Abstract
The 5′ cap of human messenger RNA consists of an inverted 7-methylguanosine linked to the first transcribed nucleotide by a unique 5′–5′ triphosphate bond followed by 2′-O-ribose methylation of the first and often the second transcribed nucleotides, likely serving to modify efficiency of transcript processing, translation and stability. We report the validation of a human enzyme that methylates the ribose of the second transcribed nucleotide encoded by FTSJD1, henceforth renamed HMTR2 to reflect function. Purified recombinant hMTr2 protein transfers a methyl group from S-adenosylmethionine to the 2′-O-ribose of the second nucleotide of messenger RNA and small nuclear RNA. Neither N7 methylation of the guanosine cap nor 2′-O-ribose methylation of the first transcribed nucleotide are required for hMTr2, but the presence of cap1 methylation increases hMTr2 activity. The hMTr2 protein is distributed throughout the nucleus and cytosol, in contrast to the nuclear hMTr1. The details of how and why specific transcripts undergo modification with these ribose methylations remains to be elucidated. The 2′-O-ribose RNA cap methyltransferases are present in varying combinations in most eukaryotic and many viral genomes. With the capping enzymes in hand their biological purpose can be ascertained.
INTRODUCTION
The messenger RNAs (mRNAs) of all metazoan organisms and most eukaryotic viruses possess a 5′ cap structure consisting of a 7-methyl guanosine (m7G) linked via an inverted 5′–5′ triphosphate bridge to the initiating nucleoside of the transcript (1–3), which is itself modified by 2′-O-ribose methylation at the first and often the second transcribed nucleotides. The inverted m7G or cap0 structure marks transcription start sites, and has multiple effects on gene expression, including enhancement of RNA stability, splicing, nucleocytoplasmic transport and translation initiation, as facilitated by interactions with nuclear and cytoplasmic cap binding proteins (4–8).
Cap0 is present on all eukaryotic mRNAs and is essential for cell growth of Saccharomyces cerevisiae (9,10) and survival of mammalian cells (8,11). The m7G capping is the earliest mRNA processing event, occurring cotranscriptionally on nascent chains synthesized by RNA polymerase II (12,13). A series of enzymatic reactions are required for cap0 formation, in which the 5′-triphosphate terminus of a nascent pre-mRNA is hydrolyzed to a diphosphate by RNA triphosphatase (Enzyme Commission number 3.1.3.33), the diphosphate RNA end is capped with GMP by RNA guanylyltransferase (GTase) (E.C. 2.7.7.50) to yield a GpppNpNp- structure, here referred to as ‘capG’, and finally capG is methylated by RNA (guanine-N7) methyltransferase (MTase) (EC 2.1.1.56) to yield a cap0 structure. The three enzymatic activities are found encoded by independent peptides or fused in bi- or tri-functional multidomain proteins [reviews: (14,15)]. In humans, the small nuclear (sn) RNAs involved in splicing receive cotranscriptional cap0 as well and continue their 5′-end maturation with hypermethylation of m7G to m2,2,7G (trimethylguanosine or TMG). The capTMG structure is formed in the cytoplasm by trimethylguanosine synthase 1 (Tgs1) (16,17). TMG formation, particularly in U1 snRNA, is required for proper snRNA trafficking and thus for efficient pre-mRNA splicing (18). The U6 snRNA is the exception, as this RNA polymerase III-derived transcript carries a γ methyl triphosphate cap (19).
In higher eukaryotes, including insects, vertebrates and their viruses, mRNA and snRNA 5′-ends are modified by the addition of methyl groups to the first and/or the second transcribed nucleotides [reviewed in (20)]. Methylation of the ribose on the first transcribed nucleotide is termed cap1 and similar methylation of the second transcribed position is termed cap2. Thirty years ago, the human cap1 and cap2 MTases (hMTr1 and hMTr2, respectively) were detected and enriched in fractionated HeLa cell extracts (21). These activities were found predominantly in the nucleus and cytoplasm, respectively. In humans, cap0 and cap1 methylations are present on all mRNA molecules, while about half of the capped poly(A) molecules contain a 2′-O-ribose methylated residue on the second transcribed nucleotide (22). The U1, U2, U4 and U5 snRNAs are methylated at the two first positions (23). Cap1 and cap2 methylations in U2 snRNA are required for spliceosomal E-complex formation and, as a consequence, for efficient pre-mRNA splicing (24). Some eukaryotic organisms, such as the kinetoplastids possess a more elaborate mRNA cap structure, with additional cap3 and cap4 methylations of riboses and additional methylations of the first and fourth base (25,26). Complete methylation on the first four nucleotides is not required for viability of Trypanosoma brucei (27,28); however, reduced methylation results in lower rates of translation (29). For clarity in this study, a cap methylated on both the 0 and 1 positions (m7GpppN1m) is referred to as cap01, and a cap methylated on the inverted guanosine and the first two riboses (m7GpppN1mN2m) is cap012; cap1 and cap2 are reserved for structures with the GpppN1N2 cap modified only on N1 or N2, respectively, without other modifications, in particular the m7G methylation.
With the extensive cap1 and cap2 methylation performed by the host enzymatic activities, the maintenance of 2′-O-ribose MTases in many viral genomes is suggestive of a vital role in gene expression. Cytosolic transcription, as used by pox viruses or RNA polymerase specificities that prevent interaction of the nuclear host cap1 MTase may explain the necessity for an independent enzyme. Alternative scenarios, such as the appropriation of host cap structures by influenza virus (30), tell the same story: the 2′-O-ribose methylations are required, if not for basic viability, for efficient translation among a capped transcript population, without which the virus faces competitive elimination. Regardless of their current genomic location, these enzymes are related, a predicted consequence of horizontal gene transfer events between the host and invading virus. Cap2 and cap3-specific MTases in trypanosomes are related to the poxvirus cap1 MTase (27), while the cap1 MTase in trypanosomes is related to the cap1-specific enzyme from nucleopolyhedroviruses (26). The viral or cellular origin of cap methylation is unclear, as is that of the cap-specific 2′-O-ribose MTases in different eukaryotic cells.
Sequence analysis of ribose MTases lead to identification of two paralogous human genes KIAA0082 and FLJ11171 (also known as FTSJD2 and FTSJD1) (31). In the course of our work on experimental validation of those results, cap1 activity of FTSJD2 was shown by another group (32). Here, we demonstrate the cap2 activity of FTSJD1 and validate FTSJD2 as the hMTr1. The hMTr2 protein is found throughout the cell, in contrast to hMTr1, which is present only in the nucleus. Both can modify substrates with the GpppN (capG) structure. Neither enzyme requires absolutely the presence of other methylations; however, the presence of cap1 modification increases the efficiency of cap2 modification. Comparative analysis of human cap1 and cap2 MTases and their homologs sheds light on their common origin and relationship to other cap-modifying enzymes.
MATERIALS AND METHODS
Cloning of hMTr1 and hMTr2
The cDNAs of the HMTR1 (a.k.a. KIAA0082, ISG95, FTSJD2) and HMTR2 (a.k.a. AFT, FLJ11171, FTSJD1) open reading frames were purchased from imaGenes GmbH (IMAGE ID 4944457 and 5267637) and amplified by PCR using oligonucleotides that introduce restriction sites compatible with the cloning sites of the indicated plasmids. To direct the synthesis of proteins with a FLAG-tag at the N-terminus, a NotI restriction site was introduced 5′ of the predicted translation start site and an XbaI site 3′ of the stop codon. PCR products were digested accordingly and inserted into p3xFLAG-CMV-10 Expression Vector (Sigma), leading to plasmids p3xFLAG-CMV-10-hMTr1 and p3xFLAG-CMV-10-hMTr2. The p3xFLAG-CMV-7-BAP control plasmid with the sequence of Escherichia coli bacterial alkaline phosphatase (BAP) with FLAG-tag was purchased from Sigma. For the cellular localization assay, HMTR1 and HMTR2 open reading frames were inserted into the GW1 plasmid (kind gift from Dr Morgan Sheng), with the use of KpnI and SalI for HMTR1, and BglI and SalI for HMTR2, to direct the expression of proteins with myc-tag.
Mutant variants of these genes were constructed by PCR. The residues targeted for alanine substitution included K239, D364 and K404 for the hMTr1 protein and K117, D235 and K275 for hMTr2. The construction for expression of the N-terminal part of hMTr2 was prepared by inserting a stop codon after 530 or 430 amino acid. The mutated genes were sequenced and found to contain only the desired changes. Sequences of all primers used in this study are listed in Supplementary Table S1.
