Abstract
DNA methylation profiling has become an important aspect of biomedical molecular analysis. Polymerase chain reaction (PCR) amplification of bisulphite-treated DNA is a processing step that is common to many currently used methods of quantitative methylation analysis. Preferential amplification of unmethylated alleles—known as PCR-bias—may significantly affect the accuracy of quantification. To date, no universal experimental approach has been reported to overcome the problem. This study presents an effective method of correcting biased methylation data. The procedure includes a calibration performed in parallel to the analysis of the samples under investigation. DNA samples with defined degrees of methylation are analysed. The observed deviation of the experimental results from the expected values is used for calculating a regression curve. The equation of the best-fitting curve is then used for correction of the data obtained from the samples of interest. The process can be applied irrespective of the locus interrogated and the number of sites analysed, avoiding an optimization of the amplification conditions for each individual locus.
INTRODUCTION
Quantification of changes in DNA methylation is becoming increasingly important in biomedical and particularly cancer research, since epigenetic biomarkers have a considerable potential for diagnostics (1). Several methods are in use to analyse DNA methylation patterns (2), ranging from measurements at individual CpG dinucleotides (3–5) to large-scale and genome-wide approaches by next-generation sequencing (6) or hybridization to DNA microarrays (7,8). Many analysis techniques are based on the common step of treating the DNA with bisulphite. Sodium bisulphite converts unmethylated cytosine to uracil, which turns into thymine upon polymerase chain reaction (PCR) amplification. In contrast, methylated cytosines remain unaffected. The C-T conversion can be picked up by any method of DNA sequence determination. For sensitivity reasons, an amplification step is required subsequent to the bisulphite treatment. Since the actual measurement occurs on the amplicon and not the original DNA, the accuracy of the PCR step strongly influences the accuracy of the analysis.
PCR amplification of bisulphite-treated DNA often results in a selective enrichment of unmethylated alleles—a phenomenon known as PCR-bias (9)—and may therefore reflect the real situation incorrectly. Preferential recovery of methylated forms is also possible although less common. The extent of such deviations is difficult to predict, however. Several studies addressed the experimental conditions in order to enable unbiased amplification. One approach suggests the use of specially designed PCR primers (10,11), which should contain CpG dinucleotides (usually 1 or 2). This is meant to facilitate primer binding to the methylated allele and could thus avoid disproportional amplification. An alternative strategy aims at inhibiting the formation of secondary structures by GC-rich and methylated regions, which is considered to be the reason of biased amplification, by increasing the primer annealing temperature during the PCR cycles (12). Finally, the use of single-molecule PCR has been proposed (13). It eliminates bias because there is no competition between DNA templates that are amplified with different efficiencies.
The reported processes for avoiding PCR-bias rely on a careful optimization of PCR conditions and their effectiveness remains contradictory (14). Although shown to be effective for particular genes, their implementation is time consuming and labour intensive, especially if multiple loci are analysed. A method for obtaining unbiased DNA methylation data irrespective of the locus under investigation would therefore be advantageous. In contrast to the approaches mentioned above, we suggest not to avoid PCR-bias by laborious optimization of the experimental conditions but to correct appropriately the results obtained after amplification. The procedure established to achieve this end includes a calibration performed on DNA samples with defined degrees of methylation in parallel to the analysis of the samples under investigation. The observed deviation from the expected results is then applied for correcting the data obtained from the samples of interest.
MATERIALS AND METHODS
Calibration DNA
Fully methylated and unmethylated human control DNA that was bisulphite-treated (EpiTect PCR control DNA; Qiagen, Hilden, Germany) was mixed in different ratios to obtain calibration samples that represent distinct methylation percentages of 0%, 12.5%, 25%, 37.5%, 50%, 62.5%, 75%, 87.5% and 100%, respectively. An additional calibration DNA of 6.25% methylation was included to the study of the SFRP1 promoter because of the extreme bias observed for amplifying methylated DNA. The fully methylated calibration DNA was produced by the manufacturer using SssI methylase. Unmethylated DNA was generated by means of whole-genome amplification. According to the manufacturer’s information, the fully methylated control is completely methylated at all CpG sites and can be used irrespective of the locus analysed. This could be verified for sites that are cleaved by restriction enzymes, whose activity depends on the methylation status. Concordantly, full methylation of different loci has been reported by independent studies, which employed this commercial control DNA (15–17).
PCR amplification
PCR was carried out in 25-µl reactions of 1.5 µl EpiTect control DNA (10 ng/µl), 1.5 mM MgCl2, 125 mM dNTP, 200 nM primers, 0.65 U HotStarTaq DNA polymerase and 1 x Q-solution (Qiagen). An amplification programme was used that had been described previously (18) with minor modification. It was started by an initial activation of the HotStarTaq DNA polymerase at 95°C for 15 min. The initial amplification cycle was denaturation at 95°C for 1 min, annealing at 62°C for 2 min and elongation at 72°C for 3 min. This procedure was continued for 20 cycles, reducing the annealing temperature by 0.5°C each cycle, followed by 25 cycles of 1 min denaturation at 95°C, 2 min annealing at 52°C and 2 min elongation at 72°C. The sequences of the PCR primers used are listed in Table 1. Three separate PCR reactions were performed for the amplification of each region of interest. About 5 µl of each reaction was examined on 2% agarose gels.
Table 1.
