family members, such as Ets1/2, Sfpi1, Evx1/2 and Elf1, were enriched in all three categories. These results are consistent with previous reports that Myb interacts with these factors to regulate target gene expression (39,44–46). Interestingly, motifs resembling a half-MARE (Maf response element) for large Maf proteins were enriched in Myb repressed or not Myb-regulated/not expressed genes. Maf has been shown to interact with Myb and repress Myb transactivation of certain targets in both myeloid and T lineages (47–49). Reciprocally, Myb can repress Mafb transactivation in a SUMO-dependant manner (50). Taken together, these results suggest that Maf/Mafb may be cofactors important for Myb repression of some genes.
p300 occupies MBRs in vivo, and is required for repression and activation of Myb targets
Cbp/p300 is a known co-activator of Myb (51,52). Mice carrying point mutations of p300 which disrupt the Myb interaction site (53,54) partially resemble the phenotypes of Myb hypomorphic mice (55,56). Furthermore, we have shown that interaction with Cbp/p300 is essential for transactivation and transformation by Myb (15). Hence we examined our set of validated MBRs for Cbp/p300 occupancy in ERMYB cells. We detected p300 occupancy at 25 of 27 MBRs, regardless of whether they were associated with Myb activated or repressed genes (Figure 6A). Moreover, the p300 occupancy followed the changes in Myb occupancy: decreased occupancy of both Myb and p300 was evident when β-E2 was withdrawn from the cells. In contrast, Cbp occupancy was only detected at few MBRs, and moreover, did not parallel Myb occupancy (Supplementary Figure S3).
Figure 6.
p300 was required for both activation and repression by Myb. (A) p300 occupancy was detected at 25 of 27 MBRs and the occupancy generally decreased when Myb was inactivated by β-E2 withdrawal. Data are presented as mean ± SD. IgG background represents the (mean + 1.64 SD) of IgG signals across all regions. ‘Asterisk’ denotes significant decrease in Myb occupancy upon β-E2 withdrawal (Student’s t-test, P < 0.05). (B) L302A.CT3-Myb did not maintain the repression or activation of direct Myb target genes by CT3-Myb. Expression of 53 direct Myb target genes was shown for L302A.CT3-Myb transduced bone marrow cells (y-axis, log2 scale) relative to CT3-Myb transduced cells (set to 1). ‘Asterisk’ denotes significant difference between the two samples (Student’s t-test, P < 0.05).
To further elucidate the role of p300 interaction in transcriptional regulation by Myb, we transduced mouse primary bone marrow cells with retroviruses encoding either CT3-Myb or L302A.CT3-Myb (15) and cultured them in GM-CSF for 7 days as in the previous validation experiment. The latter Myb mutant carries the L302A point mutation which completely abolishes the Myb-p300 interaction (51). We measured and compared the expression of 53 direct Myb target genes (validated previously; Supplementary Table S4) in cells expressing these two forms of Myb. Consistent with the established role of p300 as a co-activator for Myb (51,52), we found (Figure 6B) that expression of Myb activated genes was significantly decreased in cells expressing L302A.CT3-Myb compared to CT3-Myb (15 of 19 genes, 78.9%). Interestingly, the expression of Myb repressed genes was significantly increased in cells expressing L302A.CT3-Myb (29 of 34 genes, 85.3%), suggesting that transcriptional repression by Myb was relieved by abolishing p300 interaction. Taken together, these data suggest that p300 has a hitherto-unappreciated role in mediating transcriptional repression by Myb, in addition to its established role as coactivator.
We were also interested in whether we could detect p300-mediated activation and repression of Myb targets in primary myeloid progenitor cells expressing only endogenous Myb. To this end we purified GMPs (23) from wild-type mice and from the ‘booreana’ mutant strain. The latter carries a mutation (E308G) in Myb which greatly reduces or abolishes its association with p300 (24). qPCR analysis of a set of 11 Myb-activated and 11 Myb-repressed genes (Supplementary Figure S4) showed that expression of all of the Myb-activated genes was significantly decreased in the ‘booreana’ GMPs, reinforcing p300′s role as a physiological coactivator of wt-Myb. Expression of six of the Myb-repressed genes identified above was significantly increased in the ‘booreana’ GMPs, indicative of reduced repression, as anticipated. However, the magnitude of the increase was substantially less than that seen in the ectopic expression study of Figure 6B, and moreover, no significant increase was seen in the levels of the other five genes. The implications of these data are discussed below.