Eukaryotic overexpression of hMTr1, hMTr2 and BAP
HEK 293T cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen) supplemented with 10% fetal calf serum (FCS, Invitrogen), glutamine (2 mM), penicillin (100 U/ml) and streptomycin (100 µg/ml). For immunoaffinity purifications HEK 293T cells were grown to 50–60% confluency and transfected with 8 μg of p3xFLAG-CMV-10-hMTr1, p3xFLAG-CMV-10-hMTr2 or p3xFLAG-CMV-7-BAP vector per 100 mm dish using jetPEI (Polyplus Transfection) transfection reagent according to manufacturer instructions. After 40 h cells were harvested and frozen in liquid nitrogen for storage at −76°C. Cells from a 100 mm dish were resuspended in lysis buffer [50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, Protease Inhibitor Cocktail for use with mammalian cell and tissue extracts (Sigma)], disrupted on ice with a Pellet pestle (Sigma) and incubated for 1 h at 4°C. The lysate was centrifuged for 30 min at 20 000 g. Supernatant was incubated with 25 μl ANTI-FLAG M2 Affinity Gel (Sigma) with rotation overnight at 4°C. Beads were washed following manufacturer recommendations and resuspended in activity assay buffer. FLAG fusion proteins were used to test enzymatic activity in MTase assays. The process of purification was monitored by western blot using monoclonal ANTI-FLAG M2 antibody produced in mouse (Sigma) and Anti-Mouse IgG Peroxidase Conjugate (Sigma). Additionally, the identity of hMTr1 was confirmed with anti-FTSJD2 antibody produced in rabbit (Novus Biologicals) and anti-rabbit IgG–peroxidase (Sigma) (data not shown).
Cloning, bacterial expression and purification of TbMTr2 and ΔTgs1
The TbMTr2 open reading frame encoding the cap2 MTase from T. brucei was cloned into the pET28a vector (Novagen) to direct the expression of a His-tagged protein (27). For protein overexpression, the plasmid was transformed into E. coli BL21 (DE3) (Novagen). A recombinant TbMTr2 protein was obtained from cultures grown overnight at 24°C in the presence of 2% ethanol and 0.1 mM isopropyl-β-d-galactopyranoside (IPTG). The cDNA encoding a fragment of the Tgs1 enzyme (amino acid residues 574–853) was purchased from imaGenes GmbH (IMAGE ID 4513665) and the catalytically active portion of Tgs1 (residues 631–853) (33) was inserted into the pET41 vector to produce a His-tagged fusion protein with an N-terminal GST tag. The resulting plasmid pET41-ΔTgs1 was transformed into E. coli BL21 (DE3) and the protein was obtained from cultures grown at 18°C in the presence of 2% ethanol and 0.2 mM IPTG for 20 h. Recombinant proteins were purified by Ni-column chromatography with His-Select Nickel Affinity Gel (Sigma) and eluted with 25 mM HEPES (pH 8), 300 mM NaCl, 10 mM 2-mercaptoethanol, 10% glycerol and a gradient of imidazole ranging from 10 to 250 mM.
RNA substrate preparation
Double-stranded DNA oligonucleotides containing the T7 promoter and the sequence of desired RNA transcripts with sticky ends compatible with NcoI and XhoI were cloned into appropriately digested pTZ19R vector (Fermentas). For transcription of RNA starting with adenine, a T7 bacteriophage class II ø2.5 promoter (34) was used. pTZ19R-RNA vectors were digested with XhoI and gel purified. Three micrograms of linearized plasmid were used as template for in vitro transcription using the T7 Megashortscript Kit (Ambion). Subsequently, the reaction mixture was treated with 2 U RNase-free DNase I for 30 min at 37°C to remove the template DNA. RNA was purified by phenol/chloroform extraction and ethanol precipitation. Two variant RNA molecules of 63 nt were produced: RNA-GG that starts with guanosine (pppGpGpGpX) and RNA-AG that starts with adenine (pppApGpGpX) were X=TAACGCTATTATTACAAAGCTCTTTTATGTAGTGTGCGTACCACGGTAGCAGGTACTGCG, based on the T. brucei spliced leader RNA gene (GenBank # Z50171.1), chosen for convenience. RNA molecules were subjected to capping reactions using vaccinia virus capping enzyme (VCE) and vaccinia virus Cap1 MTase (VMT) ScriptCap (Epicentre; Supplementary Figure S1A). Reactions were carried out following the manufacturer recommendations with addition of 10 μCi [α-32P] GTP (3000 Ci/mmol; Hartman Analytic GmbH), with or without methyl donor S-adenosylmethionine (SAM) to form m7GpppN- (cap0) and GpppN- (capG) structures, respectively. The nomenclature used throughout this manuscript is summarized in Table 1. The ScriptCap 2′-O-MTase was used to create the m7GpppNm-(cap01) and GpppNm-(capG1) structures. Following the reaction, 32P-labeled capped RNA molecules were purified by extraction with phenol/chloroform, passed through mini Quick Spin Oligo Columns (Roche) to remove free radionucleotides and precipitated with ethanol.
Table 1.
The nomenclature of cap structure variants used throughout this manuscript
| capG | GpppNpNp- |
| capG1 | GpppNmpNp- |
| capG2 | GpppNpNmp- |
| capG12 | GpppNmpNmp- |
| cap0 | m7GpppNpNp- |
| cap01 | m7GpppNmpNp- |
| cap02 | m7GpppNpNmp- |
| cap012 | m7GpppNmpNmp- |
| capTMG | m7,2,2GpppNpNp- |
| capTMG1 | m7,2,2GpppNmpNp- |
| capTMG2 | m7,2,2GpppNpNmp- |
| capTMG12 | m7,2,2GpppNmpNmp- |
For generating internally, 32P-labeled G-capped transcript RNA-GA [GpppGp*ApGp(TpCp)12], transcription was carried out using pTZ19R-RNA-GA template, prepared as described above, with the addition of 10 μCi of [α-32P] ATP (3000 Ci/mmol; Hartman Analytic GmbH) and the GpppG cap analog (Epicentre) following the manufacturer instructions. After the removal of template DNA, RNA transcripts were gel purified. Subsequently, viral enzymes were used to form molecules containing cap0 or cap01 (Supplementary Figure S1B).
Templates for in vitro transcription of U1 and U2 snRNAs were prepared by a PCR-based method using the total DNA from human cells as a template, and primers that introduce the T7 bacteriophage class II ø2.5 promoter upstream of a desired sequence. Gel-purified PCR products (0.5 μg) were used for in vitro transcription using the T7 Megashortscript Kit. TMG-capped RNA was prepared in vitro by m7G cap modification to m2,2,7G by the active part (residues 631–853) of the Tgs1 enzyme (33). Reactions were carried out in 50 mM Tris–HCl (pH 8), 5 mM dithiothreitol (DTT), 100 μM SAM, 10 U Ribolock (Fermentas), 1 μg protein and 2 pmol RNA with different forms of 32P-labeled cap structures for 30 min at 37°C. Subsequently, RNA was purified by phenol/chloroform extraction and ethanol precipitation.
Methyltransferase assays
Methylation reactions with hMTr2 were carried out in 30 mM Tris–HCl (pH 7.4), 50 mM KCl, 1 mM EDTA, 10 mM DTT, 100 μM SAM, 10 U Ribolock, purified enzyme and 2 pmol substrate RNA in a total volume of 20 μl. The reaction buffer for hMTr1 differed in pH (8.4) and KCl concentration (150 mM) and the other components remained the same. Reactions were carried out for 1 h at 37°C. BAP protein was used as a negative control. For hMTr1, the VMT enzyme from the ScriptCap kit was used as a positive control. Methylation with VMT was performed following manufacturer recommendations. For hMTr2, the trypanosome enzyme TbMTr2 was used as a positive control. Methylation with TbMTr2 was carried out in 50 mM Tris–HCl (pH 7.4), 5 mM DTT, 100 μM SAM, 10 U Ribolock, 2 pmol 32P-labeled cap structure RNA and 4 pmol enzyme for 30 min at 27°C. The modified RNA was purified by phenol/chloroform extraction and ethanol precipitation. The RNA was digested with either 2 U nuclease P1 (Sigma) in 10 mM Tris–HCl (pH 7.5), 10 mM MgCl2 and 50 mM NaCl, or with 2.5 U RNase T2 (MoBiTec GmbH) in 50 mM ammonium acetate (pH 4.5) overnight at 37°C. The digestion products were resolved on a 21% polyacrylamide/8 M urea gel and visualized by PhosphorImaging (Storm 820, Amersham Bioscience).