Sequences of the PCR and pyrosequencing primers used in this study
| Gene symbol | Primer sequences | Amplicon length, bp | No. of CpGs quantified by pyrosequencing/total no. in amplicon |
|---|---|---|---|
| CDH1 | F: 5′-TTTTTTTTGATTTTAGGTTTTAGTGAG-3′ | 421 | 9/33 |
| R: 5′-bio-ACTCCAAAAACCCATAACTAACC-3′ | |||
| S: 5′-AGTTAGTTTAGATTTTAGTT-3′ | |||
| DACT1 | F: 5′-GTTTGGGAAGTGAAAGAAATTTAATT-3′ | 184 | 5/12 |
| R: 5′-bio-CTAAAACCCCAACATCCTATTACAAT-3′ | |||
| S: 5′-AGATTGTGTTGTAATTTGGT-3′ | |||
| DKK1 | F: 5′-bio-GGGGTGAAGAGTGTTAAAGGTT-3′ | 326 | 8/18 |
| R: 5′-AAACCATCATCTCAAAAAAACTCAA-3′ | |||
| S: 5′-CTACAAAAAACACAAAACTCTAC-3′ | |||
| DKK2 | F: 5′-bio-TTTTAGTAGTTGTGGGTGGAGATA-3′ | 456 | 11/27 |
| R: 5′-ATACTCCTTTTCAAAATTAACAAAC-3′ | |||
| S: 5′-CCTAACTCACAAAAAACAAC-3′ | |||
| DKK3 | F: 5′-GATTTTGTTGAGTTTAGTTTTTTTTGGT-3′ | 123 | 5/5 |
| R: 5′-bio-CAAACCTCTCTCAACCCCTACCTA-3′ | |||
| S: 5′-TTTTTTGGTGGATGTG-3′ | |||
| DKK4 | F: 5′-bio-ATAGATTTGAAGGGATTTGTTGAAGTTT-3′ | 328 | 2/11 |
| R: 5′-CAAAACCAACTCAACCCCAACAAAAC-3′ | |||
| S: 5′-CTAAACTAACAACTCAACAC-3′ | |||
| PYGO2 | F: 5′-TGAGATTTAGAGAGGTTATTTAAGT-3′ | 252 | 9/22 |
| R: 5′-ACATATAAAAATCCAAATTCCCC-3′ | |||
| S: 5′-GGTATTTTATAGATAGGTGT-3′ | |||
| SFRP1 | F: 5′-GTTTTGTTTTTTAAGGGGTGTTGAG-3′ | 412 | 8/25 |
| R: 5′-bio-CTCCGAAAACTACAAAACTAAAATAC-3′ | |||
| S: 5′-TYGGGAGTTGATTGG-3′ | |||
| SFRP2 | F: 5′-ATGTTTGGTAATTTAGTAGAAATTT-3′ | 409 | 14/30 |
| R: 5′-bio-CAACCAAAATTTTCTTAACCTTTTT-3′ | |||
| S: 5′-GATTGGGGTAAAATAAGTT-3′ | |||
| SFRP3 | F: 5′-bio-GTGATTTAGGGGAGGAGATATTTTAGA-3′ | 542 | 4/29 |
| R: 5′-TTCCAAAACAAAAACTTACACAAAA-3′ | |||
| S: 5′-CAAAATAAAACAAAATACAAC-3′ |
F, PCR forward; R, PCR reverse; S, pyrosequencing.
Bisulphite pyrosequencing
A volume of 20-µl PCR product was added to 2 µl Streptavidin Sepharose High Performance (GE Healthcare, Uppsala, Sweden), 38 µl of PyroMark binding buffer (Qiagen) and 20 µl water and mixed. The Vacuum Prep Workstation (Biotage, Uppsala, Sweden) was used to prepare single-stranded DNA according to the manufacturer’s instructions. The Sepharose beads with the single-stranded templates attached were released into a PSQ 96 Plate Low (Biotage) containing 15 µl of 0.6 µM corresponding sequencing primer in annealing buffer. Pyrosequencing reactions were performed using the Pyro Gold Reagent Kit (Biotage) in a PSQ HS 96 Pyrosequencing System (Biotage) according to the manufacturer’s protocol. Quantification of CpG site methylation was performed with the Software PyroQ-CpG v.1.0.9 (Biotage). The sequences of the pyrosequencing primers are listed in Table 1.
Bisulphite sequencing
The PCR-amplified region of DKK2 was subcloned using the TOPO TA cloning kit for sequencing (Invitrogen, Carlsbad, USA). Ten clones were picked at random and sequenced using Sanger chemistry by GATC (Constance, Germany). A representation of the sequencing data was made using the CpGviewer software (19).
Isolation and bisulphite conversion of DNA from cell lines and CD19+ B cells of healthy individuals
The chronic lymphocytic leukaemia cell lines MEC-1 (20) and EHEB (21) were grown in a medium consisting of 90% Iscove’s Modified Dulbecco’s Medium (Invitrogen) or Roswell Park Memorial Institute Medium (Invitrogen), respectively, supplemented with 10% fetal bovine serum (Invitrogen). CD19+ B cells (22) were isolated from the buffy coats of five healthy individuals provided by the Institute for Clinical Transfusion Medicine and Cell Therapy (Heidelberg, Germany) using Dynabeads CD19 pan B (Invitrogen). DNA was extracted using the QIAamp DNA Blood Mini kit (Qiagen). DNA concentration was measured in a ND-1000 spectrophotometer (Thermo Scientific, Wilmington, USA). A total of 1.9 µg DNA was treated with sodium bisulfite using the EpiTect Bisulfite kit (Qiagen). The efficiency of bisulphite conversion averaged 98.8% and was computed from the sequences of 63 cloned PCR products of CDH1, DACT1, DKK1, DKK2, DKK3, DKK4, SFRP2 (8 clones each) and SFRP3 (seven clones) using the BISMA software (23), which considers the non-CpG cytosines within the sequences. Subsequently, 2 µl of bisulphite-converted DNA was used for PCR amplification.
