Abstract
The standard genetic code is used by most living organisms, yet deviations have been observed in many genomes, suggesting that the genetic code has been evolving. In certain yeast mitochondria, CUN codons are reassigned from leucine to threonine, which requires an unusual tRNAThr with an enlarged 8-nt anticodon loop (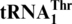 ). To trace its evolutionary origin we performed a comprehensive phylogenetic analysis which revealed that
). To trace its evolutionary origin we performed a comprehensive phylogenetic analysis which revealed that 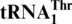 evolved from yeast mitochondrial tRNAHis. To understand this tRNA identity change, we performed mutational and biochemical experiments. We show that Saccharomyces cerevisiae mitochondrial threonyl-tRNA synthetase (MST1) could attach threonine to both
evolved from yeast mitochondrial tRNAHis. To understand this tRNA identity change, we performed mutational and biochemical experiments. We show that Saccharomyces cerevisiae mitochondrial threonyl-tRNA synthetase (MST1) could attach threonine to both 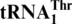 and the regular
and the regular 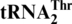 , but not to the wild-type tRNAHis. A loss of the first nucleotide (G−1) in tRNAHis converts it to a substrate for MST1 with a Km value (0.7 μM) comparable to that of
, but not to the wild-type tRNAHis. A loss of the first nucleotide (G−1) in tRNAHis converts it to a substrate for MST1 with a Km value (0.7 μM) comparable to that of 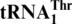 (0.3 μM), and addition of G−1 to
(0.3 μM), and addition of G−1 to 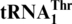 allows efficient histidylation by histidyl-tRNA synthetase. We also show that MST1 from Candida albicans, a yeast in which CUN codons remain assigned to leucine, could not threonylate
allows efficient histidylation by histidyl-tRNA synthetase. We also show that MST1 from Candida albicans, a yeast in which CUN codons remain assigned to leucine, could not threonylate 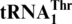 , suggesting that MST1 has coevolved with
, suggesting that MST1 has coevolved with 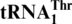 . Our work provides the first clear example of a recent recoding event caused by alloacceptor tRNA gene recruitment.
. Our work provides the first clear example of a recent recoding event caused by alloacceptor tRNA gene recruitment.
INTRODUCTION
Transfer RNAs (tRNAs) are adaptor molecules that pair each amino acid with corresponding codons on the mRNA (1). Aminoacyl-tRNA synthetases (aaRSs) attach amino acids to the 3′-terminus of tRNAs (2), and the resulting aminoacyl-tRNAs (aa-tRNAs) are delivered by elongation factors to the ribosome, where codon–anticodon recognition defines the genetic code of life (3). The genetic code was once thought to be universal and frozen (4), but later studies revealed multiple codon reassignment events in nuclear and mitochondrial genomes (5–7). These reassignments include stop-to-sense, sense-to-sense and sense-to-stop codon changes; they have been found in nuclear genomes from all three domains of life. In some bacterial, archaeal and eukaryotic species the UGA codon is reassigned to allow insertion of selenocysteine (Sec), the 21st natural amino acid, in the presence of tRNASec with a UCA anticodon (6,8). In a number of methanogenic archaea a UAG stop codon encodes the 22nd amino acid, pyrrolysine (Pyl), implemented by a tRNAPyl which recognizes UAG (9,10). A major reassignment in mitochondria is recoding UGA to tryptophan (Trp) implemented by a tRNATrp with a mutated anticodon (7). Furthermore, the mitochondria of several yeast species reassigned the AUA codon from isoleucine to methionine (Met) by the abnormal recognition of AUA by tRNAMet (7).
One last codon reassignment in the well-studied organism Saccharomyces cerevisiae is still not understood. In certain budding yeasts (including Saccharomyces, Nakaseomyces and Vanderwaltozyma) the four CUN (N denotes U, C, A or G) codons in the mitochondria are reassigned from leucine (Leu) to threonine (Thr) (11,12). This results from the loss of 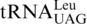 (with a UAG anticodon) that would translate CUN codons, and from the presence of an abnormal
(with a UAG anticodon) that would translate CUN codons, and from the presence of an abnormal 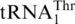 with an enlarged 8-nt anticodon loop and a UAG anticodon (Figure 1) (11,13). This codon sense change was confirmed by protein sequencing of the S. cerevisiae mitochondrial ATPase (14). Mass spectrometry studies have also validated that at least three CUU and two CUA codons in S. cerevisiae mitochondrial-encoded proteins are recoded as Thr (15). In addition to
with an enlarged 8-nt anticodon loop and a UAG anticodon (Figure 1) (11,13). This codon sense change was confirmed by protein sequencing of the S. cerevisiae mitochondrial ATPase (14). Mass spectrometry studies have also validated that at least three CUU and two CUA codons in S. cerevisiae mitochondrial-encoded proteins are recoded as Thr (15). In addition to 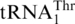 , yeast mitochondria also express a normal
, yeast mitochondria also express a normal 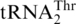 with a UGU anticodon that reads the ACN threonine codon box (Figure 1). It was shown previously that while mitochondrial extracts from a wild-type (WT) S. cerevisiae strain could attach Thr to both
with a UGU anticodon that reads the ACN threonine codon box (Figure 1). It was shown previously that while mitochondrial extracts from a wild-type (WT) S. cerevisiae strain could attach Thr to both 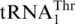 and
and 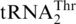 , extracts from an MST1 mutant strain could only threonylate
, extracts from an MST1 mutant strain could only threonylate 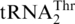 but not
but not 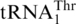 (16). Thus it was suggested that MST1 serves as a mitochondrial threonyl-tRNA synthetase (ThrRS) specific for aminoacylation of
(16). Thus it was suggested that MST1 serves as a mitochondrial threonyl-tRNA synthetase (ThrRS) specific for aminoacylation of 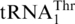 , while a different ThrRS aminoacylates
, while a different ThrRS aminoacylates 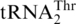 in the mitochondria. Three decades after this discovery it is still a mystery how the unusual
in the mitochondria. Three decades after this discovery it is still a mystery how the unusual 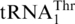 emerged in the mitochondrial genome. Previous hypotheses suggest that
emerged in the mitochondrial genome. Previous hypotheses suggest that 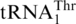 might have evolved from
might have evolved from 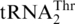 , or alternatively from the missing
, or alternatively from the missing 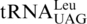 (11,17,18). However, both hypotheses lack convincing experimental evidence. To our surprise, biochemical and phylogenetic analyses demonstrate that
(11,17,18). However, both hypotheses lack convincing experimental evidence. To our surprise, biochemical and phylogenetic analyses demonstrate that 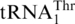 directly evolved from mitochondrial tRNAHis. Saccharomyces cerevisiae mitochondrial tRNAHis shares high (72%) sequence identity with
directly evolved from mitochondrial tRNAHis. Saccharomyces cerevisiae mitochondrial tRNAHis shares high (72%) sequence identity with 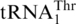 , and a single-nucleotide change converts tRNAHis to a substrate for MST1. Our work thus resolves the long-standing question regarding the origin of
, and a single-nucleotide change converts tRNAHis to a substrate for MST1. Our work thus resolves the long-standing question regarding the origin of 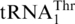 and its coding response.
and its coding response.