Internally labeled RNAs were digested with nuclease P1 as described above. Digested samples were spotted on the cellulose-coated thin layer chromatography plates (Merck), together with unlabeled 5′-monophosphate ribonucleosides: G, A, U, C, Am and m6A and developed in isobutyric acid:H2O:ammonium hydroxide (66:33:1; v:v:v) as the first-dimension solvent and isopropanol/concentrated HCl/water (68:18:14; v:v:v) as the second-dimension solvent. Detection and identification of the radiolabeled 32P-modified nucleotides in RNA digestions was performed by comparing the autoradiograph with UV pattern and reference maps established in the two TLC systems.
Immunolocalization
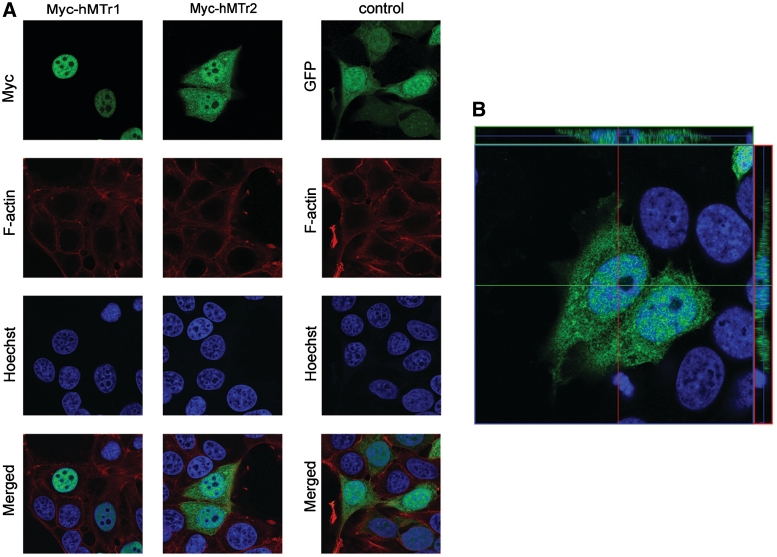
MCF-7 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen) supplemented with 10% fetal calf serum (FCS, Invitrogen), glutamine (2 mM), penicillin (100 U/ml) and streptomycin (100 µg/ml) and grown on 10 mm glass coverslips in 24-well plates to 60% confluency and transfected with 0.6 µg of GW1-myc-hMTr1, GW1-myc-hMTr2 or a control plasmid EGFP-C1 (Clontech) with the use of Lipofectamine 2000 (Invitrogen). After 24 h, cells were fixed and stained with mouse anti-myc (9E10 from Calbiochem) or rabbit anti-GFP antibodies (MBL) to visualize the recombinant protein. Hoechst 33258 (Sigma) and phalloidin 568 (Invitrogen) were used to visualize cell nuclei and F-actin (cell shape), respectively. Images of the cells were acquired with a LSM510 Exciter confocal microscope as a Z-stack of single plane images with a 63 × objective.
Bioinformatic analyses
Searches of the current version of the non-redundant protein sequence database (nr) were carried out with PSI-BLAST (35) using full-length sequences of hMTr1 and hMTr2 as queries. In order to identify subfamilies of the related sequences and to visualize sequence similarities, we chose the clustering tool CLANS (Cluster ANalysis of Sequences), which uses the P-values of high-scoring segment pairs (HSPs) obtained from the N × N BLAST search, to compute attractive and repulsive forces between each sequence pair and to move the sequences according to the force vectors resulting from all pair-wise interactions (36). The clusters of closest homologs of hMTr1 and hMTr2 were extracted. Global multiple sequence alignments were calculated for individual families using MAFFT (37) and adjusted manually to maximize the number of aligned homologous residues and preserve the continuity of predicted secondary structure elements. Incomplete sequences were discarded if the deletion spanned >30% of the alignment, or repaired using amino acid sequences predicted from the available DNA sequences of the corresponding genes.
For phylogenetic analyses, we constructed a multiple sequence alignment composed of the catalytic domains of MTases from different families, guided by superimposition of representative crystal structures or theoretical models. Structure prediction for hMTr1 and hMTr2 was carried out by the GeneSilico metaserver (38), which serves as a broker providing a comprehensive view of the results collected from various protein fold recognition methods along with the prediction of the secondary structure, protein order/disorder, solvent accessibility, potential RNA/DNA binding sites, etc. Phylogenetic trees were calculated with MEGA 4.0 (39) using the minimum evolution method with the JTT model of substitutions and pair-wise deletions. The initial tree was calculated by the NJ method, with the Closest Neighbor Search option set to level = 2. To assess confidence for each node of the phylogenetic tree, the bootstrap test was performed with 1000 replications.
RESULTS
Sequence analyses of hMTr1 and hMTr2 candidates
FTSJD1 and FTSJD2 were identified as candidates for human RNA 2′-O-ribose MTases based on the presence of a domain homologous to known bacterial and eukaryotic tRNA and rRNA MTases (31). FTSJD2 possessed an additional domain homologous to the GTase domain present in mRNA capping enzymes, which prompted us to consider both proteins as candidates for RNA cap MTases. Sequence database searches initiated with FTSJD1 and FTSJD2 yielded numerous members of the Rossmann-fold MTase (RFM) family. Sequence clustering (see ‘Materials and Methods’ section) revealed that FTSJD1 and FTSJD2 are related paralogs that form an isolated family together with a number of proteins from higher eukaryotes and eukaryotic viruses (Supplementary Figure S2). In particular, the previously characterized cap1 MTases from Autographa californica nucleopolyhedrovirus (AcNPV) (40) and T. brucei (26,41) grouped with FTSJD2, making it the primary hMTr1 candidate.
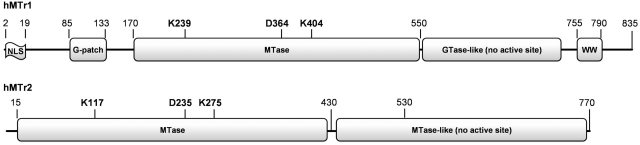
Structure prediction in combination with multiple sequence alignment analysis (data not shown) confirmed that FTSJD2, the putative hMTr1, carried a putative nuclear localization signal (NLS), a G-patch domain potentially involved in RNA binding, an RFM domain with a conserved K-D-K triad characteristic for 2′-O-ribose MTases, a GTase-like domain lacking catalytic residues (26,42) and a WW domain potentially involved in protein–protein interactions. FTSJD1, the candidate hMTr2, was composed of two RFM domains (Figure 1). A multiple sequence alignment of hMTr2 orthologs (Supplementary Figure S3) revealed that the N-terminal RFM domain had a conserved SAM-binding site (motifs I–III). Interestingly, in motif I the Asp residue present in other 2′-O-ribose MTases was replaced by His, but it reappeared elsewhere, shifted three positions toward the C-terminus. As in hMTr1, the N-terminal RFM domain of hMTr2 contained a characteristic catalytic K-D-K triad. These residues were conserved in all members of this family, with the exception of proteins from Plasmodium, where they were replaced by the variant Y-(N/D)-K. Members of Plasmodium also exhibited deletions of one residue in motif I, and lost the otherwise universally conserved E residue in motif VIII, whose counterpart in RrmJ (E199) plays a minor role in the MTase activity (43). The C-terminal RFM domain was present in all members of the hMTr2 family, with the exception of two viral and one algal proteins, and was more variable than the N-terminal domain. Despite the overall homology to enzymatically active RFM domains, this domain lacked conserved residues required to bind SAM as motifs I–III were practically undetectable, and neither the K-D-K triad nor any other obvious candidates for an active site were identified.
Figure 1.
Domain architecture of hMTr1 and hMTr2 proteins. Domain boundaries were predicted according to protein fold recognition analyses carried out via the GeneSilico metaserver (38). Amino acid residues important for the MTase activity are indicated.