RESULTS
Amplification of different DNA regions is biased to a different extent
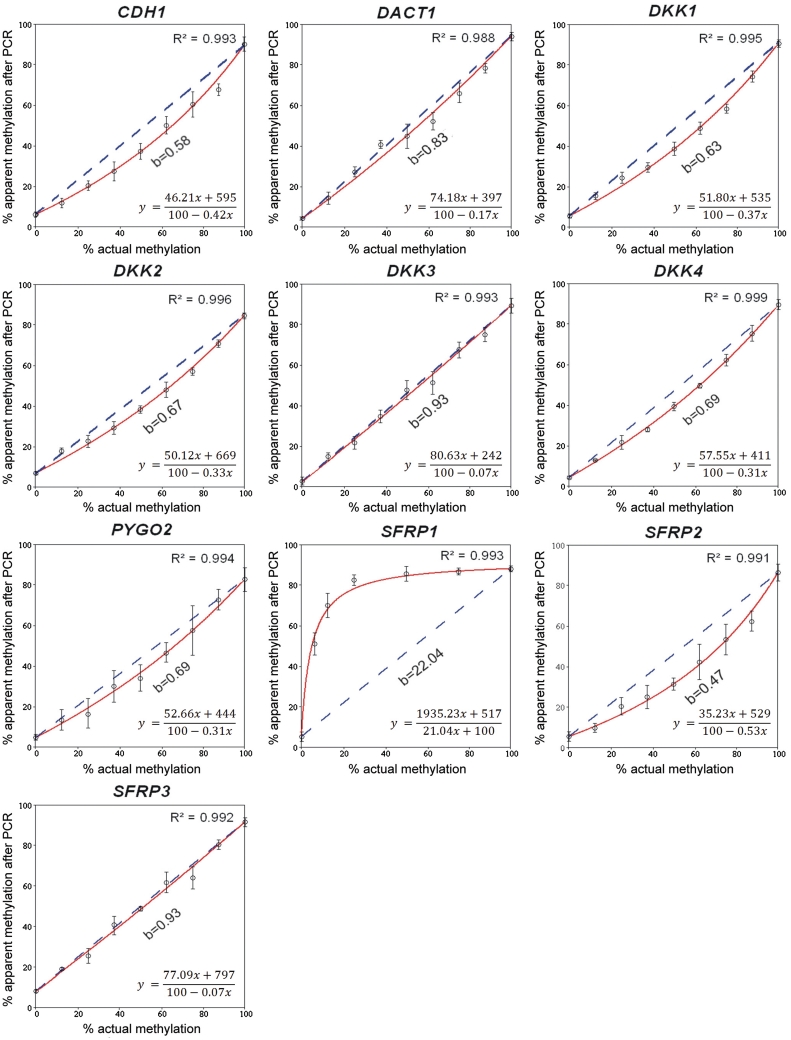
For initial analyses, calibration DNA was used. Fully methylated and unmethylated DNA was mixed to create samples of a defined methylation status. After bisulphite treatment and PCR amplification, pyrosequencing was applied to quantify the apparent degree of methylation. CpG dinucleotides were studied that are located in proximity of the transcriptional start sites of 10 human genes (Table 1). The candidate genes were selected randomly but for the fact that they are known to be epigenetically silenced in a variety of human cancers. We plotted the apparent percentage of methylation determined in the DNA amplicons produced from the calibration DNA (y-axis) as a function of the actual methylation degree (x-axis) (Figure 1). The amplification step introduced bias of a different degree to the 10 fragments. There was practically unbiased amplification of DKK3 and SFRP3, a moderate bias towards the unmethylated alleles for CDH1, DACT1, DKK1, DKK2, DKK4, PYGO2 and SFRP2 as well as an extreme deviation towards the methylated allele for SFRP1. The unusual last result might be explained as the use of a PCR primer that contained a single CpG dinucleotide and had been described in an earlier study (24) for compensating biased amplification of the unmethylated DNA, although an influence of the amplicon sequence cannot be excluded either.
Figure 1.
Degree of bias introduced by PCR amplification of 10 gene promoters from calibration DNAs. To compute a numeric value of PCR-bias, the apparent degree of methylation observed after amplification (y-axis) was plotted as a function of the actual methylation percentage (x-axis). Each value represents the average of three measurements. By regression analyses (red lines), the value b was calculated as described in the text. It reflects the difference between the actual and the measured degree of methylation. The dotted lines represent an unbiased plot (b = 1). In each panel, the equation of the best-fit curve is shown.
As a quantitative measure of PCR-bias, the value b was used (Figure 1) as suggested by Warnecke et al. (9). This factor reflects the difference between the observed and actual degree of methylation and is derived from the equation of a hyperbolic best-fit curve:
| (1) |
Balanced recovery of methylated and unmethylated alleles during amplification can be described with b = 1. Using b = 1 in Equation (1) leads to a linear function (y = x). Preferential PCR amplification of unmethylated alleles is described by 0 < b < 1 and a concave curve (Figure 1), whereas a more uncommon accumulation of methylated DNA is reflected by b > 1 and a convex curve (e.g. the SFRP1 gene in Figure 1). The value b reflects the efficiency of primer binding and polymerase elongation in amplicons derived from unmethylated or methylated DNA. Owing to sequence differences after bisulphite conversion, DNA may adopt distinct secondary structures or exhibit a different melting behaviour, which lead to amplification bias (9).
A minor modification was introduced to the equation in order to describe more adequately the data obtained from bisulphite pyrosequencing. The amplification of DNA of 0% or 100% methylation cannot be biased because there is no competition between methylated and unmethylated DNA templates. Nevertheless, differences between actual and apparent methylation of the respective calibration samples were observed. For the 10 gene loci studied, the average differences were 5% methylation instead of 0% and 89% rather than the actual 100%. These differences are the result of an additional bias introduced by the pyrosequencing and base-calling process. By including the data points obtained from samples with 0% and 100% methylation into the curve-fitting calculation, this additional bias was corrected, too. In order to find the family of hyperbolic curves, which pass through the extreme calibration points with abscissae of x = 0 and x = 100, a generalization of the above equation was used [see Supplementary Data for derivation of Equation (2)]:
| (2) |
The terms y0 and y1 represent the apparent methylation degree of DNA with 0% and 100% methylation, respectively. Equation (2) is of the same type as Equation (1) and can be transformed to it by setting y0 = 0 and y1 = 100.
Based on Equation (2), values of b were computed from the experimental data by minimizing the sum of squared errors as described (25). Application of Equation (2) was superior to that of Equation (1), leading to a significant decrease of the sum of squared errors (data not shown). The results of the amplification of each of the 10 loci in the calibration DNA could be described by such a fitted curve (Figure 1).
Correction of biased methylation values
Having demonstrated biased amplification for most genes of our set, we addressed the question, if the error could be eliminated effectively by taking advantage of the equations of the best-fit curves. Correction was done for each control sample by expressing the unknown x (the actual methylation degrees) and solving the algebraic equations substituting the variable y with the apparent methylation percentages obtained in the experiment:
| (3) |
This led to a significant improvement of the measurement accuracy and a substantial decrease of relative error (Table 2). The process was particularly effective for DNA samples of 37.5% methylation and higher. The effect was much less pronounced for low methylation degrees. For example, the 25% methylation control sample in genes DACT1, DKK1, SFRP2 and PYGO2 exhibited relative errors of 16%, 23%, 30% and 20%, respectively. Although relatively high, the corrected values produced still less error than the raw data. In consequence, we wondered if an even better regression solution may exist that would be superior to hyperbolic regression at low methylation but still of the same quality at higher values, and thus could be applied universally.
Table 2.