Myb modulation of histone modification
p300 has histone acetyltransferase activity that contributes to its function as a transcriptional coactivator (57). We therefore examined acetyl-histone 3 (H3ac) levels at our set of validated MBRs. Most of these regions showed significant H3ac levels, consistent with the fact that the corresponding genes were detectably expressed. However, changes in H3ac upon dissociation of Myb and p300 were detected at only 7 of 27 MBRs and were not consistent with changes in p300 occupancy in most cases (Supplementary Figure S5A). Moreover, changes in H3ac levels generally correlated poorly with the expression changes of corresponding genes.
We next examined the set of validated MBRs for other epigenetic markers, including two activating histone marks—trimethylated histone H3 lysine 4 (H3K4me3) and the monomethylated form (H3K4me1) (58), and two repressive marks—trimethylated form of histone H3 lysine 9 (H3K9me3) and lysine 27 (H3K27me3) (59). Most MBRs examined are ‘bivalent’ as they had high levels of both activating and repressive markers (Supplementary Figure S5B–E).
Upon dissociation of Myb, we detected significantly increased levels of the heterochromatin marker H3K9me3 at most MBRs (23 of 27, Supplementary Figure S5D), regardless of whether they are associated with Myb-activated or -repressed genes. This suggests Myb may play a general role in maintaining open chromatin structure at its target loci. We also detected a significant increase in another repressive mark H3K27me3 at 6 of 14 MBRs associated with Myb activated genes (Supplementary Figure S5E). This implies that, at least at a subset of Myb activated genes, Myb may help to inhibit the recruitment of the polycomb repressor complex PRC2 [responsible for H3K27me3; (59)], thereby activating target gene transcription. The levels of the activating promoter mark H3K4me3 upon Myb dissociation were increased at most of MBRs examined (Supplementary Figure S5B), albeit with relatively small fold changes. The levels of enhancer marker H3K4me1 were largely unchanged (Supplementary Figure S5C).
These observations raise the possibility that Myb may form similar complexes at both activated and repressed targets which result similar changes in histone modifications, and that the actual effects of these epigenetic changes may depend largely on local context, previous epigenetic memory and/or other unexamined histone modifications. It may also be that the major, immediate effects of Myb on target gene transcription may be mediated by other mechanisms or activities such as direct effects on the core transcriptional machinery.
Gene signatures for cancer, stem cells and myeloid development are overrepresented in the Myb transcriptional program
As discussed above activation of Myb is widely associated with leukemia and it is expressed in most leukemias even in the absence of genetic alteration. In agreement with this, GSEA (Table 1) demonstrated that the MLL LSC gene signature was highly enriched in Myb-regulated genes (in the appropriate directions). Gene sets comprising commonly up-regulated genes in cancer compared with normal tissue, or undifferentiated cancer compared with well differentiated cancer, were also significantly enriched for Myb-regulated genes. Consistent with an important MYB contribution to human acute myeloid leukemia (AML), gene sets associated with several subtypes of AML (such as CBFb-MYH11, MLL-fusion, AML1-ETO and other CBF subtypes), were all (negatively) enriched in Myb-repressed genes, whereas a gene set down-regulated in nucleophosmin-mutated (NPMc+) AML was positively enriched. This suggests that Myb repressed genes may be characteristic of some subtypes of AML (and is consistent with the importance of Myb-mediated repression in leukemia), while in NPMc+ AML, Myb activity may be reduced, resulting in down-regulation of its activated target genes.
Table 1.
Gene signatures for cancer, stem cells and myeloid development are overrepresented in the Myb transcriptional program
| Gene set name | NES | FDR |
|---|---|---|
| Cancer gene sets | ||
| Up-regulated gene set in MLL LSC (20) | 2.61 | 0 |
| Down-regulated gene set in MLL LSC | −2.80 | 0 |
| ALCALAY_AML_NPMC_DN (73) | 1.68 | 0.01 |
| ROSS_CBF_MYH (74) | −2.00 | 0.00 |
| ROSS_AML1_ETO | −1.58 | 0.04 |
| ROSS_MLL_FUSION | −1.55 | 0.05 |
| ROSS_CBF | −1.46 | 0.09 |
| CANCER_UNDIFFERENTIATED_ META_UP (75) | 2.16 | 0 |
| CANCER_NEOPLASTIC_META_UP | 1.94 | 0.00 |
| Stem cell gene sets | ||
| HSC_EARLYPROGENITORS_FETAL (76) | 2.07 | 0 |
| HSC_EARLYPROGENITORS_ADULT | 2.06 | 0 |
| HSC_INTERMEDIATEPROGENITORS_ FETAL | 1.85 | 0.00 |
| HSC_INTERMEDIATEPROGENITORS_ ADULT | 1.82 | 0.00 |
| HSC_MATURE_ADULT | −1.96 | 0.00 |
| HSC_MATURE_FETAL | −1.42 | 0.11 |
| Myeloid development gene sets | ||
| BROWN_MYELOID_PROLIF_AND_ SELF_RENEWAL (77) | 2.48 | 0 |
| BROWN_GRAN_MONO_DIFFERENTIATION | −2.84 | 0 |
| MYELOID_CELL_DIFFERENTIATION (78) | −1.83 | 0.01 |
| LIAN_MYELOID_DIFF_RECEPTORS (79) | −1.79 | 0.01 |
NES, normalized enrichment score; FDR, false discovery rate.