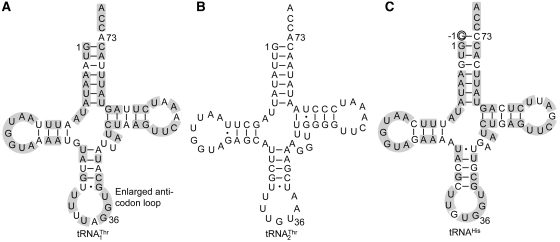
Figure 1.
Nucleotide sequences of S. cerevisiae mitochondrial tRNAs. (A) 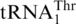 with an 8-nt anticodon loop and a UAG anticodon. (B)
with an 8-nt anticodon loop and a UAG anticodon. (B) 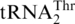 with a canonical UGU anticodon. (C) tRNAHis with a G−1 (circled, a major anti-determinant for ThrRS). The primary sequences of
with a canonical UGU anticodon. (C) tRNAHis with a G−1 (circled, a major anti-determinant for ThrRS). The primary sequences of 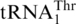 and tRNAHis are 72% identical (shaded).
and tRNAHis are 72% identical (shaded).
MATERIALS AND METHODS
Cloning, mutagenesis and general methods
Saccharomyces cerevisiae MST1, S. cerevisiae HTS1, Candida albicans MST1 and Schizosaccharomyces pombe MST1 genes were cloned into pET28a expression vector (Novagen) with an N-terminal six-His tag. Expression of recombinant proteins was induced at 37°C for 4 h with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) in Escherichia coli strain BL21-codon plus in Luria–Bertani (LB) media. His-tagged proteins were purified according to standard procedures. Mitochondrial tRNA genes were cloned into pUC18 vector (GenScript), and mutations were introduced using QuikChange Site-Directed Mutagenesis Kit (Stratagene).
In vitro assays
In vitro tRNA transcripts were obtained using the T7 RNA polymerase runoff procedure as described (19). Aminoacylation experiments were performed as described (20) in the presence of 100 mM Na-HEPES pH 7.2, 30 mM KCl, 10 mM MgCl2, 2 mM ATP, 25 µM [3H] Thr (100 μCi/ml), 25 µM [3H] His (150 μCi/ml) or 50 µM [3H] Leu (100 μCi/ml), 0.2–9 µM tRNA transcripts and 3–300 nM aaRSs.
Phylogenetic analyses
For phylogenetic analysis of mitochondrial tRNA sequences, tRNAs were predicted in mitochondrial genomes of interest with Erpin/RNAweasel (21,22) and a tRNA profile specific for fungal mitochondria that may be used via our website (http://megasun.bch.umontreal.ca/RNAweasel). Sequence columns representing the sequence positions in the variable loop between the anticodon- and T-loop were removed as the nucleotides in this region are extremely diverse in sequence and length, and phylogenetic analysis was conducted using either a Bayesian [PhyloBayes; (23)] or maximum likelihood approach [RaxML with GTR model; (24)]. For inference of a species tree, an application developed in-house (Mams) was used for automated mitochondrial protein alignment, removal of ambiguous regions in the alignment, and concatenation. Briefly, derived Cob, Cox1-3, Atp6, 9 and Nad1-6 protein sequences are pre-aligned with Muscle (25), alignments are iteratively refined with HMMalign (S. Eddy, http://hmmer.janelia.org) using E-values obtained with respective HMM models as an optimization criterion. Sequence positions that are not aligned with a posterior probability value of 1 are removed from the resulting alignment. This dataset with 22 taxa and 3728 amino acid positions was analyzed using Bayesian inference by PhyloBayes that implements the CAT + GTR model, known to be among the least sensitive to LBA artifacts [(23,26,27), and references therein].
RESULTS
Phylogenetic analysis reveals that 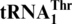 originated from tRNAHis
originated from tRNAHis
The unsolved question of the yeast mitochondrial CUN codon reassignment is the evolutionary origin of 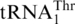 with an 8-nt anticodon loop and a UAG anticodon. To analyze the
with an 8-nt anticodon loop and a UAG anticodon. To analyze the 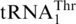 recruitment, we performed a phylogenetic analysis of all mitochondrial tRNAs of S. cerevisiae and related yeast species using Bayesian inference (Figure 2). Not surprisingly, the 10 organisms with
recruitment, we performed a phylogenetic analysis of all mitochondrial tRNAs of S. cerevisiae and related yeast species using Bayesian inference (Figure 2). Not surprisingly, the 10 organisms with 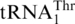 are closely related; yet this
are closely related; yet this 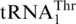 cluster is related to tRNAHis (72% sequence identity between the two respective S. cerevisiae tRNAs; Figure 1). In contrast,
cluster is related to tRNAHis (72% sequence identity between the two respective S. cerevisiae tRNAs; Figure 1). In contrast, 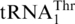 and
and 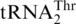 are definitively distant in the phylogeny (Figures 2, Supplementary Figure S1) and the respective S. cerevisiae tRNAs share only 52% sequence identity. As the phylogenetic signal in tRNA sequences is limited by the small number of informative sequence sites, and not a reliable marker in species phylogenies because of occasional identity shifts [e.g. (28)], we have built a yeast species tree based on mitochondrion-encoded proteins to permit mapping of evolutionary changes to this tree. The result of this phylogenetic analysis (Figure 3) is consistent with a single origin of the 10 yeast species that have a
are definitively distant in the phylogeny (Figures 2, Supplementary Figure S1) and the respective S. cerevisiae tRNAs share only 52% sequence identity. As the phylogenetic signal in tRNA sequences is limited by the small number of informative sequence sites, and not a reliable marker in species phylogenies because of occasional identity shifts [e.g. (28)], we have built a yeast species tree based on mitochondrion-encoded proteins to permit mapping of evolutionary changes to this tree. The result of this phylogenetic analysis (Figure 3) is consistent with a single origin of the 10 yeast species that have a 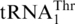 homolog with an 8-nt anticodon loop, a clade emerging close to the divergence of Kluyveromyces species and subsequent to Pichia canadensis. Together, these results strongly suggest that
homolog with an 8-nt anticodon loop, a clade emerging close to the divergence of Kluyveromyces species and subsequent to Pichia canadensis. Together, these results strongly suggest that 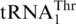 derived from mitochondrial tRNAHis in an ancestral yeast species.
derived from mitochondrial tRNAHis in an ancestral yeast species.
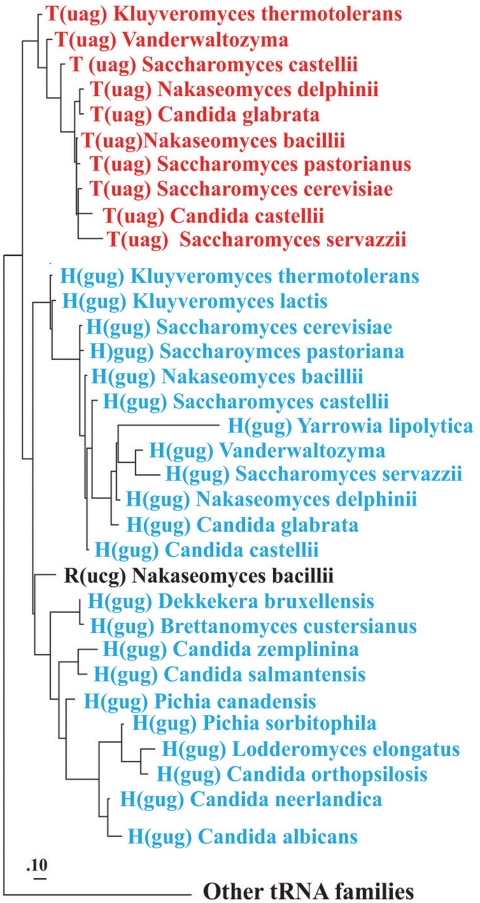
Figure 2.
