Abstract
Despite being studied for over 30 years, a consensus structure-activity relationship (SAR) that encompasses the full range peptidic and non-peptidic μ opioid receptor ligands is still not available. To achieve a consensus SAR the Conformationally Sampled Pharmacophore (CSP) method was applied to develop a predictive model of the efficacy of μ opioid receptor ligands. Emphasis was placed on predicting the efficacy of a wide range of agonists, partial agonists and antagonists as well as understanding their mode of interaction with the receptor. Inclusion of all accessible conformations of each ligand, a central feature of the CSP method, enabled structural features between diverse μ opioid receptor ligands that dictate efficacy to be identified. The models were validated against a diverse collection of peptidic and nonpeptidic ligands, including benzomorphans, fentanyl (4-anilinopiperidine), methadone (3,3-diphenylpropylamines), etonitazene (benzimidazole derivatives), funaltrexamine (C6 substituted 4,5-epoxymorphinan) and herkinorin. The model predicts 1) that interactions of ligands with the B site, as with the 19-alkyl substituents of oripavines, modulates the extent of agonism; 2) that agonists with long N-substituents, as with fentanyl and N-phenethylnormorphine, can bind in an orientation such that the N substitutent interacts with the B site that also allows the basic N-receptor Asp interaction essential for agonism and 3) that the μ agonist Herkinorin, that lacks a basic nitrogen, binds to the receptor in a manner similar to the traditional opioids via interactions mediated by water or a ion. Importantly, the proposed CSP model can be reconciled with previously published SAR models for the μ receptor.
Keywords: Quantitative structure activity relationship, QSAR, molecular modeling, molecular dynamics, analgesics, herkinorin, morphine, methadone, fentanyl
Introduction
Opioids represent the front-line clinical treatment for pain, acting mainly on the central nervous system to achieve their desirable outcome. They target three receptor subtypes μ, δ, and κ1–4 of which μ receptors are thought to mediate the majority of pain relief, as μ-specific agonists are the most effective analgesics for severe pain. However, the clinical use of agonists is accompanied by a range of adverse side effects including development of tolerance, dependence, nausea, constipation, and respiratory depression5–7. Due to these side effects μ agonists have been the object of numerous lead modification studies8–12, including a large number of structure-activity relationship (SAR) studies13–24. To date, SAR studies focused on one position on the opioid skeleton yielding predictive models for a specific class, but introduction of substituents in other positions on the opioid skeleton often abolished the SAR such that a consensus pharmacophore for all classes of μ agonists has not been achieved. For example, a number of SAR based on the 4,5-epoxymorphinans were not applicable to other series8–12 such as benzomorphans, Diels-Alder adducts of thebaine, or fentanyls. Furthermore most SAR models dealt with binding affinities. Accordingly, the need for a consensus pharmacophore model that can quantitatively predict the efficacy of the full range of structural classes of μ-opioid receptor ligands is evident. Such a model would facilitate ongoing efforts to develop novel opioids that are both μ agonists and antagonists with the goal of limiting side effects while maintaining the analgesic properties25–28.
In the present study, we develop a comprehensive μ-opioid SAR by applying the Conformationally Sampled Pharmacophore (CSP) method29–33 to a diverse collection of μ-opioid ligands. The models were constructed based on two classical opioid scaffolds, the 4,5-epoxymorphinans and Diels-Alder adducts of thebaine (i.e. oripavines), and then applied to predict efficacies of benzomorphans, fentanyls, peptidic opioids, as well as additional known μ-opioid ligands. From the SAR common structural features are identified that can account for the efficacies of the diverse classes, indicating that these ligands have the same mechanism of receptor binding and activation.
Computational methods
Empirical force field calculations were performed using the program CHARMM34 with the CGenFF35 force field parameters for the non-peptidic ligands and the C22 all-atom protein force field, including the CMAP correction for the peptidic ligands36, 37. CGenFF parameters for selected opioids were validated targeting geometries, QM vibrational spectra38 and X-ray crystal structures39. (See supporting information). For CSP model development, all compounds were initially constructed based on the internal coordinates in CGenFF for nonpeptidic opioids and the C22 force field for the peptidic ligands and assigned physiologically pertinent protonation states consistent with the experimental pH of 7.4. Compounds were first relaxed in the gas phase by energy minimization for 500 steps with the steepest decent algorithm followed by 1000 steps with adopted basis Newton-Raphson method to an RMS force of < 10−6 kcal/mol Å. In all remaining calculations the aqueous solvent environment was treated implicitly by the Generalized Born Continuum Solvent Model (GBMV)40. MD simulations were performed with a 2 fs integration time step using the Leap-Frog integrator41. Covalent bonds involving hydrogen atoms were constrained to their equilibrium bond length by the SHAKE algorithm42. For non-bonded interactions, a switching function43 was used from 16 to 18 Å and the nonbond pair list was generated out to 21 Å. Simulations to achieve comprehensive sampling of ligand conformations used the replica exchange method44 based on four MD simulations (ie. replicas) running at temperatures of 300, 330, 363, and 400K for 10 ns/replica. Exchanges were attempted every 0.5 ps, saving coordinates every 2 ps. The 5000 conformations collected from the room temperature replica were subjected to CSP analysis. With the herkinorin-water complex simulation a harmonic restraint force of 100 kcal/mol/Å with a minimum of 3.0 Å was applied to the distance between the water oxygen and respective ester oxygens.
Probability distributions were based on distances and angles calculated from the simulations between the pharmacophoric points (Figure S1) with bin sizes of 0.1 Å and 1°, respectively. The extent of overlap of the distributions for different ligands was determined by calculation of overlap coefficients (OC)45. 1D OC values are defined by equation 1,
| Eq. 1 |
which quantifies the area under the probability distributions where the two distributions overlap. represents the probability density of compound A at bin i of the respective distance or angle. OC values associated with the pharmacophoric points in Figure 1 for each molecule were calculated with respect to a reference molecule (). Morphine was initially selected as the reference compound because of its high efficacy, affinity and prototypical structure while development of the second (AB'NS) model used etorphine as the reference compound due to its greater potential interactions with the “B site” as compared to morphine. Previous studies have shown that the CSP approach is not significantly impacted by the selection of the reference compound30, though selection of a highly active species facilitates structural interpretation of the resulting model.