Myb is essential for maintaining the adult HSC pool (3). Consistently, gene sets associated with HSCs and progenitors were positively enriched, whereas gene sets up-regulated in mature blood cells were negatively enriched in the Myb transcriptional program. Moreover, consistent with the essential role Myb plays in myeloid development and myeloid transformation (1), gene sets associated with myeloid proliferation and self-renewal were significantly enriched in Myb activated genes while gene sets for myeloid differentiation were overrepresented in Myb repressed genes.
Functional analysis of identified direct Myb targets using the IPA revealed significant enrichment of genes associated with terms such as ‘hematological system development and function’ and hematopoiesis (Table 2). Other significantly overrepresented molecular function terms (Table 2) included cell death, cell growth and proliferation, cell cycle and cell signaling. Collectively, these results re-emphasized the importance of Myb in hematopoiesis and pinpointed the key cellular pathways regulated by Myb.
Table 2.
Significantly enriched pathway terms in direct Myb target genes
| Name | P-value | No. of genes |
|---|---|---|
| Physiological system development and function | ||
| Hematological system development and function | 1.47E-11–4.46E-03 | 133 |
| Hematopoiesis | 3.51E-10–4.05E-03 | 85 |
| Tissue morphology | 2.71E-08–4.19E-03 | 82 |
| Cell-mediated immune response | 5.94E-08–2.99E-03 | 54 |
| Skeletal and muscular system development and function | 2.81E-07–2.89E-03 | 22 |
| Organismal survival | 1.06E-06–1.11E-06 | 78 |
| Tissue development | 4.47E-06–4.24E-03 | 80 |
| Immune cell trafficking | 1.35E-05–4.24E-03 | 58 |
| Nervous system development and function | 5.23E-05–2.61E-03 | 35 |
| Humoral immune response | 8.54E-05–4.46E-03 | 36 |
| Molecular and cellular functions | ||
| Cell death | 1.25E-16–4.46E-03 | 230 |
| Cellular growth and proliferation | 3.74E-11–4.05E-03 | 231 |
| Cellular development | 8.90E-10–4.05E-03 | 182 |
| Cellular function and maintenance | 2.99E-08–4.46E-03 | 73 |
| Cell cycle | 4.47E-07–2.61E-03 | 101 |
| Cellular movement | 5.75E-07–3.07E-03 | 117 |
| Cellular morphology | 7.61E-07–4.18E-03 | 102 |
| Cell-to-cell signaling and interaction | 4.47E-06–4.11E-03 | 133 |
| Cell signaling | 1.43E-05–4.16E-03 | 58 |
| Carbohydrate metabolism | 5.62E-05–2.61E-03 | 10 |
Myb controls a core transcriptional regulatory network
We identified 75 transcription factors as direct Myb targets. We reasoned that such factors are likely to be critical components of the Myb transcriptional program, since in principle they can both crosstalk with each other and regulate other cascades of genes that ultimately define cellular phenotypes.
We therefore constructed a Myb-centric regulatory network comprising Myb and its target transcriptional factors with inferred regulatory interactions among these factors from IPA (Supplementary Figure S6). The most interconnected hubs were then extracted to construct a core regulatory network (illustrated in Figure 7). Interestingly, 8 of the 10 transcriptional hubs were repressed by Myb while only 2 were activated. Many of the repressed hubs are key positive regulators of myeloid differentiation, such as Runx1, Sfpi1, Junb and Cebpb (60). The activated hubs include Myc, a well established oncogene and pluripotency factor (61) and Gfi1, which is essential for HSC maintenance and neutrophil differentiation (62,63). As discussed below, this transcriptional regulatory network is likely to collectively account for several of Myb’s key activities, such as promoting self-renewal and suppressing differentiation.