Recombinant FTSJD2 and FTSJD1 exhibit cap1- and cap2-MTase activities
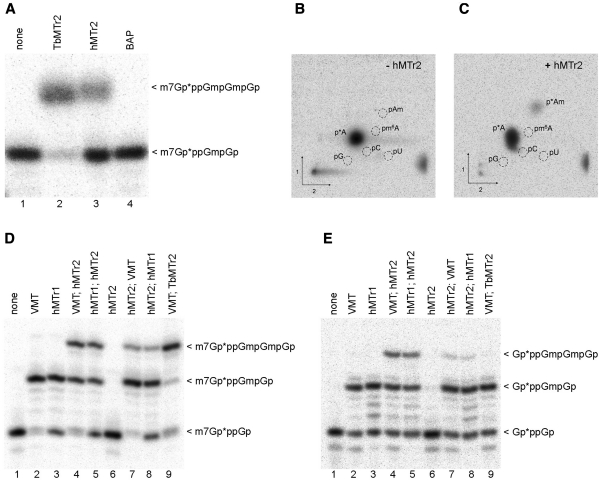
To test the cap2-MTase activity of our candidate hMTr2, in vitro transcribed RNA-GG was capped with VCE in the presence of [α-32P] GTP and modified with VMT to form a cap01 substrate (Supplementary Figure S1, Table S1). This cap01-RNA was subjected to methylation with the purified FLAG-tagged recombinant hMTr2 protein, followed by RNase T2 digestion to generate 3′-monophosphate nucleotides. Presence of the 2′-O-ribose methylation prevents hydrolysis by RNase T2 generating RNA fragments of longer length (44). Digestion products were resolved on a polyacrylamide gel, along with control samples generated by the trypanosome cap2-MTase TbMTr2 as a positive control, and immunopurified FLAG-tagged BAP protein as a negative control (Figure 2A). When cap01-RNA was treated with hMTr2, a radioactive RNA fragment that comigrated with a RNase T2-resistant fragment from a sample treated with TbMTr2 was detected (Figure 2A, lanes 2 and 3), albeit at lower efficiency, corresponding to the predicted cap012 product. No methyl incorporation was observed in a control reaction with a purified BAP protein (Figure 2A, lane 4), demonstrating that the cap2 MTase activity was specifically from overexpressed hMTr2. Analogous experiments performed with the RNA-AG substrate also yielded a methylation product (data not shown).
Figure 2.
hMTr2 activity and substrate requirements. Methyltransferase activity: In vitro transcribed RNA-GG molecules with the 32P-labeled cap01 structure (A) were incubated with enzymes as indicated in the presence of SAM. Purified product RNA was digested with RNase T2. Digestion products were resolved on 21% polyacrylamide/8 M urea gel and visualized by autoradiography. BAP protein was used as negative control. RNA with 32P-labeled cap structure created with the TbMTr2 enzyme was used as a reference. Specificity: autoradiography of two-dimensional chromatograms of 5′-phosphate nucleosides on thin layer cellulose plates. [α-32P] ATP-labeled in vitro transcribed cap01-RNA-GA was incubated with SAM in the absence, (B) or presence (C) of the hMTr2 protein. Product RNA was purified, cleaved by nuclease P1 and the resulting nucleotides were analyzed as described (44). 5′-monophosphate ribonucleosides of G, A, U, C, Am and m6A were used as standards. Substrate requirements: In vitro transcribed RNA-GG molecules with 32P-labeled cap0 (D) or capG (E) structure were incubated with one of the indicated enzymes, added in a given order, in the presence of SAM. After every modification step RNA molecules were purified by phenol/chloroform extraction and ethanol precipitation. Final products were digested and analyzed as described in legend for panel A. Asterisks indicate positions of 32P-labeled phosphates.
In order to confirm the 2′-O-ribose MTase specificity of the candidate hMTr2, we identified the radiolabeled 32P-modified nucleotides in RNA digests by two-dimensional thin layer chromatography (2D-TLC). The RNA-GA substrate used in this experiment was prepared by in vitro transcription with [α-32P] ATP and addition of the cap analog (GpppG), followed by methylation with VCE and VMT to create cap01-RNA (Supplementary Figure S1B). Digestion of the substrate RNA with nuclease P1, followed by 2D-TLC analysis, resulted in a single radiolabeled spot corresponding to pA (Figure 2B). Incubation of the substrate RNA-GA with recombinant hMTr2 in the presence of SAM yielded a new radiolabeled spot that comigrated with pAm marker (Figure 2C) and was resolved clearly from the 5′-monophosphate m6A marker that migrates to close position on a published 2D map of methylated 5′-phosphonucleosides (44). The weak signal of pAm may have resulted from the low efficiency of capG formation, as only a subset of labeled RNA molecules that had GpppG incorporated during transcription were a substrate for hMTr2 (see below), and from inefficient methylation by VMT and hMTr2. For the same experiment, we also run RNase T2/PAGE analysis and found that the efficiency of methylation seen on gel matches that seen on TLC (Supplementary Figure S4). With confirmation of the FTSJD1 gene product methylation of the 2′-hydroxyl on the second nucleotide, this protein was henceforth referred to as hMTr2. In parallel, 2D-TLC analysis were done for uncapped (pppG-ended), [α-32P] ATP-labeled RNA-GA that was incubated with recombinant hMTr2 in the presence of SAM. Only one spot that corresponds to unmodified pA was detected (Supplementary Figure S5) demonstrating that the hMTr2 MTase activity is dependent on the presence of capG.
Analogous experiments to test cap1 MTase activity of the HMTR1 gene product were performed. This protein methylated the first transcribed nucleotide (Supplementary Figure S6) in accord with parallel studies (32). For activity tests, we used RNA-GG and RNA-AG substrates and found that hMTr1 transfers a methyl group to G (32), as well as A residues at position 1 with similar effectiveness (data not shown). The precise position of the methyl group addition was analyzed using internally [α-32P] ATP-labeled RNA-GA substrate by 2D-TLC, and confirmed methylation at 2′-O-ribose position of the first transcribed nucleotide (Supplementary Figure S6).
The hMTr2 activity is not dependent on additional methylations
To broaden our knowledge about the substrate requirements of hMTr2, the methylation assay was performed using 32P-labeled capG1 that lacked the methylation on the 5′-inverted G and cap01-RNA-GG substrates (Figure 2, panels D and E). After RNase T2 digestion, an additional band was observed with both substrates (Figure 2D and E, lanes 4 and 5). Interestingly, while the trypanosome cap2-MTase activity was dependent on cap0, the hMTr2 recognized RNA with either cap0 or capG structures. This result indicated that the N7-methylation of the guanosine cap was not required for the cap2 modification.
The association of hMTr1 with RNA polymerase II in vivo (42) indicates that cap1 formation occurs early in the synthesis of nascent mRNA. As such, hMTr1 may not recognize cap02 substrates, while hMTr2 may display a preference for cap01 transcripts. To examine activities on differentially methylated substrates, the in vitro MTase assay was performed on 5′-end labeled capG and cap0-RNA-GG substrates using sequential enzymatic combinations, each performed in isolation (Figure 2D and E). hMTr1 catalyzed methylations on both cap0 (Figure 2D, lane 3) and cap02 (Figure 2D, lane 8). hMTr2 methylates both cap0 (Figure 2D, lanes 7 and 8) and cap01 (Figure 2D, lanes 4 and 5) substrates with better efficiency for cap01. The human enzymes displayed low relative efficiencies as compared with the VMT and TbMTr2 controls. The hMTr2 treatment alone did not yield a specific product after RNase T2 digestion (Figure 2D, lane 6), as the absence of the cap1 methylation resulted in separation of the label from the modified nucleotide; the secondary 2′-O-ribose methylation of position 1 by hMTr1 or VMT allowed detection of the primary cap2 modifications by hMTr2 or TbMTr2. The same regimen applied to the capG substrate yielded similar results, at reduced efficiency for hMTr2, with the exception of the TbMTr2 control, which did not recognize the capG substrate (Figure 2E, lane 9). Although neither hMTase may see this variety in vivo, the enzymes possess the capability to recognize a relatively broad array of substrates, with preferential activity suggesting a 5′ to 3′ maturation pathway.