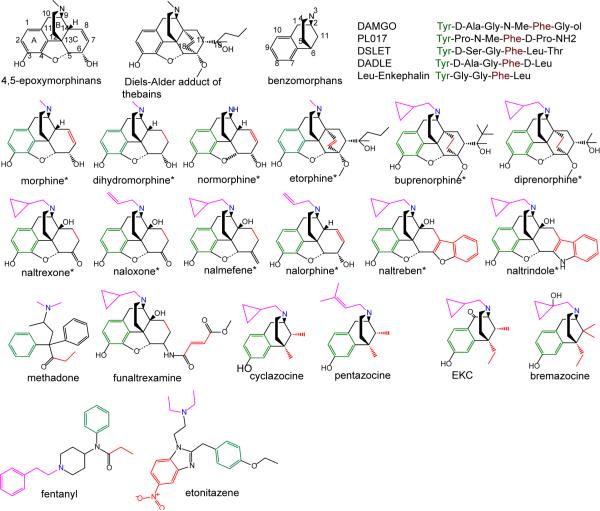
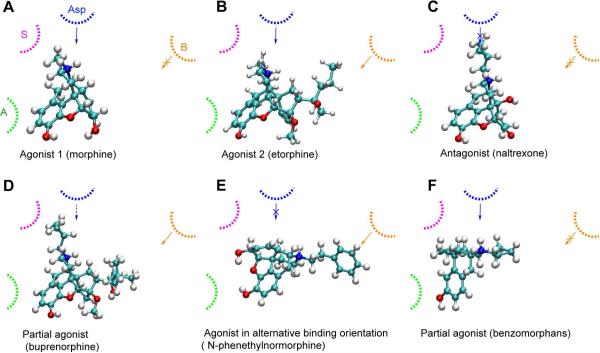
Figure 1.
Structure of opioids and representative numbering. The 12 opioids included in the training set are indicated by *. Pharmacophoric points of the ligands are shown where green represents the aromatic ring (A), red represents the hydrophobic group (B), blue represents the basic nitrogen (N), and pink represents the N substituent (S).
CSP model development was based on all possible permutations of triplets of OCs of the six distances and twelve angles for the 12 molecules in the training set (Figure 1). All permutations were iteratively treated as independent variables in multiple regression analysis with respect to the target experimental data. Statistical analysis was done using in house Perl scripts46 based on the R statistical package47. Highly correlated independent variables (correlation coefficient > 0.9, Table S3) were excluded from analysis and regression models for the individual independent variables with coefficient of determination (R2) above 0.8 were selected for the further analysis. The predictability of each model was verified by leave one out cross-validation and application of the models to the external, test set of compounds. To check convergence of the MD simulations with respect to conformational sampling was performed based on calculation of self OC values as a function of simulation time (Figure S9).
Results/Discussions
The CSP method29–33 is designed to overcome the possibility that the global minimum conformation of an unbound ligand may not be the most populated form when bound to the receptor48. This is achieved by including all accessible conformations of the ligands during model development from which probability distributions of geometries, such as distances and angles among critical functional groups, are used as the basis for SAR development. The probability distributions are converted to normalized overlap coefficients (OC) with respect to a reference compound, which may then be regressed against experimental data. As the CSP approach is ligand based, it inherently assumes that all the ligands are interacting with the same site on the receptor, although the inclusion of all accessible ligand conformations in the CSP approach may accommodate local conformational changes that may occur in the receptor upon binding to different ligands.
Model development used experimental data for 20 non-peptidic opioids and 5 peptides from Toll et al49 (Figure 1), of which 12 compounds were assigned as the training set and the remainder as the external test set (Figure 1 and Table S1 of the supporting information). Training set compounds included 4,5-epoxymorphinans and the oripavines. Experiments were conducted on CHO cells expressing the human μ-opioid receptor, which excludes issues of multi-receptor environments and/or pharmacokinetic considerations that may potentially complicate SAR development50. As the experimental data is based on homogenous μ-opioid receptor, the resulting model does not take into account the possible heterodimerization between μ, δ, and κ receptors on ligand efficacy. Efficacy was measured as the relative % stimulation with respect to that of DAMGO (100%) using a [35S]GTPγS binding assay.
Initial selection of the pharmacophoric points for model development was based on previous SAR studies of μ opioids. The aromatic ring (A), basic nitrogen (N), N substituent (S), and hydrophobic substituent on the C ring (B) of the morphine scaffold are known to play critical roles in opioid efficacy8. While the definitions of A, N, and S are clear, that of B is not obvious.15, 16 This ambiguity led to the selection of the 4,5-epoximorphans, with the classical opioid scaffold, and the oripavines (etorphine, buprenorphine and diprenorphine) as the training set compounds (Figure 1). With the 4,5-epoximorphans the 7 and 8 carbons in the C ring were assigned as the B point while in the oripavines both the 17 and 18 carbons (ABNS model) and the substituents on the 19 position were separately considered as the B group (AB'NS model). Concerning the reference compound morphine was used in the ABNS model while etorphine was used for the AB'NS model; etorphine was used in the later case as this is a full agonist with a prominent hydrophobic group at the 19 position.
Model development involved iterative fitting of individual OC values for the 6 possible pharmacophore distances and 12 angles (Tables S2 and Figure S1, supporting information) against the experimental data. Individual OCs with correlation coefficients (r2) > 0.8 were then used as independent variables in multiple regression analysis that involved all possible permutations of two and then three OCs sets, from which the top 10 models were selected for each of the ABNS and AB'NS models (Table S2). The top ABNS efficacy model had R2 = 0.96 with a P-value of 10−5 and R2 above 0.90 and Q2 above 0.5 for the 10 models (Table S3, supporting information). With the AB'NS model the top model had R2 = 0.87 with a P-value of 10−3 and in all cases R2 is above 0.77 and Q2 is above 0.5 (Table S4). In both sets of models all four pharmacophore points are well represented, consistent with the essential role of the N (though exceptions do exist (see below)51,52, 53) and A groups while the S group is known to play an important role in the activity of opioid ligands8. With the ABNS model the B point is not included in two of the models while it is present in all cases with the AB'NS model.
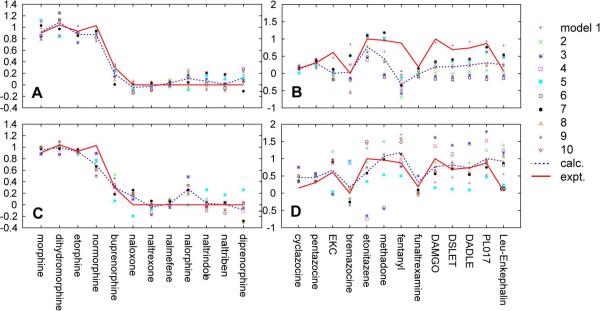
While the ABNS model yielded overall better quality fits, the model under predicts the efficacy of buprenorphine while that of diprenorphine is over predicted (Figure 2a). This is due to the B group not being included in two of the top 10 ABNS models, suggesting a limitation in the use of the 17/18 carbons of oripavines to define that group. In contrast, the AB'NS model better distinguishes the efficacy of the oripavines although the overall goodness of fit decreased (Figure 2c). This decrease is associated with the C19 substituent being unique for the oripavines such that etorphine as the reference compound was less representative for non-oripavines. This leads to some B' related OC values, such as AB', B'N, AB'N, AB'S, ANB', and ASB', being zero for non-oripavines with respect to etorphine (Table S2). Since each of the 10 AB'NS models has one of these terms as an independent variable the goodness of fit decreases. However, this degradation was deemed acceptable to predict the efficacies of oripavines and the AB'NS models more accurately predicted the efficacies of the external test set compounds than the ABNS model (see below).