DISCUSSION
The Myb transcriptional program and Myb function
In this study, we describe the transcriptional program controlled by Myb in a myeloid progenitor-like cell line derived by transformation of primary cells by Myb itself. Integration of genome-wide chromatin occupancy data and a comprehensive data set of Myb induced gene expression changes allowed us to identify direct Myb targets by correlating in vivo occupancy and transcriptional activity. A major finding reported here is the opposing regulatory effect of Myb on the transcription of different targets. This is reminiscent of the recent finding that Drosophila Myb functions as both transcriptional activator and repressor in a promoter-dependent manner (64). Consistent with its established role as a transcriptional activator, we found Myb bound and activated 418 genes (Figure 4A and Supplementary Table S7), thus extending the network of genes that Myb directly activates. Consistent with our findings, two recent studies (19,65) showed that many genes were up-regulated upon knockdown of MYB. However, this was attributed to indirect mechanisms, i.e. Myb activated other repressors to repress these genes. In direct contrast, we demonstrate, for the first time, that Myb directly repressed approximately half of its target genes (375 genes; Figure 4A and Supplementary Table S7), indicating that transcriptional repression is also a fundamental and major activity of Myb.
Knockdown of MYB induced a differentiation-like phenotype in THP-1 cells (19), as does inactivation of Myb in ERMYB cells (21). In contrast, overexpression of MEnT, a dominant negative construct which effectively inhibits Myb transactivation (41), did not induce differentiation of HL-60 cells (48). These findings pointed to the fact that simple inhibition of transactivation by Myb is not sufficient to suppress differentiation. This is consistent with our findings that Myb directly represses several key positive transcriptional regulators of hematopoietic differentiation, including Runx1, Sfpi1, Cebpb and Junb. Together these observations support the notion that repression by Myb is essential for its differentiation-suppressing and transforming activities, and conversely, that de-repression of certain Myb targets, such as the transcription factors mentioned above, might be essential to allow proper differentiation. This notion is embodied in the core regulatory network in Figure 7.
It is however certain that other transcription factors (such as Cited2), and genes other than those encoding transcription factors also contribute to transformation and suppression of differentiation by Myb. As shown in Table 2, Myb regulated genes involved in several pathways important for transformation, proliferation and differentiation, including cell survival (such as Bcl2 and Ciapin1), cellular growth and proliferation (such as Mapk3 and Tcf7l2) and cell signaling (such as Csf1 and Gnai2).
A complex relationship between Myb repression, p300 and chromatin modification
Transactivation by Myb has been relatively well characterized. It requires the transactivation domain (66) and interaction with the coactivator Cbp/p300 (51,52). However, it is far less clear how Myb represses its targets, although several models have been proposed. One model (67) suggests that subtle differences in flanking sequences of Myb binding sites could potentially lead to conformational changes in Myb upon binding to DNA and subsequent recruitment of corepressors [several of which are known to associate with Myb; (68)] instead of coactivators. However, we found that the consensus sequences immediately flanking the core ‘AACNG’ were almost identical between genes directly activated by Myb and those that were not Myb-regulated, with only a very slight difference between direct Myb activated and repressed genes (Supplementary Figure S2B). These observations do not favor the above-mentioned model.
Myb-mediated repression may also involve competition with other positive transcription regulators as in the case of ERBB2 promoter, where Myb competes with TBP (69). Alternatively, Myb may require cell type- or developmental stage-specific factors to function as a repressor. For example, repression of the Csf1r promoter by Myb occurs in macrophages but not in fibroblasts (45). This is consistent with our findings that motifs for many hematopoietic specific factors, such as Runx1, Ets factors and Maf/Mafb are found adjacent to Myb binding motifs (Supplementary Figure S2C). However, several of these are common to Myb activated and repressed genes, so cooperation with adjacently-bound factors is unlikely to explain repression in many cases.
We propose another possible mechanism here based on the fact that p300 was required for repression by Myb. This is entirely consistent with our previous finding that Cbp/p300 interaction is required for transformation by Myb (15), and the notion discussed above that repression by Myb is essential for transformation. Taken together, this would suggest the seemingly paradoxical notion that the coactivator p300 plays an essential role in repression by Myb. Although rarely, p300 might contribute directly to transcriptional repression (70). However, we suggest that the Myb-p300 complex could suppress some Myb targets by activating transcription of repressive non-coding RNAs (ncRNAs). ncRNAs have been characterized as an important feature of the eukaryotic transcriptome (71), and include microRNAs, antisense transcripts and long intergenic non-coding RNAs, all of which have known or potential suppressive effects on gene expression. At least for some repressed targets, Myb may employ the same molecular mechanism—association with the p300 coactivator—for activation and repression; however in the latter case Myb may repress these targets via activating the transcription of interfering, gene-specific suppressive ncRNAs. Indeed, we found that Myb binds to a number of genes encoding microRNAs (Supplementary Table S5), among which, the binding of Myb to two genes encoding hematopoietic specific Mir181a-1 and Mir142 respectively (72) have been validated in both ERMYB and HL-60 cells (Figure 3 and Zhao,L. and Gonda,T.J. unpublished data).