Comparison of PCR-bias correction results using two types of regression curves
| Gene symbol | Numeric value of PCR-bias (b) |
Average relative errows (%) |
Sum of squared errors |
|||||
|---|---|---|---|---|---|---|---|---|
| Raw | Hyperbolic | Polynomial | Raw | Hyperbolic | Polynomial | Hyperbolic | Polynomial | |
| CDH1 | 0.58 (1.72-fold) | 1.01 | 0.97 | 21 | 2 | 3 | 41.0 | 36.6 |
| DACT1 | 0.83 (1.20-fold) | 1.02 | 0.99 | 10 | 7 | 4 | 86.4 | 28.0 |
| DKK1 | 0.63 (1.59-fold) | 1.04 | 0.99 | 17 | 4 | 2 | 32.6 | 4.9 |
| DKK2 | 0.67 (1.49-fold) | 1.03 | 1.00 | 20 | 2 | 3 | 23.5 | 8.4 |
| DKK3 | 0.93 (1.08-fold) | 1.00 | 0.98 | 11 | 4 | 5 | 45.7 | 40.0 |
| DKK4 | 0.69 (1.45-fold) | 1.33 | 0.95 | 18 | 2 | 4 | 5.1 | 4.5 |
| PYGO2 | 0.69 (1.45-fold) | 0.99 | 0.97 | 25 | 6 | 5 | 36.32 | 31.8 |
| SFRP1 | 22.04 (22.04-fold) | 1.38 | 0.25 | 158 | 18 | 35 | 36.03 | 383.6 |
| SFRP2 | 0.47 (2.13-fold) | 1.06 | 0.95 | 28 | 6 | 4 | 49.6 | 32.8 |
| SFRP3 | 0.93 (1.08-fold) | 1.00 | 0.97 | 7 | 5 | 6 | 51.9 | 47.8 |
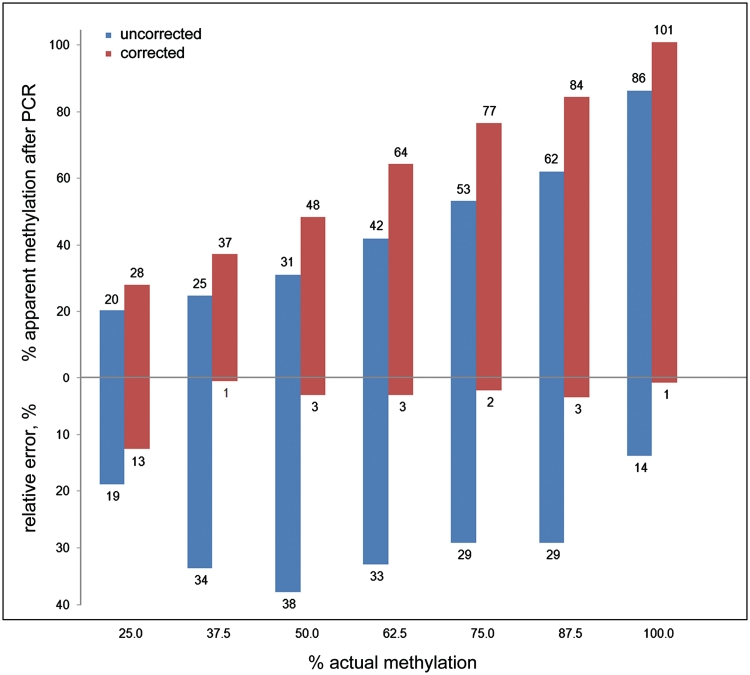
Applying the criterion of minimizing the sum of squared errors (Table 2), cubic polynomial fit curves were found to improve substantially the correction results at 25% methylation (Figure 2). The following equation was used for curve fitting:
| (4) |
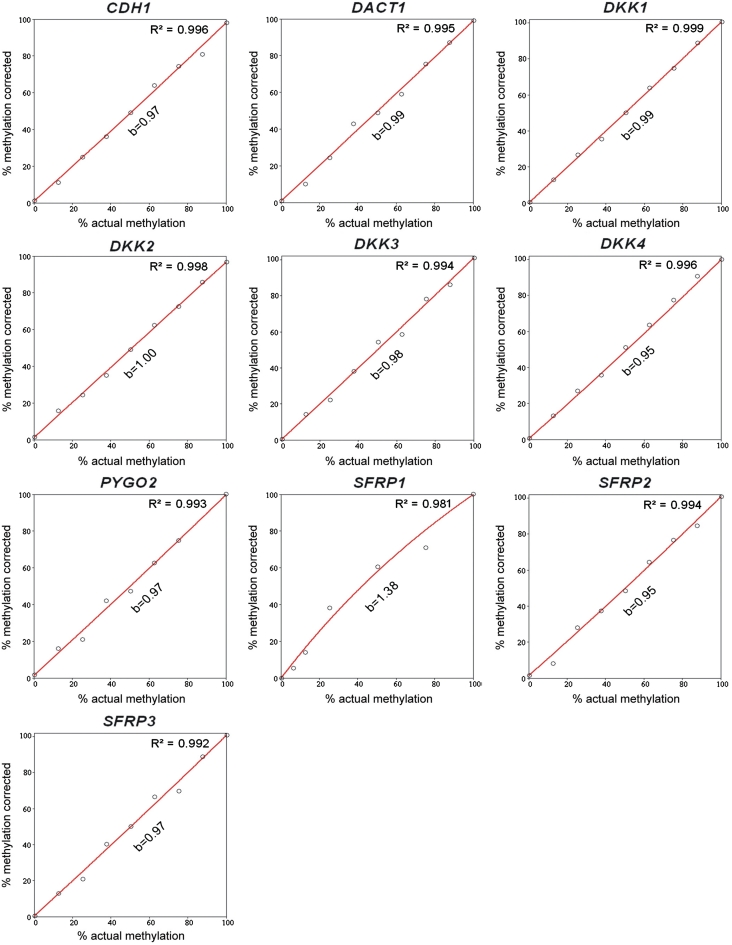
where a, c, d and e are arbitrary parameters. The fitting was performed followed by solving the cubic equations for the unknown x with Cardano’s method described elsewhere (26). Typical reductions of the relative errors were from 30% (hyperbolic) to 13% (cubic polynomial) for SFRP2 (Figure 2) and 23% (hyperbolic) to 7% (cubic polynomial) for DKK1, for example. Also, cubic polynomial fit curves could be applied generally, achieving at least the effectiveness of correction obtained with the hyperbolic fit curves (Table 2, Figure 3). The only exception was the gene SFRP1; due to the enormous bias towards the methylated allele, better correction effectiveness was achieved by using hyperbolic regression, which accommodated more readily the defined 0% and 100% values.