The hMTr2 requires both RFM domains
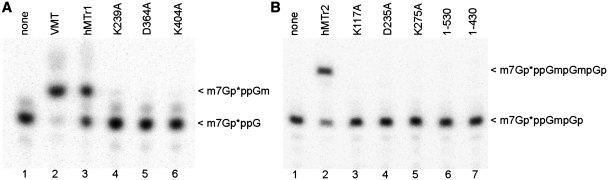
To verify the predicted K-D-K-active sites of the hMTr1 and hMTr2 enzymes, site-directed mutagenesis was performed to generate protein variants with individual residues substituted by alanine. In vitro transcribed RNA-GG with 32P-labeled cap0 or cap01 structures were incubated with the purified protein variants in the presence of SAM. The reaction products were digested with nuclease P1 or RNase T2 and separated on a polyacrylamide gel. None of the variants exhibited MTase activity (Figure 3), indicating that each residue of the K-D-K triad is essential for the activity of both hMTr1 (Figure 3A, lanes 4–6) and hMTr2 (Figure 3B, lanes 3–5).
Figure 3.
Analysis of hMTr1 and hMTr2 mutants. In vitro transcribed RNA-GG molecules with 32P-labeled cap0 (A) or cap01 (B) structures were incubated with indicated enzymes [wild-type (hMTr1, hMTr2), alanine-substituted variants of hMTr1 (K239A, D365A, K404A) and hMTr2 (K117A, D235A, K275A) or truncated forms of hMTr2 (1–530, 1–430)] in the presence of SAM. Purified product RNA was digested with nuclease P1 (A) or RNase T2 (B). Digestion products were resolved on a 21% polyacrylamide/8 M urea gel and visualized by autoradiography. Asterisks indicate positions of 32P-labeled phosphates.
Region 431–530 of hMTr2 is highly divergent, we predict that it corresponds to a degenerated SAM-binding site of the C-terminal domain, but we cannot completely exclude that it contains a linker between domains or an extension of the N-terminal domain. Therefore, we checked the activity of two truncated variants of hMTr2: 1–430 that contained just the conserved N-terminal RFM domain, and 1–530 that additionally included the diverged region, but lacked the conserved core of the C-terminal RFM domain found in the 531–770 region.
Both C-terminal deletion variants 1–530 and 1–430 were tested for the hMTr2 activity and found to be inactive (Figure 3B, lanes 6 and 7). Thus, the C-terminal MTase-like domain is essential for hMTr2 activity, despite the absence of a conserved MTase active site.
The hMTr1 and hMTr2 recognize TMG-capped snRNAs in vitro
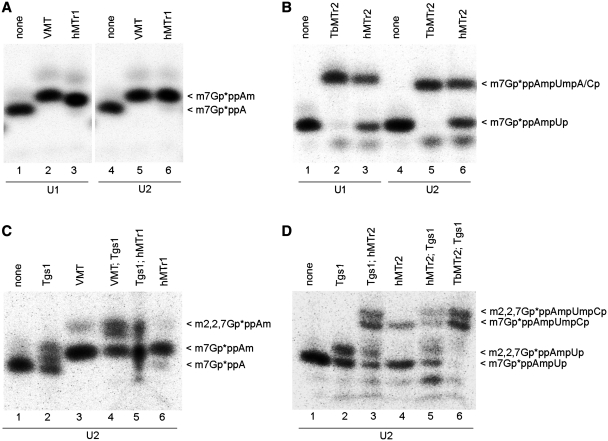
Both hMTr1 and hMTr2 were assayed on in vitro-transcribed U1 and U2 snRNAs, the endogenous RNA molecules known to possess TMG, cap1 and cap2 modifications (capTMG12). In trypanosomes, an intriguing sequence specificity was found for TbMTr1 cap1 MTase, functionally limiting activity of the enzyme to the spliced leader RNA and the cap0 form of the U1 snRNA (26,41). In vitro-transcribed human U1 and U2 snRNAs with 32P-labeled cap0 or cap01 structures representing precursor forms of the mature products were used for hMTr1 (Figure 4A) or hMTr2 (Figure 4B) activity assays. The U1 and U2 snRNA substrates initiate with AUA and AUG, respectively. Both hMTr1 and hMTr2 methylated the snRNA substrates in vitro. The hMTr1 effectively methylated RNA in that sequence context (Figure 4A, lanes 3 and 6). In contrast, hMTr2 converted approximately half of the cap01 starting material to cap012 (Figure 4B, lanes 3 and 6) as compared with the efficient conversion of the substrate by the equivalent TbMTr2 trypanosome version of the enzyme (Figure 4B, lanes 2 and 5). The reason for this difference remains to be explained.
Figure 4.
hMTr1 and hMTr2 act on the U1 and U2 snRNAs in vitro. In vitro transcribed U1 and U2 snRNA molecules with 32P-labeled cap0 (A and C) or cap01 (B and D) structures were incubated with one of the indicated enzymes, added in the order indicated and SAM. After every modification step RNA was purified by phenol/chloroform extraction and ethanol precipitation. Final products were fragmented with nuclease P1 (A and C) or RNase T2 (B and D). Digestion products were resolved on 21% polyacrylamide/8 M urea gels and visualized by autoradiography. Cap structures modified with VMT (A and C) or TbMTr2 (B and D) were used as positive controls. Asterisks indicate positions of 32P-labeled phosphates.
The relative efficiencies of specific orders of methylation events and the effects of the presence of a methyl group at additional cap0 positions were examined. To this end, capping reactions were performed with Tgs1, hMTr1 (Figure 4C) and hMTr2 (Figure 4D) on in vitro-transcribed U2 snRNA molecules with 32P-labeled cap0 or cap01 structures, respectively. The reactions differed in the order of action of capping enzymes, as indicated above the panels. Both hMTr1 and hMTr2 methylated RNA with the capTMG structure (Figure 4C, lane 5; Figure 4D, lane 3). Likewise, Tgs1 can act on RNA with methyl groups on both riboses of the first two nucleotides (Figure 4D, lanes 5 and 6) as well as on cap0 RNA (Figure 4C, lane 2). Thus, no preference was seen for any specific order of action among these enzymes. hMTr1 and hMTr2 could be responsible for snRNA 2′-O-ribose methylations in vivo.
The hMTr2 has a significant nuclear presence
Cellular localization of hMTr2 was assayed in situ by immunostaining of epitope-tagged protein. MCF7 cells were transfected with GW1-myc-hMTr2 or control plasmid EGFP-C1 that distributed throughout the cell. In parallel, hMTr1 was examined using GW1-myc-hMTr1. Twenty-four hours post-transfection, cells were fixed and immunostained for the overexpressed protein. The myc-hMTr2 was present in both the cytoplasm and the nucleus (Figure 5). Verification of labeling was done by checking the orthogonal axis views (Figure 5B). The unit volume ratio of myc-hMTr2 signal in the nucleoplasm versus cytosol was 1.25, comparable with the 1.45 ratio detected in cells expressing EGFP that was spread by passive diffusion. The myc-hMTr1 was confined to the nucleus (32,42) with almost no signal detectable in the cytosol. Both myc-hMTr1 and myc-hMTr2 were excluded from DNA-free nuclear bodies that are likely to represent nucleoli. Thus RNAs probably acquire the cap1 modification by hMTr1 during or shortly after transcription in the nucleus, followed by hMTr2 capping in either nucleus or after export to the cytoplasm. The mRNA and snRNAs may both serve as substrates for these enzymes, but their presentation may differ along with their divergent paths after acquisition of cap2.
Figure 5.
Both hMTr1 and hMTr2 have a nuclear presence. MCF7 cells were transfected with GW1-myc-hMTr1, GW1-myc-hMTr2 or EGFP-C1 (control plasmid). After 24 h, cells were fixed and stained with anti-myc or anti-GFP antibodies to visualize recombinant proteins (green). Nuclei were visualized with Hoechst 33258 staining (blue) and F-actin with phalloidin 568 (red). Images were acquired by confocal microscopy. (A) Single focal planes from the Z-stack at nucleus level are presented. (B) Cells transfected with GW1-myc-hMTr2 with respective orthogonal axis view to verify the labeling.
Evolution of cap MTases hints at additional cap variants
The identification of genes-encoding human cap MTases provided an opportunity to study their evolutionary origin. A multiple alignment of the catalytic RFM domain from cap 2′-O-ribose MTases, from humans and other species (e.g. trypanosomes and viruses), and their orthologs was constructed (Supplementary Figure S7). The family of RrmJ MTases present in the last universal common ancestor of all extant organisms was used as an outgroup. A common feature of experimentally characterized representatives is the presence of the K-D-K catalytic triad and the ability to catalyze 2′-O-ribose methylation in RNA. A group of uncharacterized proteins from fungi, which are closely related to hMTr1 and hMTr2 (Supplementary Figure S2), were excluded because they lacked the K-D-K triad (data not shown), indicating a change in substrate specificity or inactivity as enzymes.