Figure 2.
Predicted and experimental relative efficacies of the 12 training set compounds and 13 test set by the ABNS (a, b) and AB'NS (c, d) models. Red solid line: experimental values, blue dotted line: average of the predicted values from the top 10 models. The top 10 models are indicated by individual symbols.
Receptor models suggested by Casy and Beckett54, 55 include an additional binding site for the B group and Snyder et al.56, 57 postulated the presence of a specific interaction site for the C19 substituent or its equivalent which is only accessible by agonists. Probability distributions of the AB distances for these three oripavines showed significant overlap with the agonist etorphine and partial agonist buprenorphine distributions while that of the antagonist diprenorphine occurred at significantly shorter distances (Figure S2). These observations suggest an important role of the B group in differentiating efficacy considering the presence of the cyclopropylmethyl (CPM) moiety on both buprenorphine and diprenorphine, a substituent typically associated with antagonists8. The importance of the role of the B group is further supported by a previous study showing N-allyl or N-CPM substituted etorphines to have stronger agonism than morphine58, though they typically lead to antagonism in 4,5-epoxymorphinans, and that norbuprenorphine is also a potent partial agonist with higher efficacy than buprenorphine59.
The models were next challenged against the remaining compounds in Figure 1. Application of the ABNS model yielded poor agreement for selected compounds including ethylketocyclazocine (EKC), fentanyl, methadone, and the peptides (Figure 2b), even when alternate definitions of the B point were considered for fentanyl and methadone, for which the B definition is ambiguous (Figures 1, S2, and S3); this model was not considered further. In contrast, the AB'NS model yielded satisfactory predictions of efficacy for the test compounds (Figure 2d, Table S5). This was also achieved by considering multiple moieties as the S and B groups in methadone and fentanyl, a procedure that was essential for successful application of the AB'NS model to diverse opioids, as presented below.
Individually some of the AB'NS models (models 3, 4, and 5) yielded relatively poor agreement (Table S6, S7), due to B related OC values of some test compounds being higher than those in the training set thereby producing higher or lower predicted efficacies (Table S4). For example, in model 3 the ANB' OC values of peptidic opioids are five times higher than any compounds in the training set yielding high efficacies and the BS OC values of etonitazene and methadone are the cause of lower predicted efficacies. However, those errors cancelled in the average values yielding overall good predictions, including the correct prediction of the peptidic opioids. The most significant failure with the AB'NS model occurs with Leu-enkephalin, whose efficacy is systematically predicted to be too high (see below).
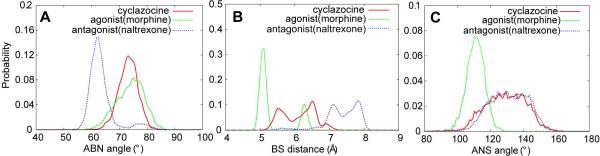
Of note is the ability of the AB'NS model to satisfactorily predict the efficacies of the benzomorphans, which range from an antagonist (bremazocine) to partial agonists (cyclazocine, pentazocine) to an agonist more active than morphine (ethylketocyclazocine (EKC)). This class of compounds lacks a C ring while the composition and configuration of their C6 and C11 substituents are crucial to their efficacy8. Given the lack of a C ring, the C6 and C11 substituents were used to define the B pharmacophoric points (Figure 1). However, it was questioned why cyclazocine and pentazocine, which contain CPM or allyl N substituents, are partial agonists while those substituents are responsible for antagonism in the 4,5-epoxymorphinans. To analyze this, selected probability distributions of cyclazocine were compared with those of the agonist morphine and the antagonist naltrexone (Figure 3). For the ABN and BS distributions, there is significant overlap of the morphine and cyclacozine distributions, while for the ANS distribution there is significant overlap with that of naltrexone. Similar overlap with naltrexone is present with the AN, AS, ANS, ASN, and NAS distributions (Table S3). Thus, with respect to the relative orientation of the A, B and N pharmacophoric points, cyclazocine mimics an agonist, while with respect to the relative orientations of the A, S and N points it mimics an antagonist. This combination is suggested to lead to cyclazocine and pentazocine being partial agonists.
Figure 3.
Probability distributions of the partial agonist cyclazocine with those of the agonist morphine and the antagonist naltrexone for the A) ABN, B) BS and C) ANS pharmacophore descriptors.
The benzomorphans, EKC and bremazocine, are structurally similar yet have significantly different efficacies. While the OCs of both compounds are generally similar to those of other antagonists (Table S3) significant differences for several angles, including BAS, BNS and NBS, allow the AB'NS model to differentiate between the two compounds, emphasizing the importance of the spatial relationship of the different pharmacophore points in the models. However, it should be emphasized that the ketone on the C1 of EKC and hydroxyl on the CPM moiety of bremazocine could directly contribute to their differential efficacies. Antagonist bremazocine's hydroxyl group is approximately 2.7 Å away from the proton on the basic nitrogen possibly affecting the interaction of the N with the receptor. Alternatively, the ketone on EKC, as an electron-withdrawing group, may enhance the basicity of the nitrogen leading to more favorable interaction with the receptor thereby favoring its agonist activity.
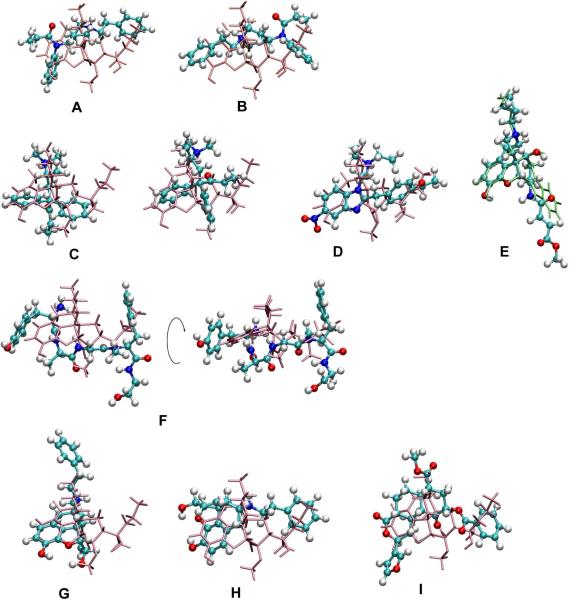
Fentanyl is an agonist for which multiple definitions of the pharmacophoric points are possible. Accordingly, in addition to the pharmacophore point definitions shown in Figure 1, alternate definitions, such as the phenethyl moiety being the B point, rather than the S point, and the N-phenyl ring being the A point were considered (Figure S3). Interestingly, classical definition of the N-substituent as S did not yield agonism activities but defining the phenethyl moiety as either the A or the B point leads to proper prediction of fentanyl efficacy. This is illustrated in Figure 4a–b showing that both orientations of the aromatic rings overlap with the A and B' regions of etorphine, as supported by probability distributions (Figure S3) showing that either aromatic ring yields significant overlap with the BN distribution of etorphine. This observation agrees with previous studies15, 16 which proposed that the N-phenethyl group is the A pharmacophoric point corresponding to A ring of 4,5-epoxymorphinans. However, the ability of the N-phenethyl moiety to also act as the B' point is particularly interesting as it suggests a model that allows compounds with long N substituents to be μ agonists, associated with the ligand reorienting in the receptor binding pocket, as will be discussed in more detail below.