Phylogeny of yeast mitochondrial tRNAs. The phylogenetic analysis with PhyloBayes (default model parameters) contained all tRNA sequences from the species shown in Figure 3. Only the section of the tRNA phylogeny covering the 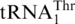 and tRNAHis clusters are shown (marked red and blue, respectively), confirming monophyly of
and tRNAHis clusters are shown (marked red and blue, respectively), confirming monophyly of 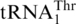 and a sister group relationship to tRNAHis. The posterior probability support for the two tRNA groups is 1.0 (note that phylogenetic analysis with tRNA sequences depends on only few informative nucleotide positions, which does not allow to resolve the branching order within these groups). Removal of the anticodon sequence positions from the dataset did not change clustering into the two tRNA groups, nor did an analysis of the same datasets with maximum likelihood and the GTR model.
and a sister group relationship to tRNAHis. The posterior probability support for the two tRNA groups is 1.0 (note that phylogenetic analysis with tRNA sequences depends on only few informative nucleotide positions, which does not allow to resolve the branching order within these groups). Removal of the anticodon sequence positions from the dataset did not change clustering into the two tRNA groups, nor did an analysis of the same datasets with maximum likelihood and the GTR model.
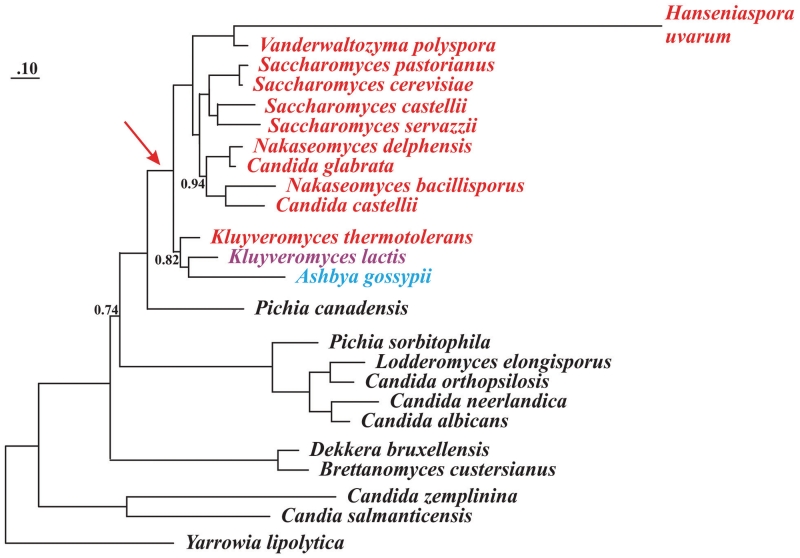
Figure 3.
Phylogeny of yeast species based on concatenated mtDNA-encoded protein sequences. The phylogenetic analysis with PhyloBayes and the CAT model is based on 13 mtDNA encoded proteins. All divergence points are supported by posterior probability values of 1.0, except where indicated. The red arrow points to the concomitant loss of all seven nad genes and the start of mitochondrial codon reassignments, including AUA methionine, CUN threonine. Species shown in black possess mitochondrial 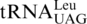 (but not
(but not 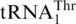 ), and in these organisms CUN codons are assigned to Leu. The yeast species marked red, such as K. thermotolerans, have lost mitochondrial
), and in these organisms CUN codons are assigned to Leu. The yeast species marked red, such as K. thermotolerans, have lost mitochondrial 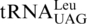 and obtained
and obtained 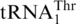 that decodes CUN codons as Thr. K. lactis is marked magenta as it has no CUN codons and no corresponding tRNA with a UAG anticodon. A. gossypii (marked blue) contains a tRNA species with a UAG anticodon, yet its identity of this tRNA and the amino acid reading CUN codons in A. gossypii remain obscure (to be discussed by BFL in Organelle Genetics: Evolution of Organelle Genomes and Gene Expression, Springer 2011).
that decodes CUN codons as Thr. K. lactis is marked magenta as it has no CUN codons and no corresponding tRNA with a UAG anticodon. A. gossypii (marked blue) contains a tRNA species with a UAG anticodon, yet its identity of this tRNA and the amino acid reading CUN codons in A. gossypii remain obscure (to be discussed by BFL in Organelle Genetics: Evolution of Organelle Genomes and Gene Expression, Springer 2011).
Saccharomyces cerevisiae MST1 threonylates both 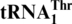 and
and 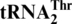
The mitochondria of all yeast species containing 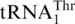 have lost the
have lost the 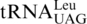 gene, leaving
gene, leaving 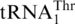 the only tRNA capable of reading CUN codons. Whereas S. cerevisiae mitochondrial LeuRS (ScmtLeuRS) efficiently attaches Leu to
the only tRNA capable of reading CUN codons. Whereas S. cerevisiae mitochondrial LeuRS (ScmtLeuRS) efficiently attaches Leu to 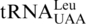 (29,30), it could not recognize
(29,30), it could not recognize 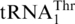 (Figure 4A), confirming that the S. cerevisiae mitochondrial CUN codons are assigned to Thr instead of Leu. It is known that the WT S. cerevisiae strain attaches Thr to
(Figure 4A), confirming that the S. cerevisiae mitochondrial CUN codons are assigned to Thr instead of Leu. It is known that the WT S. cerevisiae strain attaches Thr to 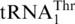 , and an MST1 deletion strain fails to threonylate
, and an MST1 deletion strain fails to threonylate 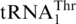 , resulting in a respiration-deficient phenotype (16). This suggests that MST1 is a putative mitochondrial ThrRS. Compared with bacterial ThrRSs, yeast MST1 enzymes lack an N-terminal editing domain, but share homologous catalytic and tRNA anticodon binding domains. To test aminoacylation of
, resulting in a respiration-deficient phenotype (16). This suggests that MST1 is a putative mitochondrial ThrRS. Compared with bacterial ThrRSs, yeast MST1 enzymes lack an N-terminal editing domain, but share homologous catalytic and tRNA anticodon binding domains. To test aminoacylation of 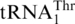 directly, the S. cerevisiae MST1 gene was cloned into pET28a for protein overexpression in E. coli. N-terminal His-tagged MST1 was purified to homogeneity and tested in aminoacylation reactions in the presence of [3H] Thr and in vitro transcribed S. cerevisiae tRNAs. Consistent with previous in vivo results (16), ScMST1 was able to charge
directly, the S. cerevisiae MST1 gene was cloned into pET28a for protein overexpression in E. coli. N-terminal His-tagged MST1 was purified to homogeneity and tested in aminoacylation reactions in the presence of [3H] Thr and in vitro transcribed S. cerevisiae tRNAs. Consistent with previous in vivo results (16), ScMST1 was able to charge 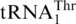 with Thr in vitro (Figure 4B). MST1 could also threonylate
with Thr in vitro (Figure 4B). MST1 could also threonylate 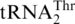 , which was unexpected as it was previously thought that a second mitochondrial ThrRS was responsible for
, which was unexpected as it was previously thought that a second mitochondrial ThrRS was responsible for 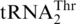 aminoacylation. Steady-state kinetic experiments revealed that ScMST1 recognized
aminoacylation. Steady-state kinetic experiments revealed that ScMST1 recognized 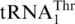 and
and 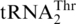 with high affinity, with Km values of 0.29 and 0.44 µM, respectively (Table 1), suggesting that tRNA modifications (17) are not critical for MST1 recognition. These data, together with previous in vivo results, establishes unequivocally that
with high affinity, with Km values of 0.29 and 0.44 µM, respectively (Table 1), suggesting that tRNA modifications (17) are not critical for MST1 recognition. These data, together with previous in vivo results, establishes unequivocally that 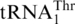 is indeed threonylated by MST1. While we favor that MST1 threonylates
is indeed threonylated by MST1. While we favor that MST1 threonylates 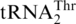 in vivo, we do not exclude the possibility that a second ThrRS activity is present in yeast mitochondria.