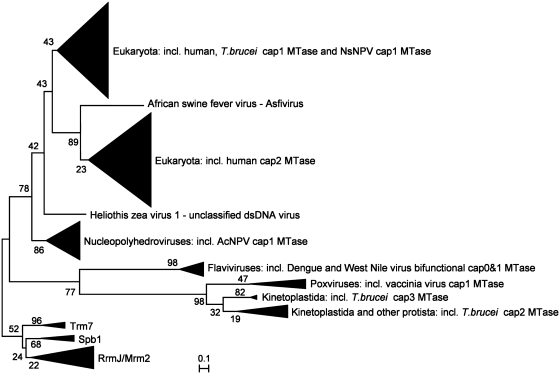
The minimal evolution phylogenetic tree (Figure 6) revealed that hMTr1 and hMTr2 were paralogs forming a subfamily with higher eukaryotic and viral members. The hMTr1 orthologs were present in metazoans and various lower eukaryotes. The remaining unicellular eukaryotes, including stramenopiles (Thalassiosira pseudonana and Phaeodactylum tricornutum), kinetoplastida (Trypanosoma and Leishmania), alveolata (Toxoplasma gondii and Perkinsus marinus, but not Plasmodium, Cryptosporidium or Theileria species), diplomonadida (Giardia lamblia and G. intestinalis), possessed a variant without the GTase-like domain. The trypanosome ortholog of hMTr1 that lacked the GTase-like domain exhibits cap1-MTase activity on spliced leader RNA and U1 snRNA (26,41), indicating that the C-terminal domain is not essential for the enzymatic activity. Interestingly, several species such as mosquitos Aedes aegypti and Culex quinquefasciatus possessed not one but two orthologs of hMTr1, products of a lineage-specific duplication. The Neodiprion sertifer nucleopolyhedrovirus (NsNPV) hMTr1 ortholog position was indicative of a horizontal gene transfer event from a eukaryotic cell to the virus, as postulated previously [(26), Figure 6 and Supplementary Figure S7]. This enzyme is distinct from another large group of AcNPVcap1-MTase orthologs (40) that formed a separate branch at the base of the clade comprising both human cap MTases.
Figure 6.
2′-O-ribose cap MTases change their specificity in the course of the evolution. A minimum evolution tree of homologs of known 2′-O-ribose mRNA cap MTases is shown. The RrmJ family of 2′-O-ribose MTases that act on rRNA (together with their close homologs Mrm2, Trm7 and SpbJ) was used as an outgroup. Branches comprising multiple sequences from the same taxon have been collapsed and are illustrated as triangles marked by the name of the taxon and an experimentally characterized representative. Bootstrap values for nodes are shown.
hMTr2 orthologs are present in metazoa as well as in some other eukaryotic taxons. Several alvaeolata possessed only hMTr2 orthologs (e.g. Plasmodium, Cryptosporidium and Theileria species), while Perkinus marinus contained two hMTr2 orthologs in addition to a hMTr1 ortholog, demonstrating that the number of hMTr1/hMTr2 family members can vary in different species of the same phylum. The hMTr2 orthologs were found in the unicellular eukaryotes Monosiga brevicollis, a choanoflagellate, and the green alga Chlamydomonas reinhardtii, both of which lacked hMTr1 orthologs. hMTr2 orthologs were present in dsDNA viruses, but limited to African swine fever virus, which formed an outgroup, and in Mimivirus, which was positioned among eukaryotic proteins. Mimivirus likely acquired its hMTr2 ortholog from a unicellular host, similar to the acquisition of the trifunctional capping enzyme comprising the TPase/GTase/cap0 MTase activities (15).
A separate branch with two groups was formed by two other families of RNA cap MTase with predominantly viral membership. One group was formed exclusively due to the presence of a bifunctional cap0/cap1 MTase domain found in flaviviral multidomain proteins, while the other comprised cap1 MTases from poxviruses, e.g. cap1 MTase from vaccinia virus (VP39) that also serves as a poly(A) polymerase stimulatory factor (45), as well as the trypanosome cap2 and cap3/cap4 MTases (27,28,46,47). The branch with the trypanosome cap2 MTase enzyme TbMTr2 contained members from other unicellular eukaryotes, including Naegleria gruberi (Heterolobosea), Paramecium tetraurelia and P. marinus (alveolata) and M. brevicollis (choanoflagellida). Hence, M. brevicollis had orthologs of both the hMTr2 and TbMTr2 cap2 MTases, while lacking a detectable ortholog of any known cap1 MTase, and P. marinus possessed as many as four orthologs of cap 2′-O-ribose MTases, including one, one, and two orthologs of hMTr1, TbMTr2, and hMTr2, respectively (Figure 6 and Supplementary Figure S7).
DISCUSSION
Here, we identify the human HMTR2 gene (formerly known as FTSJD1) that encodes cap2 MTase hMTr2 and characterize it in combination with the HMTR1-encoded cap1-MTase hMTr1 (32). The hMTr2 can methylate RNA with both m7GpppN and GpppN structures, unlike the kinetoplastid TbMTr2 (27,47). The hMTr2 is less efficient for RNA without cap1, whereas TbMTr2 shows no preference (47). hMTr1 is capable of modifying RNA that has the second transcribed nucleotide methylated (cap02-RNA and capG2-RNA) with the same effectiveness as RNA with cap0 and capG structures, as does VMT. hMTr1, like VMT and TbMTr1 (41), methylates both capG-RNA and cap0-RNA (21,32). hMTr1 shows no position 1 preference, as it can methylate A, C and G residues with the similar effectiveness (21). In contrast, TbMTr1 requires an A at position 1 with additional downstream nucleotides shared on both the spliced leader RNA and U1 snRNA (41). This discrepancy between their specificities reflects biological function, as the human enzyme methylates many diverse mRNA and snRNA molecules, whereas in Trypanosoma substrates are essentially homogeneous. No specific order of steps in cap structure formation relative to Tgs1, hMTr1 and hMTr2 action was suggested. Cellular localization of hMTr1 is nuclear, and hMTr2 is present in the cytoplasm and the nucleus. Both were excluded from DNA-free nuclear bodies that are likely to represent nucleoli, as opposed to TbMTr1 that has nucleolar localization and forms a complex with the SLA1 H/ACA snoRNP (48).
In humans, cap0 and cap1 methylations are present in all mRNAs. Diversity in cap structure is introduced by hMTr2 with about half of the capped poly(A) molecules containing a 2′-O-ribose methylated residue on the second transcribed nucleotide (22). Cap2 methylation may target mRNA for enhanced translation. Accordingly, kinetic studies found 3-fold more cap012-modified mRNA in the polysomal fraction as compared with unbound mRNA in mouse L cells, whereas the amount of cap01-modified RNA is similar in both fractions (49). Thus cap012-modified mRNA may have an increased affinity for ribosomes; alternatively, methylation of cap2 could occur after the mRNAs are loaded with ribosomes. An increase in mRNA stability may be another byproduct of cap methylation, as cap012-modified RNAs dominate among more stable mRNAs (49). The two mechanisms may act together or, depending on the sequence context, be characteristic for some mRNA classes. An important question to address is how some subclasses of mRNA are distinguished from others by the capping machinery for modification by additional methylation. For TbMTr1, the RNA sequence is the determinant of enzyme activity; this enzyme acts with substrate specificity that favors the spliced leader 5′ sequence (26). hMTr1 does not discriminate in vitro between substrates with different nucleotides at the N1 position. The hMTr2 specificity may be dictated by the sequence and/or structure of the substrate, and thus be limited to certain sequence classes. No consensus target sequence for cap2 methylation emerges from the studies of mRNAs from mammalian cells (22,49,50). Further work is required to elucidate the RNA features that direct hMTr2 activity as well as the effect of cap2 on gene expression.