Figure 4.
Superposition of selected structures with etorphine or naltrindole. Alignment of fentanyl with etorphine a) when the phenethyl group is used as the B' descriptor and b) when the phenethyl group is used as the A descriptor. In both cases the N atoms are superimposed. Note the overlap of the aromatic moiety attached to the amide N in fentanyl with the A ring of etorphine in the superpositions. c) Methadone with etorphine (blue lines) showing two possible arrangements of ethylketone group and aromatic ring with respect to the B' site. d) Superposition of etonitazene with etorphine (blue lines), based on the N, A and B points. e) Superposition of β-funaltrexamine with naltrindole (blue lines) based on the N, A and B points. f) Superimposition of DAMGO with etorphine shown in two orientations of the same overlaid structures. g) Conventional alignment of N-phenethylnormorphine (red) with etorpine (blue) and h) suggested alignment of N-phenethylnormorphine with etorpine following rotation of N-phenethylnormorphine around the AN axis, and i) the superimposition of herkinorin on etorphine.
Both methadone and etonitazene are agonists with unique structures that include a basic nitrogen. Methadone is representative of the diphenylpropylamines, while etontazene is the full agonist benzimidazole derivative; both compounds are properly predicted to be agonists. For methadone, one phenyl ring represented the A point while the ethyl ketone moiety was the B point (note that both rings were considered as the A or B groups, Figure S4). This assignment is consistent with experiments showing methadone to lose potency upon removal of one phenyl ring and an important role of ethyl ketone or ethyl alcohol groups for efficacy and/or potency8. Secondly, dimethyl or diethyl S substituents on the basic N lead to optimal analgesics but longer substitutions retain weak agonism in contrast to the 4,5-epoxymorphinans where such S groups lead to antagonism. For etonitazene, the nitro-containing benzylimidazole ring and ethoxy group acted as the A and B points, respectively, consistent with experimental studies showing them to be important for efficacy.8 Superposition of selected conformations of methadone and etonitazene with etorphine based on these definitions (Figure 4c and d) reveals the extent of overlap of the A and B moieties, emphasizing that these highly flexible molecules participate in the same ligand-receptor interaction mode as 4,5-epoxymorphinans.
The only irreversible antagonist in this study, β-funaltrexamine, has a hydrophobic group branched at C6. It has similar OC values to the oxymorphindoles including high spatial overlap with their B group distributions (Table S3). Therefore, β-funaltrexamine is predicted to interact in a similar way as naltrindole and naltriben (Figure 4e). Interestingly, such an interaction mode may orient the ester group of β-funaltrexamine such that it may form a covalent bond with a Lys in the 5th transmembrane domain (TM) of the μ opioid receptor60 based on models of the receptor that assume Asp147 in the 3rd TM is involved in an ionic interaction with the basic nitrogen of the opioids61.
The remaining compounds in the test set are the peptidic opioids (Figure 1). In these compounds the N-terminal nitrogen and tyrosine sidechain are thought to mimic the N and A ring in morphine, respectively8, and the phenylalanine ring has been hypothesized to assume a conformation similar to the C19 substituents of oripavines and take part in hydrophobic interactions with the receptors57, 62. Based on these definitions, with the N hydrogens as the S group, the AB'NS model correctly predicted the efficacies of the peptidic opioids, with the exception of the low efficacy of Leu-Enkephalin, indicating the peptidic opioids to share the same pharmacophoric features as the non-peptidic opioids. This is shown in Figure 4f where one conformation of DAMGO from the CSP simulations is superimposed on etorphine. CSP modeling made it possible to identify biologically active conformations of peptides consistent with those of the non-peptidic opioids, further validating the AB'NS model as a consensus pharmacophore.
Given that Leu-Enkephalin contains two consecutive glycines and lacks D-amino acids, a different binding mode was suspected and alternate pharmacophoric point definitions were tried. These included the sidechains of Tyr1, Phe4, and Leu5 as each the A and B points. For all these possibilities Leu-Enkephalin was predicted to be a partial agonist in the AB'NS model. Thus, despite the model correctly predicting the agonist activity of the remainder of the peptides, the efficacy of Leu-Enkephalin is over predicted.
The AB'NS model was further challenged by considering 4,5-epoxymorphinans that include N-phenethyl, C14-O-phenylpropyl, and C7β-phenylbutyl substituents (Figure S5). 4,5-epoxymorphinans with C-7β alkyl or arylalkyl groups63, 64 or C14 lipophilic chains65, 66 are highly potent agonists and there have been longstanding presumptions that their increased potency is due to a mechanism similar to that occurring in the C19 substituted oripavines, motivated by the C7β and C14 substituents being spatially similar to C19 substituents. When the efficacies of these compounds are predicted by the AB'NS they are in generally good qualitative agreement with reported potencies from a number of studies (Tables 1 and S8). Quantitative comparison was impossible between μ-opioid receptor specific cell-line efficacies and potencies from in vivo mouse assays since animal experimental results are influenced by many factors such as pharmacokinetics, affinity, and the presence of other types of opioid receptors. Of these compounds N-cyclohexylethyloxymorphindole, phenazocine and N-phenethylnormorphine are of particular interest. To be correctly predicted it was necessary to assign the B point to be the N-substituent, which is typically the S point (Figure S5). Indeed, when etorphine and N-phenethylnormorphine are aligned in the conventional fashion (Figure 4g) their putative B substituents occupy significantly different regions of space. However, if N-phenethylnormorphine is simply rotated around the AN axis the putative B moiety shows significant overlap (Figure 4h). This is analogous to the rotation applied to fentanyl and a similar effect is seen with N-cyclohexylethyloxymorphindole and phenazocine (not shown). This suggests that N-phenethylnormorphine and related analogs do NOT bind in the conventional orientation to the μ-opioid receptor, but rather reorient such that the N-phenethyl moiety occupies the B moiety binding site.