in vivo, we do not exclude the possibility that a second ThrRS activity is present in yeast mitochondria.
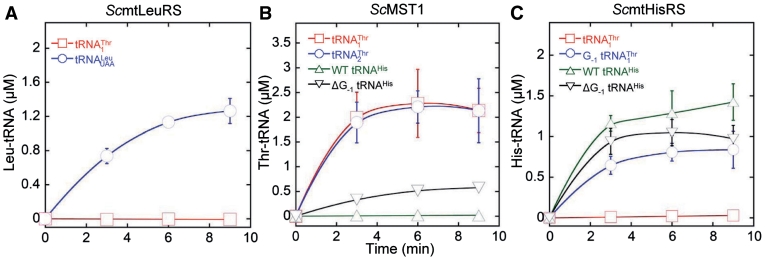
Figure 4.
Aminoacylation by S. cerevisiae MST1 and HisRS. (A) Leucylation of 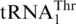 (3 µM) and
(3 µM) and 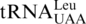 (3 µM) by ScmtLeuRS (0.3 µM). (B) Threonylation of
(3 µM) by ScmtLeuRS (0.3 µM). (B) Threonylation of 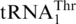 (3 µM),
(3 µM), 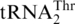 (3 µM) and tRNAHis variants (3 µM) by ScMST1 (0.3 µM). (C) Histidylation of
(3 µM) and tRNAHis variants (3 µM) by ScMST1 (0.3 µM). (C) Histidylation of 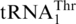 (3 µM) and tRNAHis variants (3 µM) by ScmtHisRS (0.3 µM).
(3 µM) and tRNAHis variants (3 µM) by ScmtHisRS (0.3 µM).
Table 1.
Threonylation by ScMST1
| Anticodon loop | kcat (min−1) | Km (μM) | kcat/Km (μM min−1) | Relative kcat/Km | |
|---|---|---|---|---|---|
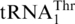 |
|||||
| WT | UUUUAGGU | 2.8 ± 0.4 | 0.29 ± 0.09 | 10.3 ± 2.3 | 100 |
| G−1 | UUUUAGGU | 0.094 ± 0.032 | 1.5 ± 0.6 | 0.065 ± 0.006 | 0.63 |
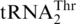 |
|||||
| WT | UUUGUAA | 2.3 ± 0.1 | 0.44 ± 0.04 | 5.4 ± 0.6 | 52 |
| tRNAHis | |||||
| WT | UUGUGGU | ND | ND | ND | ND |
| InsA35 | UUGUAGGU | ND | ND | ND | ND |
| InsA35/C73A | UUGUAGGU | ND | ND | ND | ND |
| ΔG−1 | UUGUGGU | 0.27 ± 0.10 | 0.74 ± 0.41 | 0.39 ± 0.06 | 3.8 |
| ΔG−1/InsA35 | UUGUAGGU | 0.34 ± 0.07 | 0.20 ± 0.02 | 1.7 ± 0.5 | 17 |
| ΔG−1/C73A | UUGUGGU | 0.23 ± 0.01 | 0.85 ± 0.32 | 0.29 ± 0.09 | 2.8 |
| ΔG−1/C73A/InsA35 | UUGUAGGU | 1.2 ± 0.4 | 0.57 ± 0.26 | 2.4 ± 1.0 | 23 |
ND, not determined due to low activity.
Loss of G−1 converts S. cerevisiae tRNAHis to a substrate for S. cerevisiae MST1
To provide experimental evidence for the evolution of 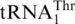 , we examined recognition of tRNAHis variants by MST1. Except in a few α-proteobacteria [(31), and references therein], all known tRNAHis species contain a G at position −1, which is a critical identity element for histidyl-tRNA synthetase (HisRS) [(32,33) and references therein]. Sequence alignments of
, we examined recognition of tRNAHis variants by MST1. Except in a few α-proteobacteria [(31), and references therein], all known tRNAHis species contain a G at position −1, which is a critical identity element for histidyl-tRNA synthetase (HisRS) [(32,33) and references therein]. Sequence alignments of 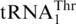 and mitochondrial tRNAHis genes revealed that G−1 addition in tRNAHis comprises one major difference between the two tRNA species (Figure 5). In vitro, ScMST1 failed to charge the WT SctRNAHis transcript with Thr, but deleting G−1 stimulated threonylation of tRNAHis by ScMST1 (Figure 4B). Steady-state kinetic data showed that ΔG−1 tRNAHis gained 4% threonylation activity of the WT
and mitochondrial tRNAHis genes revealed that G−1 addition in tRNAHis comprises one major difference between the two tRNA species (Figure 5). In vitro, ScMST1 failed to charge the WT SctRNAHis transcript with Thr, but deleting G−1 stimulated threonylation of tRNAHis by ScMST1 (Figure 4B). Steady-state kinetic data showed that ΔG−1 tRNAHis gained 4% threonylation activity of the WT 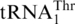 (Table 1). Compared with
(Table 1). Compared with 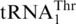 , ΔG−1 tRNAHis displayed 10-fold lower kcat and 3-fold higher Km values for threonylation by ScMST1. In addition to G−1, other major differences between tRNAHis and
, ΔG−1 tRNAHis displayed 10-fold lower kcat and 3-fold higher Km values for threonylation by ScMST1. In addition to G−1, other major differences between tRNAHis and 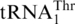 include an A insertion in
include an A insertion in 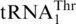 at position 35, and the discriminator base at position 73 (Figure 5). Changing C73 to A did not improve the threonylation efficiency of ΔG−1 tRNAHis, but inserting A35 in the anticodon loop of ΔG-1 tRNAHis further increased its threonylation activity 5-fold (Table 1). In the presence of G−1, A35 insertion did not allow threonylation of tRNAHis. These results suggest that G−1 is a major anti-determinant in tRNAHis for MST1. In line with this notion, addition of G−1 to
at position 35, and the discriminator base at position 73 (Figure 5). Changing C73 to A did not improve the threonylation efficiency of ΔG−1 tRNAHis, but inserting A35 in the anticodon loop of ΔG-1 tRNAHis further increased its threonylation activity 5-fold (Table 1). In the presence of G−1, A35 insertion did not allow threonylation of tRNAHis. These results suggest that G−1 is a major anti-determinant in tRNAHis for MST1. In line with this notion, addition of G−1 to 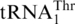 reduced its threonylation activity 150-fold (Table 1).
reduced its threonylation activity 150-fold (Table 1).