The order of capping reactions may impact gene expression. The mRNA:guanine-N7 (cap0) MTase and hMTr1 are nuclear enzymes (11,42). Creation of cap0-RNA occurs co-transcriptionally (12,13). The hMTr1 interacts with the C-terminal domain of RNA polymerase II (42), which suggests that methylation of the first transcribed nucleotide also takes place during transcription. Human cap0 MTase and hMTr1 act effectively on both non-methylated and monomethylated RNA, which does not support any specific order of action for these two enzymes. hMTr2 probably acts on cap01-RNA in vivo. This is supported by its substrate preference as it acts the most efficiently on cap01-RNA. hMTr2 is present both in the nucleus and cytoplasm, nevertheless its activity was found almost exclusively in cytoplasm with <5% of the cap2-MTase activity recovered in the nuclear fraction (21). This may suggest that the nuclear form is inactive and needs activation upon delivery to cytoplasm. Another explanation of those results is that overexpression of hMTr2 enhances its nuclear presence that may be predominantly cytoplasmic for the native protein. The TMG structure does not disturb the activity of hMTr enzymes, and the fully methylated capTMG12 structure can be formed in vitro in different ways, suggesting that the order of reactions in the cap formation pathway is determined by compartmentalization in the cell and the transport of RNAs between different cellular compartments during maturation, rather than by specific substrate requirements.
The genes encoding the human enzymes responsible for RNA 5′ cap 2′-O-ribose methylations appear to have evolved from a common ancestor present during the radiation of Eukaryota. The ancestral family from which the ancient metazoan 5′ cap 2′-O-ribose MTase most likely originated is the RrmJ family of ribose MTases, which includes bacterial rRNA-modifying enzymes and eukaryotic rRNA and tRNA-modifying enzymes, whose representative was present in the Last Universal Common Ancestor of extant organisms. The paralogous relationship of the hMTr1 and hMTr2 families implies that cap2-MTase activity in metazoans is the product of a cap1-MTase-encoding gene duplication. hMTr2 evolved independently of the TbMTr2 (cap2) MTase from trypanosomes (27). The phylogenetic trees indicate independent transfers of cap MTases from different families between unicellular eukaryotes and unrelated viruses, including those with DNA or RNA genomes. Thus, viruses played an important role in horizontal transmission of members of this family. In particular, the branches of viral MTases appear as outgroups of eukaryotic proteins, suggesting that metazoan, and consequently human, cap 2′-O-ribose MTases have a viral origin.
In addition to horizontal gene transfer, frequent duplication and gene loss events shaped the evolution of cap 2′-O-ribose MTases. As a consequence, these proteins can vary greatly in number even in closely related organisms. A question arises regarding the specificity of these enzymes along with the resulting 5′ cap structure in their hosts. The eukaryotes and viruses that possess only one (or more) hMTr2 ortholog(s), but no hMTr1 ortholog, suggest several scenarios. First, hMTr2 can act without preceding methylation by hMTr1, hence capped RNAs in some organisms and viruses may have cap02 (double methylation) or even capG2 (single methylation). The cap structures in RNAs from species that lack the hMTr1 ortholog are worth experimental examination. The second possibility is that the ‘missing’ cap1 activity may be provided by a 2′-O-ribose MTase from yet another family that remains to be discovered or groups with proteins of other specificities. The 2′-O-ribose methylation activity has evolved independently several times (31,51,52) and many members of other 2′-O-ribose MTase families (e.g. proteins from the completely unrelated SPOUT superfamily) remain to be characterized functionally. In organisms, such as P. marinus that possess multiple orthologs of one human enzyme with one activity, these proteins may have evolved toward distinct specificities. A third plausible explanation is that some of the hMTr2 orthologs are capable of both cap1 and cap2 methylation. No enzyme with such activity has been reported; however, there are many examples of MTases that modify two or more nucleosides in their substrates, including the bifunctional cap0/cap1 MTase domain from flaviviruses (53).
One striking difference between hMTr1 and hMTr2 is the C-terminal fusion of the catalytic MTase domain with an apparently inactivated variant of either a GTase or a MTase domain, respectively. It appears that the GTase-like domain is not important for the hMTr1 activity, while the MTase-like domain is essential for hMTr2. We hypothesize that it may be involved in substrate binding rather than directly in catalysis, in analogy to the rRNA MTase RsmC, which also contains two RFM domains: one involved in catalysis and the other in RNA binding (54). We plan to address the role of individual domains in the hMTr2 activity in future studies.
Our understanding of the role of the 2′-O-ribose methylations in the cap structure is still incomplete. Impact on translation rates and spliceosomal assembly have been demonstrated, but little is known about the mechanisms. The increase of translation rate by 2′-O-ribose methylations in Xenopus (5,55), together with the fact that mRNA molecules are undermethylated at position 2 in vivo suggests that the pattern of cap 2′-O-ribose methylation by hMTr2 may be a key point of translation regulation. The identification of the gene-encoding hMTr2 in humans and characterization of the enzyme provides a stepping stone toward the understanding of 2′-O-ribose methylation of RNAs in gene expression and mRNA splicing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Polish Ministry of Science and Higher Education (N301 4253 38 to J.M.B.); National Institutes of Health (AI056034 to D.A.C. and N.R.S.); the Foundation for Polish Science START fellowships (to K.H.K. and E.P.); USPHS National Research Service Award (GM07104 to J.R.Z.). Funding for open access charge: The Polish Ministry of Science and Higher Education.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Małgorzata Durawa (IIMCB) for protein preps used in this work, and Małgorzata Perycz, Mirosław Śmietański and Marcin Nowotny for their participation in the project and contribution of data that helped to plan experiments described in this manuscript. M.W., E.P. and K.H.K. have been students of the PhD school at the IBB PAS in Warsaw.
REFERENCES
- 1.Muthukrishnan S, Filipowicz W, Sierra JM, Both GW, Shatkin AJ, Ochoa S. mRNA methylation and protein synthesis in extracts from embryos of brine shrimp, Artemia salina. J. Biol. Chem. 1975;250:9336–9341. [PubMed] [Google Scholar]
- 2.Wang SP, Deng L, Ho CK, Shuman S. Phylogeny of mRNA capping enzymes. Proc. Natl Acad. Sci. USA. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 2002;3:619–625. doi: 10.1038/nrm880. [DOI] [PubMed] [Google Scholar]
- 4.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 5.Kuge H, Brownlee GG, Gershon PD, Richter JD. Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 1998;26:3208–3214. doi: 10.1093/nar/26.13.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schroeder SC, Zorio DA, Schwer B, Shuman S, Bentley D. A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol. Cell. 2004;13:377–387. doi: 10.1016/s1097-2765(04)00007-3. [DOI] [PubMed] [Google Scholar]
- 7.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 8.Chu C, Shatkin AJ. Apoptosis and autophagy induction in mammalian cells by small interfering RNA knockdown of mRNA capping enzymes. Mol. Cell Biol. 2008;28:5829–5836. doi: 10.1128/MCB.00021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsukamoto T, Shibagaki Y, Imajoh-Ohmi S, Murakoshi T, Suzuki M, Nakamura A, Gotoh H, Mizumoto K. Isolation and characterization of the yeast mRNA capping enzyme beta subunit gene encoding RNA 5′-triphosphatase, which is essential for cell viability. Biochem. Biophys. Res. Commun. 1997;239:116–122. doi: 10.1006/bbrc.1997.7439. [DOI] [PubMed] [Google Scholar]
- 10.Mao X, Schwer B, Shuman S. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol. Cell Biol. 1995;15:4167–4174. doi: 10.1128/mcb.15.8.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shafer B, Chu C, Shatkin AJ. Human mRNA cap methyltransferase: alternative nuclear localization signal motifs ensure nuclear localization required for viability. Mol. Cell Biol. 2005;25:2644–2649. doi: 10.1128/MCB.25.7.2644-2649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jove R, Manley JL. In vitro transcription from the adenovirus 2 major late promoter utilizing templates truncated at promoter-proximal sites. J. Biol. Chem. 1984;259:8513–8521. [PubMed] [Google Scholar]