Table 1.
Prediction of test set 2 compounds
| compound | relative % stimulation | prediction | |
|---|---|---|---|
| ABNS | AB'NS | ||
| 1 N-cyclohexylethyloxymorphindole | 0.582,83 | 0.0 | 1.8 |
| 2 N-phenethylnormorphine | more potent than morphine84–86 | 0.6 | 1.9 |
| 3 N-methyl-C14-O-phenylpropyl substituent | 1.0a)65, 66, 83 | 1.1 | 0.4 |
| 4 N-CPM-C14-O-phenylpropyl substituent | more potent than morphine65, 66, 83 | 0.1 | 1.0 |
| 5 metazocine | 0.785 | 1.0 | 0.8 |
| 6 phenazocine | 0.887 | 0.5 | 2.1 |
| 7 N-CPM-C7-β-phenylbutyl substituent | as potent as morphine63,64 | 0.0 | 0.4 |
Compound 3 lacks methyl group at C5 compared to the one in experiment but it is reported that C5 methyl group shows no significant effect on activities65.
Relative % stimulation is based on DAMGO's efficacy 1.00 (100%) in [35S]GTPγS binding assay
The benzomorphans, phenazocine and metazocine (Figure S5, Table 1), which are both indicated to be agonists, are an interesting example of the importance of the alternate binding modes as defined by the rotation about the N-A vector leading to the traditional N substituent occupying the B site. With phenazocine, the phenethyl N substituent is also predicted to interact with the B site on the receptor, allowing the AB'NS model to predict it to be an agonist. In contrast, metazocine, with a methyl N substituent, is predicted to interact in the traditional orientation as occurs with the potent 4,5-epoxymorphinans. Accordingly, the AB'NS model predicts metazocine to have higher efficacy than cyclazocine or pentazocine.
Recently, herkinorin52, an analog of the non-nitrogeneous κ-opioid selective agonist, Salvinorin A51, 67, was discovered and shown to be the first selective full μ agonist that lacks the basic nitrogen. There is evidence that herkinorin may have an alternate binding site as indicated by its differential cellular signaling53 and based on studies on Salvinorin A showing it to have unique binding epitopes on the κ opioid receptor68 and have allosteric effects on the μ opioid receptor.67 However, the differential cellular signaling of herkinorin is lost following subtle structural modifications53 and even morphine, etorphine, fentanyl, and DAMGO display differential responses at the cellular level69. Therefore we assumed that herkinorin binds to the same site as the other ligands. Based on this assumption herkinorin was selected as a rigorous test of the developed CSP model. Obviously, direct electrostatic interactions between herkinorin and an acidic group on the receptor, presumably Asp147, thought to interact with the prototypical μ opioids is absent. However, the compound contains a number of hydrogen bond acceptors groups (Figure S4) that led us to hypothesize that water or Na+ is acting to mediate the interaction with the receptor acidic group. This is consistent with X-ray crystal structures of GPCRs (for example, rhodopsins (PDB ID: 1U19) and A2A adenosine receptor (PDB ID: 3EML)), in which ordered waters70, 71 are located in the vicinity of Asp147 or its equivalent residue as well as the importance of sodium ions in the interaction between opioid receptor and ligands72, 73. With the assumption that a water or ion bridge is formed between Asp147 and the ester oxygens of herkinorin, an explicit water was placed near one of either the carbonyl or ether oxygens and the complexes subjected to replica exchange simulations with the water restrained to be in the vicinity of the respective ester oxygens; CSP analysis was then performed with the N pharmocophore point modeled as the water oxygen; the remaining pharmacophore points are shown in Figure S5. Based on those definitions herkinorin is predicted to be an agonist with an efficacy of 84 % when the bridging water is interacting with the carbonyl oxygen and 92 % with the ether oxygen, in excellent agreement with the experimental % stimulation in human μ-opioid receptor CHO cells of 112%.74 This success further supports the consensus quality of the model. Figure 4i shows one of the conformations of herkinorin aligned with etorphine. The overlap of the A and B pharmacophore points is evident while the ester moiety is in the vicinity of the nitrogen of etorphine.
Consensus Ligand-opioid receptor efficacy model
Based on the presented AB'NS CSP model in combination with the large body of experimental μ-opioid SAR data, we propose the following consensus pharmacophore for the efficacy of opioids targeting the μ receptor (Figure 5). Traditional agonists (e.g. morphine) lacking or having short N substituents have NH+-receptor Asp interactions that lead to the receptor assuming an active conformation and do not interact with the B binding site (Figure 5a). In agonists with large B substituents, such as etorphine, interactions occur with both the N and B sites to maximize efficacy over that of morphine (Figure 5b). In contrast, traditional antagonists cannot interact with the essential Asp as required for the receptor to assume a conformation required for agonism due to their bulky N-substituents and they also cannot interact with the B site (Figure 5c). Partial agonists, such as buprenorphine, have a favorable interaction with the B site, which partially overcomes the negative impact of bulky N substituents, leading to partial efficacy (Figure 5d). In this model, the shorter C19 substituent of the antagonist diprenorphine disallows interactions with the B site, thereby not being able to overcome the presence of the CPM N substituent. Thus, interactions with the B site modulate the extent of agonism associated with N-receptor Asp interactions. This can lead to enhanced efficacy when the basic N is not blocked, as with etorphine, or to partial agonism where interactions with the B site partially overcome the presence of large N substituents, yielding partial agonists, as with buprenorphine.
Figure 5.
Hypothetical binding states of μ opioid receptor ligands.
Importantly, the proposed model may be applied to non-traditional non-peptidic opioids. For agonists with long N substituents, such as N-phenethylnormorphine, the molecule can orient in the binding pocket such that the large N substituent (traditional S pharmacophoric point) interacts with the B site (Figure 5e). In this alternate binding orientation, the traditional AS distance mimics the AB distance leading to the receptor assuming an active state. It is suggested that this alternate binding orientation only occurs with molecules whose N-substituent is longer than N-CPM (Figure S7). Benzomorphans are unique due to the lack of a C ring and the presence of a N-CPM substituent while being partial agonists. If they are assumed to bind as other antagonists that contain an N-CPM substituent, as in Figure 5c, their partial agonism cannot be explained. However, if their binding orientation is similar to that in Figure 5e and shifted such that they move further toward the position of dihydrofuran and C ring of 4,5-epoxymorphinans (Figure S8), then the basic nitrogen is more accessible to Asp residues (Figure 5f) leading to partial agonism.