Figure 5.
Sequence alignment of mitochondrial 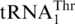 and tRNAHis. Three major differences between
and tRNAHis. Three major differences between 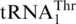 and tRNAHis sequences are indicated by boxes. Cc, Candida castellii; Cg, Candida glabrata; Kt, Kluyveromyces thermotolerans; Nb, Nakaseomyces bacillisporus; Nd, Nakaseomyces delphensis; Sca, Saccharomyces castellii; Sc, Saccharomyces cerevisiae; Sp, Saccharomyces pastorianus; Ss, Saccharomyces servazzii; Vp, Vanderwaltozyma polyspora.
and tRNAHis sequences are indicated by boxes. Cc, Candida castellii; Cg, Candida glabrata; Kt, Kluyveromyces thermotolerans; Nb, Nakaseomyces bacillisporus; Nd, Nakaseomyces delphensis; Sca, Saccharomyces castellii; Sc, Saccharomyces cerevisiae; Sp, Saccharomyces pastorianus; Ss, Saccharomyces servazzii; Vp, Vanderwaltozyma polyspora.
G−1 addition allows 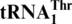 to be histidylated by S. cerevisiae HisRS
to be histidylated by S. cerevisiae HisRS
ScMST1 efficiently aminoacylates mitochondrial tRNAHis variants lacking G−1. We next investigated the recognition of tRNAHis and 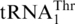 variants by mitochondrial HisRS. In S. cerevisiae, a single nuclear gene HTS1 containing two open reading frames encodes both the cytoplasmic and mitochondrial forms of HisRS (34). The mitochondrial HisRS (mtHisRS) harbors an extra mitochondria-targeting sequence, yet the predicted mature form of the mitochondrial enzyme is identical to the cytoplasmic HisRS. Previous studies revealed that ScHisRS recognizes both the G−1 and the anticodon of the cytoplasmic tRNAHis (35,36). We overexpressed mature ScmtHisRS in E. coli and purified the enzyme to homogeneity. In vitro aminoacylation reactions showed that ScmtHisRS efficiently charged His to the WT mitochondrial tRNAHis (Figure 4C, Table 2), with a Km value comparable to that of the cytoplasmic tRNAHis (35). Surprisingly, neither deleting G−1 nor inserting A35 in the anticodon loop of mitochondrial tRNAHis significantly affected the aminoacylation activity. However, when both changes were introduced into mitochondrial tRNAHis, the histidylation activity decreased by 80-fold. The discriminator base also appeared to be important for recognition by ScmtHisRS. These results indicate that in the context of S. cerevisiae mitochondrial tRNAHis, G−1 is dispensable for HisRS recognition and the anticodon can alternatively serve as the major identity element. ScmtHisRS was unable to aminoacylate WT
variants by mitochondrial HisRS. In S. cerevisiae, a single nuclear gene HTS1 containing two open reading frames encodes both the cytoplasmic and mitochondrial forms of HisRS (34). The mitochondrial HisRS (mtHisRS) harbors an extra mitochondria-targeting sequence, yet the predicted mature form of the mitochondrial enzyme is identical to the cytoplasmic HisRS. Previous studies revealed that ScHisRS recognizes both the G−1 and the anticodon of the cytoplasmic tRNAHis (35,36). We overexpressed mature ScmtHisRS in E. coli and purified the enzyme to homogeneity. In vitro aminoacylation reactions showed that ScmtHisRS efficiently charged His to the WT mitochondrial tRNAHis (Figure 4C, Table 2), with a Km value comparable to that of the cytoplasmic tRNAHis (35). Surprisingly, neither deleting G−1 nor inserting A35 in the anticodon loop of mitochondrial tRNAHis significantly affected the aminoacylation activity. However, when both changes were introduced into mitochondrial tRNAHis, the histidylation activity decreased by 80-fold. The discriminator base also appeared to be important for recognition by ScmtHisRS. These results indicate that in the context of S. cerevisiae mitochondrial tRNAHis, G−1 is dispensable for HisRS recognition and the anticodon can alternatively serve as the major identity element. ScmtHisRS was unable to aminoacylate WT 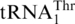 (Figure 4C). Interestingly, addition of G−1 to
(Figure 4C). Interestingly, addition of G−1 to 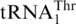 restored 7% histidylation activity of the WT tRNAHis (Figure 4C, Table 2), further strengthening our argument that the mitochondrial tRNAHis and
restored 7% histidylation activity of the WT tRNAHis (Figure 4C, Table 2), further strengthening our argument that the mitochondrial tRNAHis and 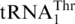 in yeast are closely related during evolution.
in yeast are closely related during evolution.
Table 2.
Histidylation by ScmtHisRS
| Anticodon loop | kcat (min−1) | Km (μM) | kcat/Km (μM min−1) | Relative kcat/Km | |
|---|---|---|---|---|---|
 |
|||||
| WT | UUUUAGGU | ND | ND | ND | ND |
| G−1 | UUUUAGGU | 1.8 ± 0.6 | 0.43 ± 0.20 | 4.3 ± 0.6 | 7.2 |
| tRNAHis | |||||
| WT | UUGUGGU | 25 ± 7 | 0.43 ± 0.18 | 60 ± 10 | 100 |
| InsA35 | UUGUAGGU | 27 ± 8 | 0.60 ± 0.13 | 45 ± 4 | 74 |
| InsA35/C73A | UUGUAGGU | 38 ± 3 | 0.28 ± 0.01 | 130 ± 16 | 220 |
| ΔG−1 | UUGUGGU | 14 ± 3 | 0.40 ± 0.22 | 45 ± 25 | 75 |
| ΔG−1/InsA35 | UUGUAGGU | 1.8 ± 0.1 | 2.4 ± 0.8 | 0.78 ± 0.22 | 1.3 |
| ΔG−1/C73A | UUGUGGU | 1.2 ± 0.2 | 0.54 ± 0.28 | 2.6 ± 1.0 | 4.3 |
| ΔG−1/C73A/InsA35 | UUGUAGGU | 0.75 ± 0.07 | 2.2 ± 0.5 | 0.35 ± 0.06 | 0.58 |
ND, not determined due to low activity.