Figure 2.
Typical results of bias correction. The experimental methylation results obtained from the promoter region of the SFRP2 gene were corrected using a cubic polynomial fit curve. The blue bars represent raw data; the red bars show the corrected values. The actual methylation degrees were 25, 37.5, 50, 62.5, 75, 87.5 and 100%. In addition to each methylation percentage value, also the relative error is shown.
Figure 3.
The result of PCR-bias correction by means of cubic polynomial regression. The corrected methylation degree (y-axis) is plotted as a function of the actual percentage of methylation (x-axis) for the set of genes analysed (for comparison see Figure 1). The red lines represent the corrected plots. The essentially linear function of y(x) and the fact that the values of b are close to 1 demonstrate effective elimination of PCR-bias from the experimental data. The data of the SFRP1 gene were corrected using hyperbolic regression (see text for details).
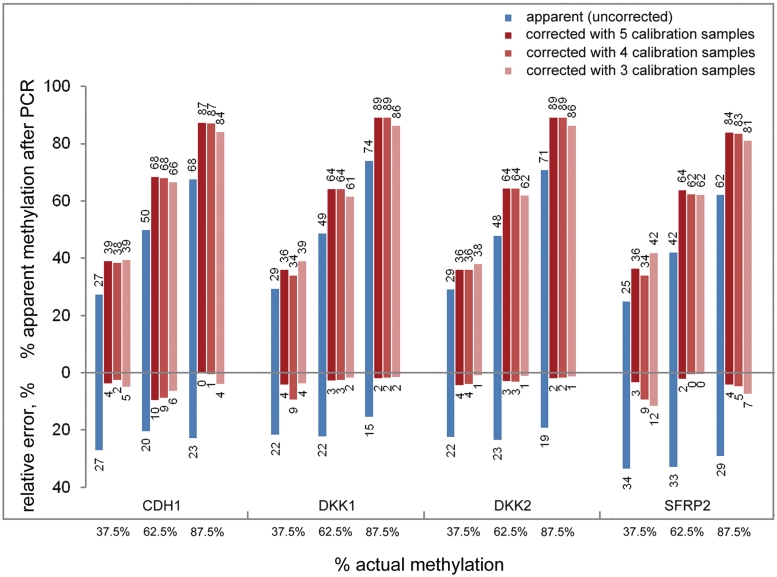
Reduction of the number of calibration samples
The PCR-bias correction described so far was based on nine calibration samples. For simplifying the correction process, we examined if the number of controls could be reduced while still providing consistent correction power. Using the fit curves based on only five (0%, 25%, 50%, 75% and 100% methylation), four (0%, 25%, 75% and 100%) or three (0%, 50% and 100%) calibration samples, we investigated if we were able accurately to predict methylation degrees for the calibration samples of 37.5%, 62.5% and 87.5% methylation, using the genes with the largest bias (SFRP2, CDH1, DKK1 and DKK2). The corrected values were very similar by using down to three calibration samples (Figure 4). No major difference was observed in average relative errors compared to using nine calibration samples (Supplementary Figure S1).
Figure 4.
Influence of the number of calibration samples on the accuracy of bias correction. An analysis which was corrected on the basis of only five (0, 25, 50, 75 and 100% methylation), four (0, 25, 75 and 100%) or three (0, 50 and 100%) DNA samples resulted in essentially similarly correct data. The blue bars represent the raw data; the bars of different shades of red show the corrected values. In addition to each methylation percentage value, also the relative error is indicated. The actual methylation percentages are listed at the bottom.
Accuracy of correction does not depend on the degree of PCR-bias
Correction of PCR-bias is only of use if the accuracy of correction is not affected by its degree. In order to test this, DNA fragments with differently biased PCR amplification yields were artificially produced of the same loci. Preferential amplification of unmethylated alleles is thought to be due to a dissimilar ability of unmethylated and methylated DNA to form secondary structures (9). Therefore, we hypothesized that a PCR additive, which modifies the melting behaviour of DNA should affect the extent of PCR-bias at least in some cases. Q-solution of Qiagen has such an effect and was actually routinely used for amplification reactions in this study. For four genes—DACT1, DKK2, DKK3 and DKK4—we produced PCR products without the additive. This affected negatively the value of PCR-bias for DACT1 (b value 0.57 instead of 0.83) and DKK3 (0.68 versus 0.93), had a minor influence on DKK2 (0.56 versus 0.66) and did not influence amplification of DKK4 (0.71 versus 0.71). However, the values obtained after correction using five calibration DNA samples were very similar irrespective of the numeric value of initial PCR-bias (Table 3).
Table 3.