This hypothesis is supported by our and other results. First, p300, a well established transcriptional coactivator, appears to co-occupy MBRs of both activated and repressed genes with Myb (Figure 6A). Second, Myb dissociation caused similar changes in H3K9me3 (and H3K4me3) with both activated and repressed genes (Supplementary Figure S5B and D). This argues in favor of the idea that Myb recruits the same set of proteins to its target genes regardless of whether they are to be activated or repressed. Third, the distribution patterns of MBRs near activated or repressed Myb targets show an apparent difference (Figure 5B): generally, activated genes have significantly higher enrichment of MBRs in the proximity of TSS whereas repressed genes tend to have more MBRs at locations relatively distal to TSS. These distal sites may be potential promoters that are activated upon Myb binding and initiate transcription of the suppressive ncRNAs.
We also found that in primary GMPs, p300-dependent repression of those genes identified as repressed by activated or ectopically-expressed Myb was less apparent. This is consistent with the notion that transcriptional repression of critical targets is a key activity of Myb in a transforming/leukemic context, which as we discuss is marked by suppression of differentiation. This is also in line with our findings that gene signatures of several AML subtypes are enriched in Myb repressed genes (Table 1). One might expect less suppression of differentiation by Myb in normal progenitors as these cells do in fact differentiate.
To summarize, our data provide a genome-wide analysis of Myb chromatin occupancy and target gene expression. We demonstrated for the first time that, in addition to its established role as a transactivator, Myb directly repressed a substantial number of genes including key regulators of myeloid differentiation. This may shed some light on the mechanisms of how Myb, and possibly other oncogenes, transform hematopoietic cells and cause malignances. We also provide evidence that the interaction with p300 is required for both transcriptional activation and repression by Myb, and suggest that the latter may quantitatively distinguish Myb activity in transformed versus normal contexts. Finally, this data set should provide a valuable resource for further study of the transcriptional regulation of hematopoietic development and leukemic transformation.
ACCESSION NUMBERS
The ChIP-Seq and expression profiling data have been deposited in the Gene Expression Omnibus (GEO) database under accession numbers GSE22095 and GSE22486, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council of Australia (grant 569639 to T.J.G.); Australian Cancer Research Foundation, Infrastructure grant; Leukemia Foundation (Australia), PhD scholarship (to D.R.P.). Funding for open access charge: Diamantina Institute central funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Michael Tallack and Andrew Perkins (Institute for Molecular Bioscience, University of Queensland, Australia) for advice about ChIP assays, Dr Shunsuke Ishii (RIKEN, Tsukuba, Japan) for the CT5 Myb antibody and Dr Michael Rist (University of Queensland Diamantina Institute) for assistance with cell sorting.
REFERENCES
- 1.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat. Rev. Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 2.Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, Potter SS. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 3.Lieu YK, Reddy EP. Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc. Natl Acad. Sci. USA. 2009;106:21689–21694. doi: 10.1073/pnas.0907623106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolff L, Koller R, Bies J, Nazarov V, Hoffman B, Amanullah A, Krall M, Mock B. Retroviral insertional mutagenesis in murine promonocytic leukemias: c-myb and Mml1. Curr. Top. Microbiol. Immunol. 1996;211:191–199. doi: 10.1007/978-3-642-85232-9_19. [DOI] [PubMed] [Google Scholar]
- 5.Lipsick JS, Wang DM. Transformation by v-Myb. Oncogene. 1999;18:3047–3055. doi: 10.1038/sj.onc.1202745. [DOI] [PubMed] [Google Scholar]
- 6.Lahortiga I, De Keersmaecker K, Van Vlierberghe P, Graux C, Cauwelier B, Lambert F, Mentens N, Beverloo HB, Pieters R, Speleman F, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat. Genet. 2007;39:593–595. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 7.Murati A, Gervais C, Carbuccia N, Finetti P, Cervera N, Adelaide J, Struski S, Lippert E, Mugneret F, Tigaud I, et al. Genome profiling of acute myelomonocytic leukemia: alteration of the MYB locus in MYST3-linked cases. Leukemia. 2009;23:85–94. doi: 10.1038/leu.2008.257. [DOI] [PubMed] [Google Scholar]