The possible interaction of long N substituents in the B binding site was first proposed by Snyder and coworkers in 1976.57 Their model involved two conformational states of the receptor, with only one being accessible to agonists. The presence of C19 substituents of the oripavines or long N substituents was suggested to favor the active state, due to these moieties interacting at the “F” site, which corresponds to our B site. For the long N substituents to interact with the F site, it was proposed that they assume an axial configuration, although this configuration is now known to not be populated based on crystallographic and NMR studies of a number of non-peptidic opioids75, 76. It should be noted that Snyder et al. thought that every N substituent interacted with the F/B site, with the exception of the N-CPM moiety in the case of antagonists due to the presence of a hydroxyl at the C14 position. The present model clarifies that of Synder in that the interactions of the long N substituents with the F/B site can occur due to reorientation of the ligands in the receptor-binding pocket. Thus, while details of their model were not correct, Synder and coworkers correctly predicted the presence of an F/B site on the receptor that modulates its efficacy.
Conclusions
In the present study the subtle relationship of N substituents and of the C19 substituents of the oripavines to efficacy were explored. Relationships between the composition and conformation of N substituents and biological activity are intriguing SAR. Antagonist properties tend to be associated with allyl, dimethylallyl, and CPM substituents, whereas shorter N-methyl and longer N-phenethyl substitutions afford agonist properties. To date it has not been understood why the change from methyl to CPM abolishes agonist effects while the change from CPM to phenethyl recovers the agonist activity. Previous studies, including those based on homology models of the receptor61, 77–81 approached this problem simply by presuming different binding sites for each ligands and there is no published SAR model including the effect of N substituents. However, the CSP model developed in this work presents a hypothesis to explain these observations. The receptor-binding site, which can accommodate ligands of up to 10 amino acids residues, allows different ligands to bind in different orientations, such that the extended N substituents are interacting with the B site in the receptor, the site where C19 substituents usually interact. This allows compounds with long N substituents such as fentanyl, cyclohexylethyl-oxymorphindole, phenethylnormorphine and phenazocine to act as agonists. While more detailed structural studies, including experimental elucidation of the 3D structure of the μ-opioid receptor in the presence of ligands, are required to verify this hypothesis the present results based on the ligand-based CSP approach, that includes all accessible ligand conformations in model development while not requiring ligand alignment, presents an intriguing solution to an issue that has confounded the opioid community for over 30 years.
Supplementary Material
Acknowledgment
Financial support from the NIH (DA13583 and DA19634) and computational support from the Computer-Aided Drug Design Center, School of Pharmacy, University of Maryland, Baltimore are acknowledged.
Footnotes
Supporting Information Available. Tables of (1) experimental data used in model construction, (2) overlap coefficients, (3, 4) top 10 regression models, (5) RMSD of models, (6) efficacies of fentanyl and methadone with various pharmacophore point definitions, (7) prediction of test set 1, and (8) antinociceptive potency of test set. Figures of (1) pharmacophore point and descriptors, (2) probability distribution of AB' distances of orvinols, (3), (4) pharmacophore point definitions for fentanyl and methadone, (5) test set compounds, (6) probability distributions of AB' distances of test set, (7) probability distribution of distances between aromatic ring and N-substituents or C19 substituents, (8) reorientation of cyclazocine, (9) convergence of conformational sampling, and (10) conformation energy of the ensemble, and details on parametrization methods and results. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Evans CJ, Keith DE, Morrison H, Magendzo K, Edwards RH. Science. 1992;258:1952–1955. doi: 10.1126/science.1335167. [DOI] [PubMed] [Google Scholar]
- 2.Kieffer BL, Befort K, Gaveriaux-Ruff C, Hirth CG. Proc. Natl. Acad. Sci. U. S. A. 1992;89:12048–12052. doi: 10.1073/pnas.89.24.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mansson E, Bare L, Yang D. Biochem. Biophys. Res. Commun. 1994;202:1431–1437. doi: 10.1006/bbrc.1994.2091. [DOI] [PubMed] [Google Scholar]
- 4.Wang JB, Johnson PS, Persico AM, Hawkins AL, Griffin CA, Uhl GR. FEBS Lett. 1994;338:217–222. doi: 10.1016/0014-5793(94)80368-4. [DOI] [PubMed] [Google Scholar]
- 5.Kieffer BL, Evans CJ. Cell. 2002;108:587–590. doi: 10.1016/s0092-8674(02)00666-9. [DOI] [PubMed] [Google Scholar]
- 6.McNicol E, Horowicz-Mehler N, Fisk RA, Bennett K, Gialeli-Goudas M, Chew PW, Lau J, Carr D. J. Pain. 2003;4:231–256. doi: 10.1016/s1526-5900(03)00556-x. [DOI] [PubMed] [Google Scholar]
- 7.von Zastrow M, Svingos A, Haberstock-Debic H, Evans C. Curr. Opin. Neurobiol. 2003;13:348–353. doi: 10.1016/s0959-4388(03)00069-2. [DOI] [PubMed] [Google Scholar]