Accuracy of bias correction does not depend on its degree
| Actual methylation (%) | Raw and corrected methylation values (%) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
DACT1 |
DKK2 |
DKK3 |
||||||||||
| +Q-sol. |
−Q-sol. |
+Q-sol. |
−Q-sol. |
+Q-sol. |
−Q-sol. |
|||||||
| raw | corr. | raw | corr. | raw | corr. | raw | corr. | raw | corr. | raw | corr. | |
| 0 | 4 | 0 | 5 | 0 | 7 | 0 | 12 | 0 | 2 | 0 | 5 | 0 |
| 25 | 27 | 25 | 22 | 25 | 23 | 25 | 28 | 22 | 21 | 24 | 23 | 23 |
| 50 | 45 | 49 | 36 | 50 | 38 | 50 | 39 | 55 | 48 | 52 | 41 | 54 |
| 75 | 66 | 74 | 54 | 75 | 57 | 75 | 48 | 73 | 68 | 74 | 55 | 74 |
| 100 | 94 | 98 | 87 | 100 | 85 | 100 | 80 | 100 | 89 | 100 | 87 | 100 |
Correcting methylation measurements in leukaemic cell lines MEC-1 and EHEB and CD19+ B cells from healthy individuals
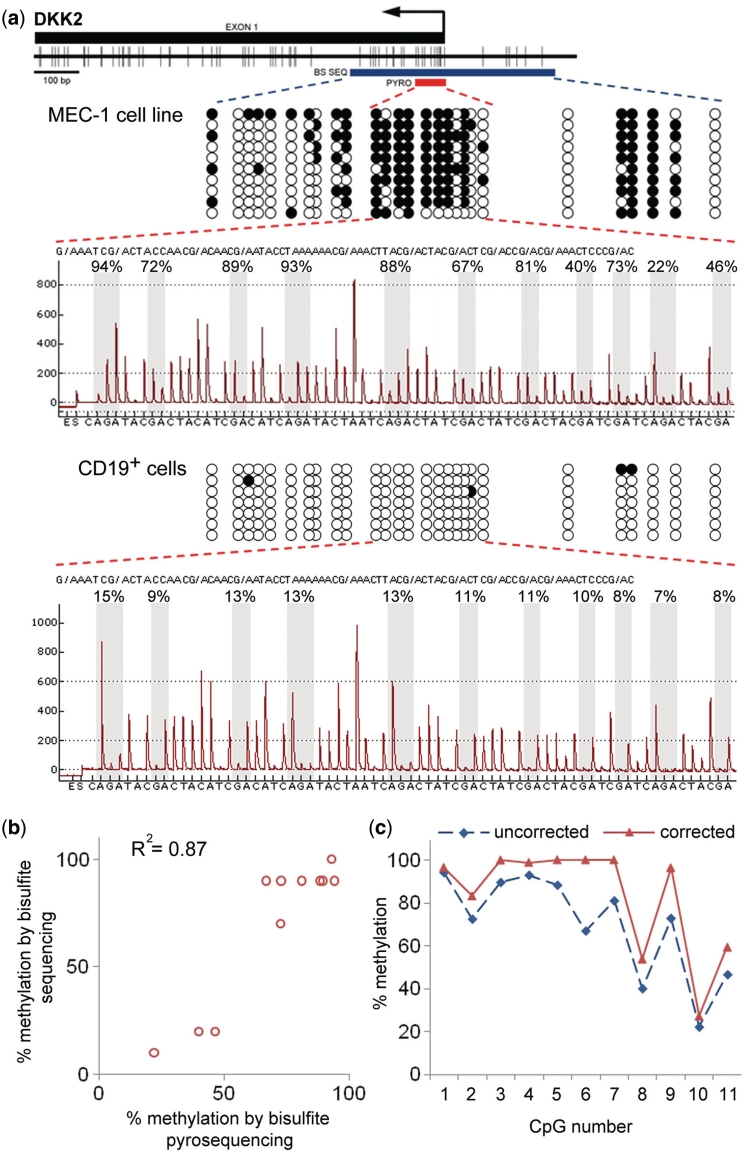
As an example of PCR-bias correction with a DNA from a biological sample, we analysed in the chronic lymphocytic leukaemia cell line MEC-1 (20) the methylation of 11 CpG dinucleotides located in close proximity to the DKK2 transcriptional start site. Bisulphite-treated DNA was analysed by pyrosequencing as well as Sanger sequencing. For the latter, 10 randomly picked plasmid clones with subcloned PCR products were analysed (Figure 5a). The raw data derived from both sequencing methods were in remarkably good agreement (Figure 5b), which might be explained by a relatively heterogeneous methylation pattern of the region. In parallel, an analysis of calibration DNA was performed. Using this information, the methylation values for each of the 11 CpG dinucleotides had to be corrected (Figure 5c), indicating the frequency of biased results if DNA from natural sources is being studied. The absolute value of the difference between corrected and uncorrected data differs among sites. As can be inferred from the regression curves (Figure 1), the difference between ascertained (uncorrected) and actual methylation values depends on the absolute percentage of methylation. The difference should be minimal at either hypo- or hypermethylated CpGs and reach a maximum for intermediate methylation levels (around 50% methylation). This could be the reason for smaller differences between the corrected and uncorrected values at CpG sites 1 and 10 of Figure 5c. However, superposition of an extra bias introduced by the pyrosequencing readout may also happen and lead to the unexpectedly high difference at CpG 6, for example.
Figure 5.
Correction of the DKK2 methylation degree in leukaemic cell line MEC-1 using cubic polynomial regression. (a) CpG map of the interrogated region (top). Vertical bars indicate the positions of CpG dinucleotides. The position of the first exon is shown as a black rectangle. The arrow indicates the DKK2 transcriptional start site. The red bar (denoted ‘PYRO’) specifies the CpG sites quantified by pyrosequencing; the blue bar (marked ‘BS SEQ’) indicates the region analysed by Sanger sequencing. In the DNA methylation patterns shown below, each row of circles represents the CpG dinucleotides of an individual clone sequence. The open and filled circles stand for unmethylated and methylated CpGs, respectively. The pyrogram at the bottom indicates the methylation degrees of 11 CpG dinucleotides in proximity of the DKK2 transcriptional start site. Grey bars highlight the signals corresponding to CpG sites analysed. The percent values above the pyrogram reflect the methylation degree determined for each CpG site. The sequence of nucleotides at the bottom is the dispensation order of the dNTPs during the pyrosequencing process. The lower panel provides the corresponding conformation for CD19+ B cells of a healthy individual. (b) A scatterplot comparison of the methylation degree of each of the 11 CpGs as determined by Sanger sequencing (vertical axis) and pyrosequencing (horizontal axis). (c) The diagram shows uncorrected (blue) and corrected (cubic polynomial regression with nine control DNA samples; red) methylation values of the CpG sites.
As an additional illustration of PCR-bias correction, methylation degrees of CDH1, DACT1, DKK1, DKK2, DKK3, DKK4, SFRP2 and SFRP3 were quantified in the leukaemic cell lines MEC-1 and EHEB as well as CD19+ B cells from healthy individuals (Supplementary Figure S2). Most of the loci were aberrantly hypermethylated in both cell lines, whereas essentially no methylation was observed in the CD19+ B cells, with few exceptions. Also in this experiment, the methylation percentages had to be corrected for nearly every gene. As expected, the differences between the apparent and corrected methylation levels were more pronounced for those genes, which exhibited a higher degree of PCR-bias (Table 2) and a higher percentage of methylation (for example, CDH1, DKK1 and DKK2).
DISCUSSION
The occurrence of PCR-bias in DNA methylation studies, which are based on bisulphite treatment, and the lack of processes to predict its extent ask for the development of correction procedures in order to produce accurate data, since erroneous measurements may strongly hamper the interpretation of the results. To date, no universally applicable solution had been reported to overcome the problem.