- 8.Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela J-M, Dik WA, Langerak AW, Montpellier B, Nadel B, Walrafen P, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–1261. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 9.O’Neil J, Tchinda J, Gutierrez A, Moreau L, Maser RS, Wong KK, Li W, McKenna K, Liu XS, Feng B, et al. Alu elements mediate MYB gene tandem duplication in human T-ALL. J. Exp. Med. 2007;204:3059–3066. doi: 10.1084/jem.20071637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castaneda VL, Parmley RT, Saldivar VA, Cheah MS. Childhood undifferentiated leukemia with early erythroid markers and c-myb duplication. Leukemia. 1991;5:142–149. [PubMed] [Google Scholar]
- 11.Gorello P, La Starza R, Varasano E, Chiaretti S, Elia L, Pierini V, Barba G, Brandimarte L, Crescenzi B, Vitale A, et al. Combined interphase fluorescence in situ hybridization elucidates the genetic heterogeneity of T-cell acute lymphoblastic leukemia in adults. Haematologica. 2010;95:79–86. doi: 10.3324/haematol.2009.010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomita A, Watanabe T, Kosugi H, Ohashi H, Uchida T, Kinoshita T, Mizutani S, Hotta T, Murate T, Seto M, et al. Truncated c-Myb expression in the human leukemia cell line TK-6. Leukemia. 1998;12:1422–1429. doi: 10.1038/sj.leu.2401113. [DOI] [PubMed] [Google Scholar]
- 13.Hess JL, Bittner CB, Zeisig DT, Bach C, Fuchs U, Borkhardt A, Frampton J, Slany RK. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108:297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lidonnici MR, Corradini F, Waldron T, Bender TP, Calabretta B. Requirement of c-Myb for p210BCR/ABL-dependent transformation of hematopoietic progenitors and leukemogenesis. Blood. 2008;111:4771–4779. doi: 10.1182/blood-2007-08-105072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pattabiraman DR, Sun J, Dowhan DH, Ishii S, Gonda TJ. Mutations in multiple domains of c-Myb disrupt interaction with CBP/p300 and abrogate myeloid transforming ability. Mol. Cancer. Res. 2009;7:1477–1486. doi: 10.1158/1541-7786.MCR-09-0070. [DOI] [PubMed] [Google Scholar]
- 16.Patel G, Kreider B, Rovera G, Reddy EP. v-myb blocks granulocyte colony-stimulating factor-induced myeloid cell differentiation but not proliferation. Mol. Cell. Biol. 1993;13:2269–2276. doi: 10.1128/mcb.13.4.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke MF, Kukowska-Latallo JF, Westin E, Smith M, Prochownik EV. Constitutive expression of a c-myb cDNA blocks Friend murine erythroleukemia cell differentiation. Mol. Cell. Biol. 1988;8:884–892. doi: 10.1128/mcb.8.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonda TJ, Buckmaster C, Ramsay RG. Activation of c-myb by carboxy-terminal truncation: relationship to transformation of murine haemopoietic cells in vitro. EMBO J. 1989;8:1777–1783. doi: 10.1002/j.1460-2075.1989.tb03571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The FANTOM Consortium and the Riken Omics Science Center. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somervaille TCP, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, Chang HY, Shurtleff SA, Downing JR, Cleary ML. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogg A, Schirm S, Nakagoshi H, Bartley P, Ishii S, Bishop JM, Gonda TJ. Inactivation of a c-Myb/estrogen receptor fusion protein in transformed primary cells leads to granulocyte/macrophage differentiation and down regulation of c-kit but not c-myc or cdc2. Oncogene. 1997;15:2885–2898. doi: 10.1038/sj.onc.1201472. [DOI] [PubMed] [Google Scholar]
- 22.Zhao L, Neumann B, Murphy K, Silke J, Gonda TJ. Lack of reproducible growth inhibition by Schlafen1 and Schlafen2 in vitro. Blood Cells Mol. Dis. 2008;41:188–193. doi: 10.1016/j.bcmd.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 23.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 24.Papathanasiou P, Tunningley R, Pattabiraman DR, Ye P, Gonda TJ, Whittle B, Hamilton AE, Cridland SO, Lourie R, Perkins AC. A recessive screen for genes regulating hematopoietic stem cells. Blood. 2010;116:5849–5858. doi: 10.1182/blood-2010-04-269951. [DOI] [PubMed] [Google Scholar]
- 25.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 26.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 27.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodge D, Coghill E, Keys J, Maguire T, Hartmann B, McDowall A, Weiss M, Grimmond S, Perkins A. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107:3359–3370. doi: 10.1182/blood-2005-07-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanei-Ishii C, Nomura T, Tanikawa J, Ichikawa-Iwata E, Ishii S. Differential Sensitivity of v-Myb and c-Myb to Wnt-1-induced Protein Degradation. J. Biol. Chem. 2004;279:44582–44589. doi: 10.1074/jbc.M407831200. [DOI] [PubMed] [Google Scholar]