MST1 has coevolved with 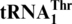 to establish CUN codon reassignment in yeast mitochondria
to establish CUN codon reassignment in yeast mitochondria
The biochemical and phylogenetic evidence above suggests that 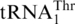 has evolved from mitochondrial tRNAHis, at a time point close to the divergence of Kluyveromyces species. To understand whether
has evolved from mitochondrial tRNAHis, at a time point close to the divergence of Kluyveromyces species. To understand whether 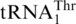 could be recognized by MST1 enzymes from other fungal species, we overexpressed and purified MST1s from C. albicans and S. pombe [a non-hemiascomycete ‘fission yeast’ belonging to Taphrinomycotina (37)], and tested them for threonylation of
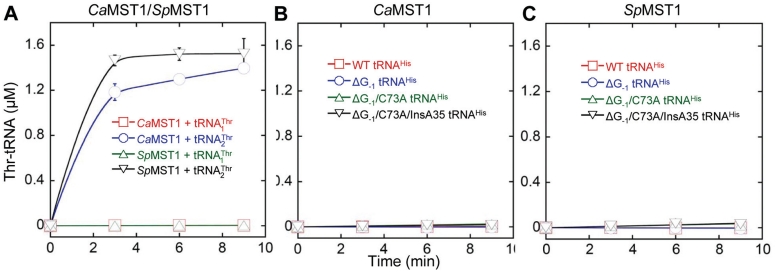
could be recognized by MST1 enzymes from other fungal species, we overexpressed and purified MST1s from C. albicans and S. pombe [a non-hemiascomycete ‘fission yeast’ belonging to Taphrinomycotina (37)], and tested them for threonylation of 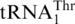 . CaMST1 and SpMST1 share 49 and 43% sequence identity with ScMST1, respectively, and both enzymes were able to aminoacylate S. cerevisiae mitochondrial
. CaMST1 and SpMST1 share 49 and 43% sequence identity with ScMST1, respectively, and both enzymes were able to aminoacylate S. cerevisiae mitochondrial 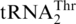 (Figure 6A). However, neither CaMST1 nor SpMST1 recognized
(Figure 6A). However, neither CaMST1 nor SpMST1 recognized 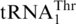 or the tRNAHis variants tested above (Figure 6). These results strongly suggest that MST1 specifically evolved to recognize
or the tRNAHis variants tested above (Figure 6). These results strongly suggest that MST1 specifically evolved to recognize 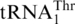 in a group of yeasts comprising S. cerevisiae. Therefore, CUN codon reassignment was completed following the coevolution of MST1 and
in a group of yeasts comprising S. cerevisiae. Therefore, CUN codon reassignment was completed following the coevolution of MST1 and 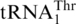 , which established specific protein–tRNA interactions.
, which established specific protein–tRNA interactions.
Figure 6.
Threonylation of 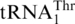 and tRNAHis variants (3 µM) by C. albicans and S. pombe MST1 (0.3 µM). (A) CaMST1 and SpMST1 threonylate
and tRNAHis variants (3 µM) by C. albicans and S. pombe MST1 (0.3 µM). (A) CaMST1 and SpMST1 threonylate 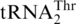 but not
but not 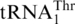 . (B and C) CaMST1 and SpMST1 are unable to threonylate tRNAHis variants.
. (B and C) CaMST1 and SpMST1 are unable to threonylate tRNAHis variants.
DISCUSSION
Reassignment of CUN codons from Leu to Thr in yeast mitochondria
CUN codons in the mitochondria of Saccharomyces, Nakaseomyces and Vanderwaltozyma have been previously assigned to Thr (5,11,12). This recoding event is accompanied with the loss of 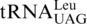 and the appearance of
and the appearance of 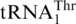 with an unmodified UAG anticodon (17). Analysis of a mass spectrometry database (PeptideAtlas) confirms that CUU and CUA codons indeed encode Thr in S. cerevisiae mitochondria. CUG and CUC codons are rarely used in yeast mitochondria, and the nature of the amino acid translated by such codons has not been verified experimentally. Given that the unmodified U at the first anticodon position is able to recognize all 4 nt (6,38), and no tRNA bearing GAG or CAG anticodons have been shown to be encoded by or imported into yeast mitochondria (39), it is plausible that CUG and CUC are also decoded by
with an unmodified UAG anticodon (17). Analysis of a mass spectrometry database (PeptideAtlas) confirms that CUU and CUA codons indeed encode Thr in S. cerevisiae mitochondria. CUG and CUC codons are rarely used in yeast mitochondria, and the nature of the amino acid translated by such codons has not been verified experimentally. Given that the unmodified U at the first anticodon position is able to recognize all 4 nt (6,38), and no tRNA bearing GAG or CAG anticodons have been shown to be encoded by or imported into yeast mitochondria (39), it is plausible that CUG and CUC are also decoded by 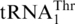 as Thr.
as Thr. 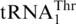 possesses an enlarged anticodon loop with 8 nt (UUUUAGGU), whereas a canonical tRNA anticodon loop consists of 7 nt with the triplet anticodon centered at positions 34–36. In addition to
possesses an enlarged anticodon loop with 8 nt (UUUUAGGU), whereas a canonical tRNA anticodon loop consists of 7 nt with the triplet anticodon centered at positions 34–36. In addition to 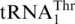 , several natural tRNAs have been found to contain eight bases in the anticodon loop, yet such tRNAs decode quadruplet codons instead of triplet codons (40). A question that arises is whether
, several natural tRNAs have been found to contain eight bases in the anticodon loop, yet such tRNAs decode quadruplet codons instead of triplet codons (40). A question that arises is whether 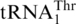 can also decode quadruplet codons. In yeast mitochondria, several protein-encoding genes, such as cox1, harbor in-frame CUAA sequences. Decoding of CUAA by a quadruplet anticodon could cause detrimental frameshift translation of these critical mitochondrial proteins. It is thus reasonable to think that
can also decode quadruplet codons. In yeast mitochondria, several protein-encoding genes, such as cox1, harbor in-frame CUAA sequences. Decoding of CUAA by a quadruplet anticodon could cause detrimental frameshift translation of these critical mitochondrial proteins. It is thus reasonable to think that 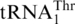 decodes all four CUN triplet codons but not quadruplet codons.
decodes all four CUN triplet codons but not quadruplet codons.
Evolutionary origin of 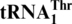
Our analyses reveal that 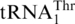 was recruited from mitochondrial tRNAHis. It has been long considered that tRNAs accepting the same amino acid (isoacceptors) evolved by gene duplication from the same common ancestor. However, studies in bacteria demonstrate that several tRNA genes may derive from different amino acid accepting groups (alloacceptors) (41). Naturally-occurring recruitment of alloacceptor tRNA genes was later reported in sponge mitochondria based on phylogenetic studies (28). Our work combines both biochemical and phylogenetic approaches to provide the first clear evidence that alloacceptor tRNA gene recruitment is directly responsible for a recent recoding event, suggesting that such a mechanism might have played an important role in the establishment of the genetic code during evolution.
was recruited from mitochondrial tRNAHis. It has been long considered that tRNAs accepting the same amino acid (isoacceptors) evolved by gene duplication from the same common ancestor. However, studies in bacteria demonstrate that several tRNA genes may derive from different amino acid accepting groups (alloacceptors) (41). Naturally-occurring recruitment of alloacceptor tRNA genes was later reported in sponge mitochondria based on phylogenetic studies (28). Our work combines both biochemical and phylogenetic approaches to provide the first clear evidence that alloacceptor tRNA gene recruitment is directly responsible for a recent recoding event, suggesting that such a mechanism might have played an important role in the establishment of the genetic code during evolution.