- 13.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 15.Benarroch D, Smith P, Shuman S. Characterization of a trifunctional mimivirus mRNA capping enzyme and crystal structure of the RNA triphosphatase domain. Structure. 2008;16:501–512. doi: 10.1016/j.str.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Mattaj IW. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986;46:905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- 17.Plessel G, Luhrmann R, Kastner B. Electron microscopy of assembly intermediates of the snRNP core: morphological similarities between the RNA-free (E.F.G) protein heteromer and the intact snRNP core. J. Mol. Biol. 1997;265:87–94. doi: 10.1006/jmbi.1996.0713. [DOI] [PubMed] [Google Scholar]
- 18.Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonne R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell. 2002;9:891–901. doi: 10.1016/s1097-2765(02)00484-7. [DOI] [PubMed] [Google Scholar]
- 19.Singh R, Reddy R. Gamma-monomethyl phosphate: a cap structure in spliceosomal U6 small nuclear RNA. Proc. Natl Acad. Sci. USA. 1989;86:8280–8283. doi: 10.1073/pnas.86.21.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langberg SR, Moss B. Post-transcriptional modifications of mRNA. Purification and characterization of cap I and cap II RNA- (nucleoside-2′-) methyltransferases from HeLa cells. J. Biol. Chem. 1981;256:10054–10060. [PubMed] [Google Scholar]
- 22.Furuichi Y, Morgan M, Shatkin AJ, Jelinek W, Salditt-Georgieff M, Darnell JE. Methylated, blocked 5 termini in HeLa cell mRNA. Proc. Natl Acad. Sci. USA. 1975;72:1904–1908. doi: 10.1073/pnas.72.5.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massenet S, Mougin A, Branlant C. Posttranscriptional modifications in the U small nuclear RNAs. In: Grosjean H, Benne R, editors. The Modification and Editing of RNA. Washington, DC: ASM Press; 1998. pp. 201–227. [Google Scholar]
- 24.Donmez G, Hartmuth K, Luhrmann R. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA. 2004;10:1925–1933. doi: 10.1261/rna.7186504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bangs JD, Crain PF, Hashizume T, McCloskey JA, Boothroyd JC. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 1992;267:9805–9815. [PubMed] [Google Scholar]
- 26.Zamudio JR, Mittra B, Foldynova-Trantirkova S, Zeiner GM, Lukes J, Bujnicki JM, Sturm NR, Campbell DA. The 2′-O-ribose methyltransferase for cap 1 of spliced leader RNA and U1 small nuclear RNA in Trypanosoma brucei. Mol. Cell. Biol. 2007;27:6084–6092. doi: 10.1128/MCB.00647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zamudio JR, Mittra B, Zeiner GM, Feder M, Bujnicki JM, Sturm NR, Campbell DA. Complete cap 4 formation is not required for viability in Trypanosoma brucei. Eukaryot Cell. 2006;5:905–915. doi: 10.1128/EC.00080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arhin GK, Li H, Ullu E, Tschudi C. A protein related to the vaccinia virus cap-specific methyltransferase VP39 is involved in cap 4 modification in Trypanosoma brucei. RNA. 2006;12:53–62. doi: 10.1261/rna.2223406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zamudio JR, Mittra B, Campbell DA, Sturm NR. Hypermethylated cap 4 maximizes Trypanosoma brucei translation. Mol. Microbiol. 2009;72:1100–1110. doi: 10.1111/j.1365-2958.2009.06696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plotch SJ, Bouloy M, Krug RM. Transfer of 5′-terminal cap of globin mRNA to influenza viral complementary RNA during transcription in vitro. Proc. Natl Acad. Sci. USA. 1979;76:1618–1622. doi: 10.1073/pnas.76.4.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feder M, Pas J, Wyrwicz LS, Bujnicki JM. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene. 2003;302:129–138. doi: 10.1016/s0378-1119(02)01097-1. [DOI] [PubMed] [Google Scholar]
- 32.Belanger F, Stepinski J, Darzynkiewicz E, Pelletier J. Characterization of hMTr1, a human cap1 2′O-ribose methyltransferase. J. Biol. Chem. 2010;285:33037–33044. doi: 10.1074/jbc.M110.155283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hausmann S, Zheng S, Costanzo M, Brost RL, Garcin D, Boone C, Shuman S, Schwer B. Genetic and biochemical analysis of yeast and human cap trimethylguanosine synthase: functional overlap of 2,2,7-trimethylguanosine caps, small nuclear ribonucleoprotein components, pre-mRNA splicing factors, and RNA decay pathways. J. Biol. Chem. 2008;283:31706–31718. doi: 10.1074/jbc.M806127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coleman TM, Wang G, Huang F. Superior 5′ homogeneity of RNA from ATP-initiated transcription under the T7 phi 2.5 promoter. Nucleic Acids Res. 2004;32:e14. doi: 10.1093/nar/gnh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 37.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurowski MA, Bujnicki JM. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31:3305–3307. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 40.Wu X, Guarino LA. Autographa californica nucleopolyhedrovirus orf69 encodes an RNA cap (nucleoside-2′-O)-methyltransferase. J. Virol. 2003;77:3430–3440. doi: 10.1128/JVI.77.6.3430-3440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mittra B, Zamudio JR, Bujnicki JM, Stepinski J, Darzynkiewicz E, Campbell DA, Sturm NR. The TbMTr1 spliced leader RNA cap 1 2′-O-ribose methyltransferase from Trypanosoma brucei acts with substrate specificity. J. Biol. Chem. 2008;283:3161–3172. doi: 10.1074/jbc.M707367200. [DOI] [PubMed] [Google Scholar]
- 42.Haline-Vaz T, Silva TC, Zanchin NI. The human interferon-regulated ISG95 protein interacts with RNA polymerase II and shows methyltransferase activity. Biochem. Biophys. Res. Commun. 2008;372:719–724. doi: 10.1016/j.bbrc.2008.05.137. [DOI] [PubMed] [Google Scholar]
- 43.Hager J, Staker BL, Bugl H, Jakob U. Active site in RrmJ, a heat shock-induced methyltransferase. J. Biol. Chem. 2002;277:41978–41986. doi: 10.1074/jbc.M205423200. [DOI] [PubMed] [Google Scholar]
- 44.Grosjean H, Droogmans L, Roovers M, Keith G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 2007;425:55–101. doi: 10.1016/S0076-6879(07)25003-7. [DOI] [PubMed] [Google Scholar]
- 45.Schnierle BS, Gershon PD, Moss B. Cap-specific mRNA- (nucleoside-O2′-) methyltransferase and poly(A) polymerase stimulatory activities of vaccinia virus are mediated by a single protein. Proc. Natl Acad. Sci. USA. 1992;89:2897–2901. doi: 10.1073/pnas.89.7.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arhin GK, Ullu E, Tschudi C. 2′-O-methylation of position 2 of the trypanosome spliced leader cap 4 is mediated by a 48 kDa protein related to vaccinia virus VP39. Mol. Biochem. Parasitol. 2006;147:137–139. doi: 10.1016/j.molbiopara.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 47.Hall MP, Ho CK. Functional characterization of a 48 kDa Trypanosoma brucei cap 2 RNA methyltransferase. Nucleic Acids Res. 2006;34:5594–5602. doi: 10.1093/nar/gkl573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zamudio JR, Mittra B, Chattopadhyay A, Wohlschlegel JA, Sturm NR, Campbell DA. Trypanosoma brucei spliced leader RNA maturation by the cap 1 2′-O-ribose methyltransferase and SLA1 H/ACA snoRNA pseudouridine synthase complex. Mol. Cell. Biol. 2009;29:1202–1211. doi: 10.1128/MCB.01496-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perry RP, Kelley DE. Kinetics of formation of 5′ terminal caps in mRNA. Cell. 1976;8:433–442. doi: 10.1016/0092-8674(76)90156-2. [DOI] [PubMed] [Google Scholar]
- 50.Friderici K, Kaehler M, Rottman F. Kinetics of Novikoff cytoplasmic messenger RNA methylation. Biochemistry. 1976;15:5234–5241. doi: 10.1021/bi00669a006. [DOI] [PubMed] [Google Scholar]
- 51.Tkaczuk KL, Obarska A, Bujnicki JM. Molecular phylogenetics and comparative modeling of HEN1, a methyltransferase involved in plant microRNA biogenesis. BMC Evol. Biol. 2006;6:6. doi: 10.1186/1471-2148-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tkaczuk KL, Dunin-Horkawicz S, Purta E, Bujnicki JM. Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinformatics. 2007;8:73. doi: 10.1186/1471-2105-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 2006;80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sunita S, Purta E, Durawa M, Tkaczuk KL, Swaathi J, Bujnicki JM, Sivaraman J. Functional specialization of domains tandemly duplicated within 16S rRNA methyltransferase RsmC. Nucleic Acids Res. 2007;35:4264–4274. doi: 10.1093/nar/gkm411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuge H, Richter JD. Cytoplasmic 3′ poly(A) addition induces 5′ cap ribose methylation: implications for translational control of maternal mRNA. EMBO J. 1995;14:6301–6310. doi: 10.1002/j.1460-2075.1995.tb00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.