- 30.Ramsay RG, Ciznadija D, Mantamadiotis T, Anderson R, Pearson R. Expression of stress response protein glucose regulated protein-78 mediated by c-Myb. Int. J. Biochem. Cell Biol. 2005;37:1254–1268. doi: 10.1016/j.biocel.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Liu T, Meyer C, Eeckhoute J, Johnson D, Bernstein B, Nussbaum C, Myers R, Brown M, Li W, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat. Methods. 2009;6:S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey TL, Elkan C. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. Menlo Park, California: AAAI Press; 1994. pp. 28–36. [PubMed] [Google Scholar]
- 35.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahony S, Benos PV. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 2007;35:W253–W258. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Britos-Bray M, Friedman AD. Core binding factor cannot synergistically activate the myeloperoxidase proximal enhancer in immature myeloid cells without c-Myb. Mol. Cell. Biol. 1997;17:5127–5135. doi: 10.1128/mcb.17.9.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oelgeschlager M, Nuchprayoon I, Luscher B, Friedman AD. C/EBP, c-Myb, and PU.1 cooperate to regulate the neutrophil elastase promoter. Mol. Cell. Biol. 1996;16:4717–4725. doi: 10.1128/mcb.16.9.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salomoni P, Perrotti D, Martinez R, Franceschi C, Calabretta B. Resistance to apoptosis in CTLL-2 cells constitutively expressing c-Myb is associated with induction of BCL-2 expression and Myb-dependent regulation of bcl -2 promoter activity. Proc. Natl Acad. Sci. USA. 1997;94:3296–3301. doi: 10.1073/pnas.94.7.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor D, Badiani P, Weston K. A dominant interfering Myb mutant causes apoptosis in T cells. Genes Dev. 1996;10:2732–2744. doi: 10.1101/gad.10.21.2732. [DOI] [PubMed] [Google Scholar]
- 42.Drabsch Y, Robert R, Gonda T. MYB suppresses differentiation and apoptosis of human breast cancer cells. Breast Cancer Res. 2010;12:R55. doi: 10.1186/bcr2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shapiro LH. Myb and ets proteins cooperate to transactivate an early myeloid gene. J. Biol. Chem. 1995;270:8763–8771. doi: 10.1074/jbc.270.15.8763. [DOI] [PubMed] [Google Scholar]
- 45.Reddy MA, Yang BS, Yue X, Barnett CJ, Ross IL, Sweet MJ, Hume DA, Ostrowski MC. Opposing actions of c-ets/PU.1 and c-myb protooncogene products in regulating the macrophage-specific promoters of the human and mouse colony-stimulating factor-1 receptor (c-fms) genes. J. Exp. Med. 1994;180:2309–2319. doi: 10.1084/jem.180.6.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lutz PG, Houzel-Charavel A, Moog-Lutz C, Cayre YE. Myeloblastin is an Myb target gene: mechanisms of regulation in myeloid leukemia cells growth-arrested by retinoic acid. Blood. 2001;97:2449–2456. doi: 10.1182/blood.v97.8.2449. [DOI] [PubMed] [Google Scholar]
- 47.Peng S, Lalani S, Leavenworth JW, Ho IC, Pauza ME. c-Maf interacts with c-Myb to down-regulate Bcl-2 expression and increase apoptosis in peripheral CD4 cells. Eur. J. Immunol. 2007;37:2868–2880. doi: 10.1002/eji.200636979. [DOI] [PubMed] [Google Scholar]
- 48.Hegde SP, Zhao J, Ashmun RA, Shapiro LH. c-Maf induces monocytic differentiation and apoptosis in bipotent myeloid progenitors. Blood. 1999;94:1578–1589. [PubMed] [Google Scholar]
- 49.Hedge SP, Kumar A, Kurschner C, Shapiro LH. c-Maf interacts with c-Myb to regulate transcription of an early myeloid gene during differentiation. Mol. Cell. Biol. 1998;18:2729–2737. doi: 10.1128/mcb.18.5.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tillmanns S, Otto C, Jaffray E, DuRoure C, Bakri Y, Vanhille L, Sarrazin S, Hay RT, Sieweke MH. SUMO-modification regulates MafB driven macrophage differentiation by enabling Myb dependent transcriptional repression. Mol. Cell. Biol. 2007;27:5554–5564. doi: 10.1128/MCB.01811-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parker D, Rivera M, Zor T, Henrion-Caude A, Radhakrishnan I, Kumar A, Shapiro LH, Wright PE, Montminy M, Brindle PK. Role of secondary structure in discrimination between constitutive and inducible activators. Mol. Cell. Biol. 1999;19:5601–5607. doi: 10.1128/mcb.19.8.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dai P, Akimaru H, Tanaka Y, Hou DX, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 53.Kasper LH, Boussouar F, Ney PA, Jackson CW, Rehg J, van Deursen JM, Brindle PK. A transcription-factor-binding surface of coactivator p300 is required for haematopoiesis. Nature. 2002;419:738–743. doi: 10.1038/nature01062. [DOI] [PubMed] [Google Scholar]