Phylogenetic analysis suggests that 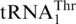 likely occurred after the split between Candida and Saccharomyces, subsequent to the divergence of P. canadensis. Interestingly, the C. albicans and C. parapsilosis mitochondrial genomes each contains two copies of the tRNAHis gene but no
likely occurred after the split between Candida and Saccharomyces, subsequent to the divergence of P. canadensis. Interestingly, the C. albicans and C. parapsilosis mitochondrial genomes each contains two copies of the tRNAHis gene but no 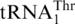 , and the CUN codons in these mitochondria remain assigned to Leu. In Kluvyeromyces lactis, the mitochondrial genome lacks both CUN codons and any tRNA reading such codons (7). Previous studies also suggest that current CUN codons decoded as Thr did not originate from ancestral Leu codons, but instead are derived from codons for other amino acids (18). Although we do not exclude the possibility that the original CUN codons could be ambiguously assigned to both Leu and Thr in certain intermediate yeast species, we favor that the emergence of
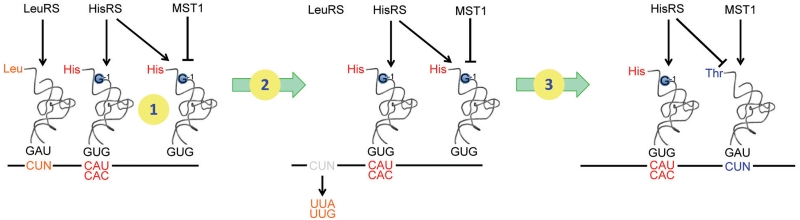
, and the CUN codons in these mitochondria remain assigned to Leu. In Kluvyeromyces lactis, the mitochondrial genome lacks both CUN codons and any tRNA reading such codons (7). Previous studies also suggest that current CUN codons decoded as Thr did not originate from ancestral Leu codons, but instead are derived from codons for other amino acids (18). Although we do not exclude the possibility that the original CUN codons could be ambiguously assigned to both Leu and Thr in certain intermediate yeast species, we favor that the emergence of 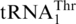 is a late event following the loss of original CUN codons, and such codons have reemerged in present-day yeast mitochondria. We propose that CUN codon reassignment initiated with duplication of the tRNAHis gene, followed by the loss of CUN codons and
is a late event following the loss of original CUN codons, and such codons have reemerged in present-day yeast mitochondria. We propose that CUN codon reassignment initiated with duplication of the tRNAHis gene, followed by the loss of CUN codons and 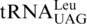 (Figure 7). Next, one copy of the tRNAHis gene evolved to the present-day
(Figure 7). Next, one copy of the tRNAHis gene evolved to the present-day 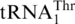 , and MST1 coevolved with
, and MST1 coevolved with 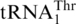 to form a cognate aaRS/tRNA pair. Finally, the CUN codons reappeared from various other codons and were reassigned to Thr. Our biochemical studies suggest that HisRS could still recognize the intermediate tRNA during the evolution from tRNAHis to
to form a cognate aaRS/tRNA pair. Finally, the CUN codons reappeared from various other codons and were reassigned to Thr. Our biochemical studies suggest that HisRS could still recognize the intermediate tRNA during the evolution from tRNAHis to 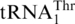 (Table 2). However, the mitochondrial proteome remained largely unaffected as the CUN codons were not present in the mitochondrial genome during this period.
(Table 2). However, the mitochondrial proteome remained largely unaffected as the CUN codons were not present in the mitochondrial genome during this period.
Figure 7.
Proposed model for CUN codon reassignment in yeast mitochondria. (1) tRNAHis duplicated in an ancestral yeast species while CUN codons remain assigned to Leu. (2) CUN codons were changed to UUA or UUG decoded by 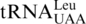 , and
, and 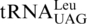 was lost. (3) tRNAHis evolved to
was lost. (3) tRNAHis evolved to 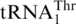 , and CUN codons reemerged from various codons.
, and CUN codons reemerged from various codons.
Several mechanisms have been proposed to explain the codon reassignment processes, including: (i) codon capture mechanism (42), hypothesizing that a specific tRNA and corresponding codons completely disappear from a genome before a novel tRNA evolves to read such codons; and (ii) ambiguous intermediate mechanism (43), postulating that a tRNA mutant is recognized by more than one aminoacyl-tRNA synthetases, and a codon may be assigned to multiple amino acids in an intermediate state. The latter mechanism is supported by the discovery that in several extant Candida species, the CUG codon is ambiguously decoded by both Leu and Ser due to the presence of a tRNACAG charged by both leucyl- and seryl-tRNA synthetases (12,44). Our work suggests that CUN codons and 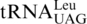 are lost in yeast mitochondria prior to the emergence of
are lost in yeast mitochondria prior to the emergence of 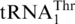 , which provides a paradigm for the codon capture mechanism and lends support to the evolving genetic code theory.
, which provides a paradigm for the codon capture mechanism and lends support to the evolving genetic code theory.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of General Medical Sciences (grant GM022854 to D.S.); Canadian Research Chair Program (to B.F.L.); Brown-Coxe Post-doctoral Fellowship (to J.L.). Funding for open access charge: National Institute of General Medical Sciences (grant GM022854).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Susan Martinis and Jaya Sarkar (University of Illinois at Urbana-Champaign) for the kind gifts of ScmtLeuRS and tRNALeu and Dr Markus Englert for the gift of S. pombe genomic DNA. We thank Dr Lennart Randau (MPI for Terrestrial Microbiology, Marburg, Germany), Patrick O’Donoghue and Ilka Heinemann (Yale University) for insightful discussion and comments.