- 8.Casy AF, Parfitt RT. Opioid analgesics: Chemistry and receptors. Plenum Press; New York: 1986. [Google Scholar]
- 9.Goodman AJ, Le Bourdonnec B, Dolle RE. ChemMedChem. 2007;2:1552–1570. doi: 10.1002/cmdc.200700143. [DOI] [PubMed] [Google Scholar]
- 10.Janeckah A, Fichna J, Janecki T. Curr. Top. Med. Chem. 2004;4:1–17. doi: 10.2174/1568026043451618. [DOI] [PubMed] [Google Scholar]
- 11.Kaczor A, Matosiuk D. Curr. Med. Chem. 2002;9:1567–1589. doi: 10.2174/0929867023369394. [DOI] [PubMed] [Google Scholar]
- 12.Kaczor A, Matosiuk D. Curr. Med. Chem. 2002;9:1591–1603. doi: 10.2174/0929867023369358. [DOI] [PubMed] [Google Scholar]
- 13.Cheng J, Liu G, Zhang J, Xu Z, Tang Y. J. Mol. Model. 2011;17:477–493. doi: 10.1007/s00894-010-0745-1. [DOI] [PubMed] [Google Scholar]
- 14.Dong N, Lu WC, Chen NY, Zhu YC, Chen KX. Acta Pharmacol. Sin. 2005;26:107–112. doi: 10.1111/j.1745-7254.2005.00014.x. [DOI] [PubMed] [Google Scholar]
- 15.Filizola M, Villar HO, Loew GH. Bioorg. Med. Chem. 2001;9:69–76. doi: 10.1016/s0968-0896(00)00223-6. [DOI] [PubMed] [Google Scholar]
- 16.Filizola M, Villar HO, Loew GH. J. Comput.-Aided Mol. Des. 2001;15:297–307. doi: 10.1023/a:1011187320095. [DOI] [PubMed] [Google Scholar]
- 17.Heyl DL, Schullery SE, Renganathan K, Jayamaha MN, Rodgers DW, Traynor JR. Bioorg. Med. Chem. 2003;11:3761–3768. doi: 10.1016/s0968-0896(03)00329-8. [DOI] [PubMed] [Google Scholar]
- 18.Huang XQ, Jiang HL, Luo XM, Rong SB, Gu JD, Tan XJ, Zhu YC, Chen KX, Ji RY, Cao Y. Acta Pharmacol. Sin. 2000;21:46–54. [PubMed] [Google Scholar]
- 19.Johnson H. NIDA Res. Mono. 1978;22:146–158. [PubMed] [Google Scholar]
- 20.Peng Y, Keenan SM, Zhang Q, Kholodovych V, Welsh WJ. J. Med. Chem. 2005;48:1620–1629. doi: 10.1021/jm049117e. [DOI] [PubMed] [Google Scholar]
- 21.Ramírez-Galicia G, Garduño-Juárez R, Deeb O, Hemmateenejad B. Chem. Biol. Drug Des. 2008;71:260–270. doi: 10.1111/j.1747-0285.2008.00626.x. [DOI] [PubMed] [Google Scholar]
- 22.Schullery SE, Mohammedshah T, Makhlouf H, Marks EL, Wilenkin BS, Escobar S, Mousigian C, Heyl DL. Bioorg. Med. Chem. 1997;5:2221–2234. doi: 10.1016/s0968-0896(97)00163-6. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian G, Ferguson DM. Drug Des. Discov. 2000;17:55–67. [PubMed] [Google Scholar]
- 24.Wang XH, Tang Y, Xie Q, Qiu ZB. Eur. J. Med. Chem. 2006;41:226–232. doi: 10.1016/j.ejmech.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 25.Abdelhamid EE, Sultana M, Portoghese PS, Takemori AE. J. Pharmacol. Exp. Ther. 1991;258:299–303. [PubMed] [Google Scholar]
- 26.Hepburn MJ, Little PJ, Gingras J, Kuhn CM. J. Pharmacol. Exp. Ther. 1997;281:1350–1356. [PubMed] [Google Scholar]
- 27.Wells JL, Bartlett JL, Ananthan S, Bilsky EJ. J. Pharmacol. Exp. Ther. 2001;297:597–605. [PubMed] [Google Scholar]
- 28.Schiller PW, Fundytus ME, Merovitz L, Weltroska G, Nguyen TMD, Lemieux C, Chung NN, Coderre TJ. J. Med. Chem. 1999;42:3520–3526. doi: 10.1021/jm980724+. [DOI] [PubMed] [Google Scholar]
- 29.Bernard D, Coop A, MacKerell AD., Jr. J. Am. Chem. Soc. 2003;125:3101–3107. doi: 10.1021/ja027644m. [DOI] [PubMed] [Google Scholar]
- 30.Bernard D, Coop A, MacKerell AD., Jr. J. Med. Chem. 2007;50:1799–1809. doi: 10.1021/jm0612463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernard D, Coop A, MacKerell AD., Jr. J. Med. Chem. 2005;48:7773–7780. doi: 10.1021/jm050785p. [DOI] [PubMed] [Google Scholar]
- 32.González PM, Acharya C, MacKerell AD, Jr., Polli JE. Pharm. Res. 2009;26:1665–1678. doi: 10.1007/s11095-009-9877-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rais R, Acharya C, Tririya G, MacKerell AD, Jr., Polli JE. J. Med. Chem. 2010;53:4749–4760. doi: 10.1021/jm1003683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brooks BR, Brooks CL, MacKerell AD, Jr., Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, MacKerell AD., Jr. J. Comput. Chem. 2009;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacKerell AD, Jr., Feig M, Brooks CL. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 37.MacKerell AD, Jr., Brooks B, Brooks CLI, Nilsson L, Roux B, Won Y, Karplus M. CHARMM: The Energy Function and Its Parameterization with an Overview of the Program. Vol. 1. John Wiley & Sons; Chichester: 1998. pp. 271–277. [Google Scholar]
- 38.Frisch MJT,GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr., Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision C.02. Gaussian, Inc.; Wallingford CT: 2004. [Google Scholar]
- 39.Allen FH. Acta Crystallogr., Sect. B: Struct. Sci. 2002;58:380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 40.Lee MS, Feig M, Salsbury FR, Brooks CL. J. Comput. Chem. 2003;24:1348–1356. doi: 10.1002/jcc.10272. [DOI] [PubMed] [Google Scholar]
- 41.Hockney RW. The potential calculation and some applications Methods. Vol. 9 B. Academic Press; New York and London: 1969. [Google Scholar]
- 42.Ryckaert JP, Ciccotti G, Berendsen HJC. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 43.Steinbach PJ, Brooks BR. J. Comput. Chem. 1994;15:667–683. [Google Scholar]
- 44.Sugita Y, Okamoto Y. Chem. Phys. Lett. 1999;314:141–151. [Google Scholar]
- 45.Bradley EL. Overlapping coefficient. Vol. 6. Wiley; New York: 1985. [Google Scholar]
- 46.The Perl Programming Language. http://www.perl.org.
- 47.The R Project for Statistical Computing. http://www.r-project.org.