In most cases analysed, an over-amplification of the unmethylated alleles was observed; this is in agreement with earlier reports interrogating other loci (9,10,12). Interestingly, amplification of SFRP1 demonstrated an inverse and rather large (22-fold) deviation towards the methylated allele. In consequence, amplification of genomic DNA of only 6.25% and 12.5% real methylation already yielded a strongly exaggerated experimental result of 51% and 70%, respectively. This deviation could be explained by the presence of a CpG dinucleotide at the 5′-end of the reverse PCR primer used for amplification. Rather than balancing amplification bias, the presence of the CpG site in the primer sequence may have led to over-compensation, thereby introducing an inverse bias. This observation is important in the context of recent reports (10,11,14), which propose the use of this primer design scheme for a more unbiased amplification of bisulphite-treated DNA on a routine basis. Although the approach could be effective for particular genes, wider applicability may be hampered by effects such as demonstrated for the biased amplification of SFRP1. However, earlier studies (e.g. 9) have also reported a few other examples of preferential enrichment of methylated alleles. While the presence of CpGs in the primer-annealing sites could be a factor which causes PCR-bias of the methylated allele, also other reasons may be possible. In any case, each primer pair needs to be checked to assure its effectiveness.
In this context, non-CpG methylation as found in embryonic cells (27,28) represents a special case and possibly a challenge to both PCR-primer design and calibration-based correction. For primer design, the number of non-CpG cytosines in the sequences should be limited in order to avoid binding variation, although their effect should be less pronounced compared to CpGs, because the latter influence DNA structure stronger. If a possibly methylated cytosine within the primer sequence cannot be avoided, a mismatch to both the methylated and unmethylated sequence should be incorporated into the primer at the respective position as has been suggested for CpG dinucleotides (29). For calibration, the problem arises on how the actual degree of methylation in the calibration samples could be confirmed.
As opposed to refining experimental conditions, we approached the correction of amplification bias by accepting its occurrence during experimentation but adjusting the initial amplification result by a comparison to calibration data and the application of regression curves for deriving correction factors. Two types of regression—hyperbolic and cubic polynomial—were applied. The correction process based on cubic polynomial regression was found to be superior overall. A reason for this could be the fact that the experimental background noise contributes maximally to the uncorrected data if the signal intensities (for both unmethylated and methylated alleles) are low. Interestingly, the methylation values for a 50% methylation control (the point of minimal background influence) belong to both the hyperbolic and cubic fit curves for all the genes.
The method is applicable irrespective of the locus that is interrogated or the number of sites analysed. Based on curve fitting, the method is not influenced by the type of bias–preferential recovery of methylated or unmethylated alleles—and works equally well for both as documented by SFRP1. Furthermore, any bias that was additionally introduced by the pyrosequencing readout could be compensated by the very process. The method is also automatable for high-throughput analyses.
On the down side of the method, concomitant measurements of at least three calibration DNA samples are required. However, given the fact that no optimization of the reagents is needed, such as the use of modified primers for example, and that sequencing technologies are currently rapidly getting cheaper and higher in throughput, this fact should not be a limiting factor in the long run. However, also many currently employed methods of quantitative DNA methylation analysis may benefit.
Obviously, the relative monetary and time effect of our correction method in comparison to conventional protocol-optimizing approaches depends on the scale (number of loci), throughput (number of test samples) and the particular analytical technique that is applied. For a bisulphite pyrosequencing analysis of one locus in 96 patients, for example, the regression algorithm is estimated to save about 80% of time and actual cost, while losing only 3 out of 96 wells and thus samples (3%) to calibration.
The situation is similar in more high-throughput approaches based on microarray analysis (30) or massively parallel bisulphite sequencing (31). The latter publication, for instance, describes a DNA methylation analysis of 12 amplified loci from 69 breast cancer samples. The resulting 828 PCR products were tagged with sample-specific sequences, pooled and sequenced with a depth of coverage of 880 reads per amplicon and sample. Inclusion of three calibration samples to the pools would increase the number of amplification reactions by about 4.5% (36 in addition to 828) and reduce the sequence coverage per amplicon and sample insignificantly to about 840 instead of 880. The only cost increase would be an extra 4.5% for PCR, while sequencing cost would not be affected.
Finally, for genome-wide analyses, there is actually no feasible alternative to the calibration approach. For instance, when a sequencing library is prepared by fragmentation of genomic DNA using a cocktail of restriction enzymes (32), one could not reasonably perform with current technology an optimisation of the amplification conditions for all possible methylation sites of the genome. Genome-wide calibration, however, can be done. Admittedly, this would require the sequencing of three calibration genome copies, while only one sequence would represent the real epigenetic status of the respective genome. However, no alternative exists to get exact data. In addition, the calibration sequences can be used for all additional genome sequences. Still, accurate profiling of DNA methylation does come at a cost.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The German Federal Ministry of Education and Research (BMBF) as part of the NGFN-2 and NGFNplus programmes (01GR490 and 01GS08117 to J.D.H.); and a research scholarship of the German Academic Exchange Service (DAAD) (to E.A.M., A/08/81018). Funding for open access charge: DKFZ.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Yasser Riazalhosseini for helpful discussions.