REFERENCES
- 1.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 3.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 4.Crick FH. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 5.Moura GR, Paredes JA, Santos MA. Development of the genetic code: insights from a fungal codon reassignment. FEBS Lett. 2010;584:334–341. doi: 10.1016/j.febslet.2009.11.066. [DOI] [PubMed] [Google Scholar]
- 6.Ambrogelly A, Palioura S, Söll D. Natural expansion of the genetic code. Nat. Chem. Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 7.Sengupta S, Yang X, Higgs PG. The mechanisms of codon reassignments in mitochondrial genetic codes. J. Mol. Evol. 2007;64:662–688. doi: 10.1007/s00239-006-0284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schön A, Böck A, Ott G, Sprinzl M, Söll D. The selenocysteine-inserting opal suppressor serine tRNA from E. coli is highly unusual in structure and modification. Nucleic Acids Res. 1989;17:7159–7165. doi: 10.1093/nar/17.18.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krzycki JA. The direct genetic encoding of pyrrolysine. Curr. Opin. Microbiol. 2005;8:706–712. doi: 10.1016/j.mib.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Nozawa K, O’Donoghue P, Gundllapalli S, Araiso Y, Ishitani R, Umehara T, Söll D, Nureki O. Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality. Nature. 2009;457:1163–1167. doi: 10.1038/nature07611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Tzagoloff A. Assembly of the mitochondrial membrane system: sequences of yeast mitochondrial valine and an unusual threonine tRNA gene. Cell. 1979;18:47–53. doi: 10.1016/0092-8674(79)90352-0. [DOI] [PubMed] [Google Scholar]
- 12.Miranda I, Silva R, Santos MA. Evolution of the genetic code in yeasts. Yeast. 2006;23:203–213. doi: 10.1002/yea.1350. [DOI] [PubMed] [Google Scholar]
- 13.Macino G, Tzagoloff A. Assembly of the mitochondrial membrane system: two separate genes coding for threonyl-tRNA in the mitochondrial DNA of Saccharomyces cerevisiae. Mol. Gen. Genet. 1979;169:183–188. doi: 10.1007/BF00271669. [DOI] [PubMed] [Google Scholar]
- 14.Sebald W, Wachter E, Tzagoloff A. Identification of amino acid substitutions in the dicyclohexylcarbodiimide-binding subunit of the mitochondrial ATPase complex from oligomycin-resistant mutants of Saccharomyces cerevisiae. Eur. J. Biochem. 1979;100:599–607. doi: 10.1111/j.1432-1033.1979.tb04207.x. [DOI] [PubMed] [Google Scholar]
- 15.Deutsch EW, Lam H, Aebersold R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pape LK, Koerner TJ, Tzagoloff A. Characterization of a yeast nuclear gene (MST1) coding for the mitochondrial threonyl-tRNA1 synthetase. J. Biol. Chem. 1985;260:15362–15370. [PubMed] [Google Scholar]
- 17.Sibler AP, Dirheimer G, Martin RP. Nucleotide sequence of a yeast mitochondrial threonine-tRNA able to decode the C-U-N leucine codons. FEBS Lett. 1981;132:344–348. doi: 10.1016/0014-5793(81)81194-5. [DOI] [PubMed] [Google Scholar]
- 18.Osawa S, Collins D, Ohama T, Jukes TH, Watanabe K. Evolution of the mitochondrial genetic code. III. Reassignment of CUN codons from leucine to threonine during evolution of yeast mitochondria. J. Mol. Evol. 1990;30:322–328. doi: 10.1007/BF02101886. [DOI] [PubMed] [Google Scholar]
- 19.Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy H, Ling J, Irnov M, Ibba M. Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase. EMBO J. 2004;23:4639–4648. doi: 10.1038/sj.emboj.7600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang BF, Laforest MJ, Burger G. Mitochondrial introns: a critical view. Trends Genet. 2007;23:119–125. doi: 10.1016/j.tig.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Gautheret D, Lambert A. Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J. Mol. Biol. 2001;313:1003–1011. doi: 10.1006/jmbi.2001.5102. [DOI] [PubMed] [Google Scholar]
- 23.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 24.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 25.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lartillot N, Philippe H. Improvement of molecular phylogenetic inference and the phylogeny of Bilateria. Philos. Trans. Roy. Soc. Lond. B. Biol. Sci. 2008;363:1463–1472. doi: 10.1098/rstb.2007.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez-Ezpeleta N, Brinkmann H, Roure B, Lartillot N, Lang BF, Philippe H. Detecting and overcoming systematic errors in genome-scale phylogenies. Syst. Biol. 2007;56:389–399. doi: 10.1080/10635150701397643. [DOI] [PubMed] [Google Scholar]
- 28.Lavrov DV, Lang BF. Transfer RNA gene recruitment in mitochondrial DNA. Trends Genet. 2005;21:129–133. doi: 10.1016/j.tig.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Nawaz MH, Pang YL, Martinis SA. Molecular and functional dissection of a putative RNA-binding region in yeast mitochondrial leucyl-tRNA synthetase. J. Mol. Biol. 2007;367:384–394. doi: 10.1016/j.jmb.2006.12.051. [DOI] [PubMed] [Google Scholar]
- 30.Hsu JL, Martinis SA. A Flexible peptide tether controls accessibility of a unique C-terminal RNA-binding domain in leucyl-tRNA synthetases. J. Mol. Biol. 2008;376:482–491. doi: 10.1016/j.jmb.2007.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan J, Gogakos T, Babina AM, Söll D, Randau L. Change of tRNA identity leads to a divergent orthoganol histidyl-tRNA synthetase/tRNAHis pair. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1176. doi:10.1093/nar/gkq1176e [Epub ahead of print, 17 November 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosen AE, Brooks BS, Guth E, Francklyn CS, Musier-Forsyth K. Evolutionary conservation of a functionally important backbone phosphate group critical for aminoacylation of histidine tRNAs. RNA. 2006;12:1315–1322. doi: 10.1261/rna.78606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiu MI, Mason TL, Fink GR. HTS1 encodes both the cytoplasmic and mitochondrial histidyl-tRNA synthetase of Saccharomyces cerevisiae: mutations alter the specificity of compartmentation. Genetics. 1992;132:987–1001. doi: 10.1093/genetics/132.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nameki N, Asahara H, Shimizu M, Okada N, Himeno H. Identity elements of Saccharomyces cerevisiae tRNA(His) Nucleic Acids Res. 1995;23:389–394. doi: 10.1093/nar/23.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rudinger J, Florentz C, Giegé R. Histidylation by yeast HisRS of tRNA or tRNA-like structure relies on residues -1 and 73 but is dependent on the RNA context. Nucleic Acids Res. 1994;22:5031–5037. doi: 10.1093/nar/22.23.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Leigh JW, Brinkmann H, Cushion MT, Rodriguez-Ezpeleta N, Philippe H, Lang BF. Phylogenomic analyses support the monophyly of Taphrinomycotina, including Schizosaccharomyces fission yeasts. Mol. Biol. Evol. 2009;26:27–34. doi: 10.1093/molbev/msn221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heckman JE, Sarnoff J, Alzner-DeWeerd B, Yin S, RajBhandary UL. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc. Natl Acad. Sci. USA. 1980;77:3159–3163. doi: 10.1073/pnas.77.6.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duchêne AM, Pujol C, Maréchal-Drouard L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr. Genet. 2009;55:1–18. doi: 10.1007/s00294-008-0223-9. [DOI] [PubMed] [Google Scholar]
- 40.Bossi L, Roth JR. Four-base codons ACCA, ACCU and ACCC are recognized by frameshift suppressor sufJ. Cell. 1981;25:489–496. doi: 10.1016/0092-8674(81)90067-2. [DOI] [PubMed] [Google Scholar]
- 41.Saks ME, Sampson JR, Abelson J. Evolution of a transfer RNA gene through a point mutation in the anticodon. Science. 1998;279:1665–1670. doi: 10.1126/science.279.5357.1665. [DOI] [PubMed] [Google Scholar]
- 42.Osawa S, Jukes TH. Codon reassignment (codon capture) in evolution. J. Mol. Evol. 1989;28:271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- 43.Schultz DW, Yarus M. Transfer RNA mutation and the malleability of the genetic code. J. Mol. Biol. 1994;235:1377–1380. doi: 10.1006/jmbi.1994.1094. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki T, Ueda T, Watanabe K. The `polysemous' codon–a codon with multiple amino acid assignment caused by dual specificity of tRNA identity. EMBO J. 1997;16:1122–1134. doi: 10.1093/emboj/16.5.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.