- 54.Kauppi M, Murphy JM, de Graaf CA, Hyland CD, Greig KT, Metcalf D, Hilton AA, Nicola NA, Kile BT, Hilton DJ, et al. Point mutation in the gene encoding p300 suppresses thrombocytopenia in Mpl-/- mice. Blood. 2008;112:3148–3153. doi: 10.1182/blood-2007-10-119677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, Cooke MP. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev. Cell. 2005;8:153–166. doi: 10.1016/j.devcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 56.Carpinelli MR, Hilton DJ, Metcalf D, Antonchuk JL, Hyland CD, Mifsud SL, Di Rago L, Hilton AA, Willson TA, Roberts AW, et al. Suppressor screen in Mpl-/- mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc. Natl Acad. Sci. USA. 2004;101:6553–6558. doi: 10.1073/pnas.0401496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalkhoven E. CBP and p300: HATs for different occasions. Biochem. Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 58.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 59.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 60.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 61.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hock H, Hamblen MJ, Rooke HM, Schindler JW, Saleque S, Fujiwara Y, Orkin SH. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 63.Karsunky H, Zeng H, Schmidt T, Zevnik B, Kluge R, Schmid KW, Duhrsen U, Moroy T. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 2002;30:295–300. doi: 10.1038/ng831. [DOI] [PubMed] [Google Scholar]
- 64.Georlette D, Ahn S, MacAlpine DM, Cheung E, Lewis PW, Beall EL, Bell SP, Speed T, Manak JR, Botchan MR. Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes Dev. 2007;21:2880–2896. doi: 10.1101/gad.1600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomaru Y, Simon C, Forrest A, Miura H, Kubosaki A, Hayashizaki Y, Suzuki M. Regulatory interdependence of myeloid transcription factors revealed by Matrix RNAi analysis. Genome Biol. 2009;10:R121. doi: 10.1186/gb-2009-10-11-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weston K, Bishop JM. Transcriptional activation by the v-myb oncogene and its cellular progenitor, c-myb. Cell. 1989;58:85–93. doi: 10.1016/0092-8674(89)90405-4. [DOI] [PubMed] [Google Scholar]
- 67.Ganter B, Lipsick JS. Myb binding sites within the N-ras promoter repress transcription. Oncogene. 1997;15:193–202. doi: 10.1038/sj.onc.1201173. [DOI] [PubMed] [Google Scholar]
- 68.Nomura T, Tanikawa J, Akimaru H, Kanei-Ishii C, Ichikawa-Iwata E, Khan MM, Ito H, Ishii S. Oncogenic activation of c-Myb correlates with a loss of negative regulation by TIF1beta and Ski. J. Biol. Chem. 2004;279:16715–16726. doi: 10.1074/jbc.M313069200. [DOI] [PubMed] [Google Scholar]
- 69.Mizuguchi G, Kanei-Ishii C, Takahashi T, Yasukawa T, Nagase T, Horikoshi M, Yamamoto T, Ishii S. c-Myb Repression of c- erbB-2 Transcription by Direct Binding to the c- erbB-2 Promoter. J. Biol. Chem. 1995;270:9384–9389. doi: 10.1074/jbc.270.16.9384. [DOI] [PubMed] [Google Scholar]
- 70.Lee K, Crowe A, Barton M. p53-mediated repression of alpha-fetoprotein gene expression by specific DNA binding. Mol. Cell. Biol. 1999;19:1279–1288. doi: 10.1128/mcb.19.2.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 72.Chen C-Z, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 73.Alcalay M, Tiacci E, Bergomas R, Bigerna B, Venturini E, Minardi SP, Meani N, Diverio D, Bernard L, Tizzoni L, et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005;106:899–902. doi: 10.1182/blood-2005-02-0560. [DOI] [PubMed] [Google Scholar]
- 74.Ross ME, Mahfouz R, Onciu M, Liu H-C, Zhou X, Song G, Shurtleff SA, Pounds S, Cheng C, Ma J, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104:3679–3687. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 75.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc. Natl Acad. Sci. USA. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 77.Brown AL, Wilkinson CR, Waterman SR, Kok CH, Salerno DG, Diakiw SM, Reynolds B, Scott HS, Tsykin A, Glonek GF, et al. Genetic regulators of myelopoiesis and leukemic signaling identified by gene profiling and linear modeling. J. Leukoc. Biol. 2006;80:433–447. doi: 10.1189/jlb.0206112. [DOI] [PubMed] [Google Scholar]
- 78.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lian Z, Wang L, Yamaga S, Bonds W, Beazer-Barclay Y, Kluger Y, Gerstein M, Newburger PE, Berliner N, Weissman SM. Genomic and proteomic analysis of the myeloid differentiation program. Blood. 2001;98:513–524. doi: 10.1182/blood.v98.3.513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.