- 48.Perola E, Charifson PS. J. Med. Chem. 2004;47:2499–2510. doi: 10.1021/jm030563w. [DOI] [PubMed] [Google Scholar]
- 49.Toll L, Berzetei-Gurske IP, Polgar WE, Brandt SR, Adapa ID, Rodriguez L, Schwartz RW, Haggart D, O'Brien A, White A, Kennedy JM, Craymer K, Farrington L, Auh JS. NIDA Res. Mono. 1998;178:440–466. [PubMed] [Google Scholar]
- 50.Malatynska E, Wang Y, Knapp RJ, Waite S, Calderon S, Rice K, Hruby VJ, Yamamura HI, Roeske WR. J. Pharmacol. Exp. Ther. 1996;278:1083–1089. [PubMed] [Google Scholar]
- 51.Roth BL, Baner K, Westkaemper R, Siebert D, Rice KC, Steinberg S, Ernsberger P, Rothman RB. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11934–11939. doi: 10.1073/pnas.182234399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harding WW, Tidgewell K, Byrd N, Cobb H, Dersch CM, Butelman ER, Rothman RB, Prisinzano TE. J. Med. Chem. 2005;48:4765–4771. doi: 10.1021/jm048963m. [DOI] [PubMed] [Google Scholar]
- 53.Tidgewell K, Groer CE, Harding WW, Lozama A, Schmidt M, Marquam A, Hiemstra J, Partilla JS, Dersch CM, Rothman RB, Bohn LM, Prisinzano TE. J. Med. Chem. 2008;51:2421–2431. doi: 10.1021/jm701162g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beckett AH, Casy AF. J. Pharm. Pharmacol. 1954;6:986–1001. doi: 10.1111/j.2042-7158.1954.tb11033.x. [DOI] [PubMed] [Google Scholar]
- 55.Lewis JW, Bentley KW, Cowan A. Annu. Rev. Pharmacol. 1971;11:241–270. doi: 10.1146/annurev.pa.11.040171.001325. [DOI] [PubMed] [Google Scholar]
- 56.Brandt W, Barth A, Höltje HD. Drug Des. Discov. 1993;10:257–283. [PubMed] [Google Scholar]
- 57.Feinberg AP, Creese I, Snyder SH. Proc. Natl. Acad. Sci. U. S. A. 1976;73:4215–4219. doi: 10.1073/pnas.73.11.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gorin FA, Marshall GR. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5179–5183. doi: 10.1073/pnas.74.11.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang P, Kehner GB, Cowan A, Liu-Chen LY. J. Pharmacol. Exp. Ther. 2001;297:688–695. [PubMed] [Google Scholar]
- 60.Chen C, Yin J, de Riel JK, DesJarlais RL, Raveglia LF, Zhu J, Liu-Chen LY. J. Biol. Chem. 1996;271:21422–21429. doi: 10.1074/jbc.271.35.21422. [DOI] [PubMed] [Google Scholar]
- 61.Pogozheva ID, Przydzial MJ, Mosberg HI. AAPS J. 2005;7:434–448. doi: 10.1208/aapsj070243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loew GH, Toll L, Uyeno E, Cheng A, Judd A, Lawson J, Keys C, Amsterdam P, Polgar W. NIDA Res. Mono. 1986;69:231–265. [PubMed] [Google Scholar]
- 63.Kotick MP, Leland DL, Polazzi JO, Howes JF, Bousquet A. J. Med. Chem. 1983;26:1050–1056. doi: 10.1021/jm00361a019. [DOI] [PubMed] [Google Scholar]
- 64.Leland DL, Kotick MP. J. Org. Chem. 1983;48:1813–1819. [Google Scholar]
- 65.Lattanzi R, Spetea M, Schüllner F, Rief SB, Krassnig R, Negri L, Schmidhammer H. J. Med. Chem. 2005;48:3372–3378. doi: 10.1021/jm040894o. [DOI] [PubMed] [Google Scholar]
- 66.Schütz J, Spetea M, Koch M, Aceto MD, Harris LS, Coop A, Schmidhammer H. J. Med. Chem. 2003;46:4182–4187. doi: 10.1021/jm030878b. [DOI] [PubMed] [Google Scholar]
- 67.Rothman RB, Murphy DL, Xu H, Godin JA, Dersch CM, Partilla JS, Tidgewell K, Schmidt M, Prisinzano TE. J. Pharmacol. Exp. Ther. 2007;320:801–810. doi: 10.1124/jpet.106.113167. [DOI] [PubMed] [Google Scholar]
- 68.Kane BE, McCurdy CR, Ferguson DM. J. Med. Chem. 2008;51:1824–1830. doi: 10.1021/jm701040v. [DOI] [PubMed] [Google Scholar]
- 69.Zheng H, Loh HH, Law PY. IUBMB life. 2010;62:112–119. doi: 10.1002/iub.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Angel TE, Chance MR, Palczewski K. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8555–8560. doi: 10.1073/pnas.0903545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Orban T, Gupta S, Palczewski K, Chance MR. Biochemistry. 2010;49:827–834. doi: 10.1021/bi901889t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Horstman DA, Brandon S, Wilson AL, Guyer CA, Cragoe EJ, Limbird LE. J. Biol. Chem. 1990;265:21590–21595. [PubMed] [Google Scholar]
- 73.Selley DE, Cao CC, Liu Q, Childers SR. Br. J. Pharmacol. 2000;130:987–996. doi: 10.1038/sj.bjp.0703382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu H, Wang X, Partilla JS, Bishop-Mathis K, Benaderet TS, Dersch CM, Simpson DS, Prisinzano TE, Rothman RB. Brain Res. Bull. 2008;77:49–54. doi: 10.1016/j.brainresbull.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berger JG, Davidson F, Langford GE. J. Med. Chem. 1977;20:600–602. doi: 10.1021/jm00214a035. [DOI] [PubMed] [Google Scholar]
- 76.Jimeno ML, Alkorta I, Cano C, Jagerovic N, Goya P, Elguero J, Foces-Foces C. Chem. Pharm. Bull. 2003;51:929–934. doi: 10.1248/cpb.51.929. [DOI] [PubMed] [Google Scholar]
- 77.Filizola M, Laakkonen L, Loew GH. Protein Eng. 1999;12:927–942. doi: 10.1093/protein/12.11.927. [DOI] [PubMed] [Google Scholar]
- 78.Liu X, Kai M, Jin L, Wang R. Bioorg. Med. Chem. Lett. 2009;19:5387–5391. doi: 10.1016/j.bmcl.2009.07.121. [DOI] [PubMed] [Google Scholar]
- 79.Sagara T, Egashira H, Okamura M, Fujii I, Shimohigashi Y, Kanematsu K. Bioorg. Med. Chem. 1996;4:2151–2166. doi: 10.1016/s0968-0896(96)00219-2. [DOI] [PubMed] [Google Scholar]
- 80.Strahs D, Weinstein H. Protein Eng. 1997;10:1019–1038. doi: 10.1093/protein/10.9.1019. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Y, Sham YY, Rajamani R, Gao J, Portoghese PS. ChemBioChem. 2005;6:853–859. doi: 10.1002/cbic.200400207. [DOI] [PubMed] [Google Scholar]
- 82.McLamore S, Ullrich T, Rothman RB, Xu H, Dersch C, Coop A, Davis P, Porreca F, Jacobson AE, Rice KC. J. Med. Chem. 2001;44:1471–1474. doi: 10.1021/jm000511w. [DOI] [PubMed] [Google Scholar]
- 83.Drug Evaluation Committee (DEC) http://www3.pharmacy.umaryland.edu/faculty/acoop/decfolder/DEC.htm.
- 84.Eddy NB. J. Chronic. Dis. 1956;4:59–71. doi: 10.1016/0021-9681(56)90007-8. [DOI] [PubMed] [Google Scholar]
- 85.Small L, Eddy N, Ager J, May E. J. Org. Chem. 1958;23:1387–1388. [Google Scholar]
- 86.DeGraw JI, Lawson JA, Crase JL, Johnson HL, Ellis M, Uyeno ET, Loew GH, Berkowitz DS. J. Med. Chem. 1978;21:415–422. doi: 10.1021/jm00203a002. [DOI] [PubMed] [Google Scholar]
- 87.Rothman L, Becker E. J. Org. Chem. 1959;24:294–294. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.