REFERENCES
- 1.Mikeska T, Candiloro ILM, Dobrovic A. The implications of heterogeneous DNA methylation for the accurate quantification of methylation. Epigenomics. 2010;2:561–573. doi: 10.2217/epi.10.32. [DOI] [PubMed] [Google Scholar]
- 2.Moskalev EA, Eprintsev AT, Hoheisel JD. DNAmethylation profiling in cancer: from single nucleotides towards the methylome. Mol. Biol. 2007;41:723–736. [PubMed] [Google Scholar]
- 3.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dupont JM, Tost J, Jammes H, Gut IG. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal. Biochem. 2004;333:119–127. doi: 10.1016/j.ab.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Bormann Chung CA, Boyd VL, McKernan KJ, Fu Y, Monighetti C, Peckham HE, Barker M. Whole methylome analysis by ultra-deep sequencing using two-base encoding. PLoS ONE. 2010;5:e9320. doi: 10.1371/journal.pone.0009320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mund C, Beier V, Bewerunge P, Dahms M, Lyko F, Hoheisel JD. Array-based analysis of genomic DNA methylation patterns of the tumour suppressor gene p16INK4A promoter in colon carcinoma cell lines. Nucleic Acids Res. 2005;33:e73. doi: 10.1093/nar/gni072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoheisel JD. Microarray technology: beyond transcript profiling and genotype analysis. Nat. Rev. Genet. 2006;7:200–210. doi: 10.1038/nrg1809. [DOI] [PubMed] [Google Scholar]
- 9.Warnecke PM, Stirzaker C, Melki JR, Millar DS, Paul CL, Clark SJ. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 1997;25:4422–4426. doi: 10.1093/nar/25.21.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wojdacz TK, Hansen LL, Dobrovic A. A new approach to primer design for the control of PCR bias in methylation studies. BMC Res. Notes. 2008;1:54. doi: 10.1186/1756-0500-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wojdacz TK, Borgbo T, Hansen LL. Primer design versus PCR bias in methylation independent PCR amplifications. Epigenetics. 2009;4:231–234. doi: 10.4161/epi.9020. [DOI] [PubMed] [Google Scholar]
- 12.Shen L, Guo Y, Chen X, Ahmed S, Issa JP. Optimizing annealing temperature overcomes bias in bisulfite PCR methylation analysis. BioTechniques. 2007;42:48–58. doi: 10.2144/000112312. [DOI] [PubMed] [Google Scholar]
- 13.Chhibber A, Schroeder BG. Single-molecule polymerase chain reaction reduces bias: application to DNA methylation analysis by bisulfite sequencing. Anal. Biochem. 2008;377:46–54. doi: 10.1016/j.ab.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 14.Wojdacz KT, Hansen LL. Reversal of PCR bias for improved sensitivity of the DNA methylation melting curve assay. BioTechniques. 2006;41:274–278. doi: 10.2144/000112240. [DOI] [PubMed] [Google Scholar]
- 15.Archer KL, Mas VR, Maluf DG, Fisher RA. High-throughput assessment of CpG site methylation for distinguishing between HCV-cirrhosis and HCV-associated hepatocellular carcinoma. Mol. Genet. Genomics. 2010;283:341–349. doi: 10.1007/s00438-010-0522-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wojdacz TK, Dobrovic A, Hansen LL. Methylation-sensitive high-resolution melting. Nat. Protoc. 2008;3:1903–1908. doi: 10.1038/nprot.2008.191. [DOI] [PubMed] [Google Scholar]
- 17.Wiesmann F, Veeck J, Galm O, Hartmann A, Esteller M, Knüchel R, Dahl E. Frequent loss of endothelin-3 (EDN3) expression due to epigenetic inactivation in human breast cancer. Breast Cancer Res. 2009;11:R34. doi: 10.1186/bcr2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melki JR, Vincent PC, Clark SJ. Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Res. 1999;59:3730–3740. [PubMed] [Google Scholar]
- 19.Carr IM, Valleley EM, Cordery SF, Markham AF, Bonthron DT. Sequence analysis and editing for bisulphite genomic sequencing projects. Nucleic Acids Res. 2007;35:e79. doi: 10.1093/nar/gkm330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stacchini A, Aragno M, Vallario A, Alfarano A, Circosta P, Gottardi D, Faldella A, Rege-Cambrin G, Thunberg U, Nilsson K, et al. MEC1 and MEC2: two new cell lines derived from B-chronic lymphocytic leukaemia in prolymphocytoid transformation. Leuk. Res. 1999;23:127–136. doi: 10.1016/s0145-2126(98)00154-4. [DOI] [PubMed] [Google Scholar]
- 21.Saltman D, Bansal NS, Ross FM, Ross JA, Turner G, Guy K. Establishment of a karyotypically normal B-chronic lymphocytic leukemia cell line; evidence of leukemic origin by immunoglobulin gene rearrangement. Leuk. Res. 1990;14:381–387. doi: 10.1016/0145-2126(90)90167-8. [DOI] [PubMed] [Google Scholar]
- 22.Liu T, Raval A, Chen SS, Matkovic JJ, Byrd JC, Plass C. CpG island methylation and expression of the secreted frizzled-related protein gene family in chronic lymphocytic leukemia. Cancer Res. 2006;66:653–658. doi: 10.1158/0008-5472.CAN-05-3712. [DOI] [PubMed] [Google Scholar]
- 23.Rohde C, Zhang Y, Reinhardt R, Jeltsch A. BISMA – fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010;11:230. doi: 10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujikane T, Nishikawa N, Toyota M, Suzuki H, Nojima M, Maruyama R, Ashida M, Ohe-Toyota M, Kai M, Nishidate T, et al. Genomic screening for genes upregulated by demethylation revealed novel targets of epigenetic silencing in breast cancer. Breast Cancer Res. Treat. 2010;122:699–710. doi: 10.1007/s10549-009-0600-1. [DOI] [PubMed] [Google Scholar]
- 25.John EG. Simplified curve fitting using spreadsheet add-ins. Int. J. Engg. Ed. 1998;14:375–380. [Google Scholar]
- 26.Jacobson N. Basic Algebra. Mineola, NY: Dover Pubn Inc; 2009. [Google Scholar]
- 27.Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woodcock DM, Lawler CB, Linsenmeyer ME, Doherty JP, Warren WD. Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. J. Biol. Chem. 1997;272:7810–7816. doi: 10.1074/jbc.272.12.7810. [DOI] [PubMed] [Google Scholar]
- 29.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gitan RS, Shi H, Chen CM, Yan PS, Huang TH. Methylation-specific oligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome Res. 2002;12:158–164. doi: 10.1101/gr.202801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korshunova Y, Maloney RK, Lakey N, Citek RW, Bacher B, Budiman A, Ordway JM, McCombie WR, Leon J, Jeddeloh JA, et al. Massively parallel bisulphite pyrosequencing reveals the molecular complexity of breast cancer-associated cytosine-methylation patterns obtained from tissue and serum DNA. Genome Res. 2008;18:19–29. doi: 10.1101/gr.6883307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeschnigk M, Martin M, Betzl G, Kalbe A, Sirsch C, Buiting K, Gross S, Fritzilas E, Frey B, Rahmann S, et al. Massive parallel bisulfite sequencing of CG-rich DNA fragments reveals that methylation of many X-chromosomal CpG islands in female blood DNA is incomplete. Hum. Mol. Genet. 2009;18:1439–1448. doi: 10.1093/hmg/ddp054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.