Polyalanine expansions can result in aggregation and cause cytotoxicity. We have created the first yeast model of polyalanine-expansion aggregation and toxicity using the poly(Ade)-binding protein Pab1.
Abstract
Nine human disorders result from the toxic accumulation and aggregation of proteins with expansions in their endogenous polyalanine (polyA) tracts. Given the prevalence of polyA tracts in eukaryotic proteomes, we wanted to understand the generality of polyA-expansion cytotoxicity by using yeast as a model organism. In our initial case, we expanded the polyA tract within the native yeast poly(Adenine)-binding protein Pab1 from 8A to 13A, 15A, 17A, and 20A. These expansions resulted in increasing formation of Pab1 inclusions, insolubility, and cytotoxicity that correlated with the length of the polyA expansion. Pab1 binds mRNA as part of its normal function, and disrupting RNA binding or altering cytoplasmic mRNA levels suppressed the cytotoxicity of 17A-expanded Pab1, indicating a requisite role for mRNA in Pab1 polyA-expansion toxicity. Surprisingly, neither manipulation suppressed the cytotoxicity of 20A-expanded Pab1. Thus longer expansions may have a different mechanism for toxicity. We think that this difference underscores the potential need to examine the cytotoxic mechanisms of both long and short expansions in models of expansion disorders.
INTRODUCTION
In the past two decades, the expansion of homopolymeric amino acid tracts has emerged as a common etiological factor in 17 human neurodegenerative and developmental disorders (Ross, 2002; Albrecht and Mundlos, 2005). The most widely known of these are nine age-dependent, neurodegenerative disorders associated with the expansion of polyglutamine (polyQ) tracts. Huntington's disease is the most prominent example of the polyQ-expansion class, which also includes spinocerebellar ataxias 1, 2, 3, 6, 7, and 17; spinobulbar muscular atrophy; and dentatorubral-pallidoluysian atrophy (Ross, 2002). In addition to the polyQ-expansion disorders, nine developmental disorders are associated with expansions of polyalanine (polyA) tracts. The polyA-expansions disorders include oculopharyngeal muscular dystrophy (OPMD), syndactyly type II, cleidocranial dysplasia, holoproseccephaly, hand-foot-genital syndrome, blepharophimosis ptosis and epicanthus inversus, X-linked mental retardation, X-linked infantile spasm syndrome, and congenital central hypoventilation syndrome (Albrecht and Mundlos, 2005). Because the human proteome contains nearly 400 polyQ tract-containing proteins and >600 polyA tract-containing proteins (Faux et al., 2005), advancing our understanding of polyQ and polyA tracts’ functional roles in proteins and how certain expansions lead to cellular dysfunction should provide insight into key aspects of human physiology and disease.
Despite the fact that polyQ and polyA expansions are commonly shared among a number of disorders, the molecular mechanisms by which the expansions cause disease are still not clear. Of the two different classes of expansions, the polyQ class has been the more extensively studied, and some important clues about its molecular pathology have been gleaned from studies in patients as well as in yeast, fly, worm, mouse, and cell culture models (Jana and Nukina, 2003; Riley and Orr, 2006; van Ham et al., 2009). Although the length of the endogenous polyQ tract and the pathogenic expansion is specific to each particular polyQ protein, longer tract expansions are generally correlated with greater aggregation and intracellular inclusion formation, increased cellular toxicity, and an earlier age of disease onset (Zoghbi and Orr, 2000). In all cases, the polyQ expansions result in a dominant gain of function associated with aggregation and inclusion formation (Zoghbi and Orr, 2000). PolyQ-expansion proteins typically form inclusions within the nucleus of cells (Zoghbi and Orr, 2000), and there are indications that this inclusion may lead to defects in transcription (Riley and Orr, 2006). The inclusions are also typically associated with proteasome subunits, ubiquitin, and chaperones (Jana and Nukina, 2003), suggesting that they might interfere with the cell's normal protein quality control machineries and alter the burden of other misfolded proteins in the cell. The nature of cytotoxicity due to polyQ-expansion remains unknown, however.
In contrast to the polyQ-expansion diseases, the polyA-expansion class is less well studied. Similar to polyQ expansions, the length of the polyA expansion is correlated with the severity of the developmental malformations (Amiel et al., 2004). In most cases, expansion of the polyA tract causes the affected proteins to accumulate in intracellular inclusions due to an increased propensity for aggregation (Fan et al., 2001; Albrecht et al., 2004; Caburet et al., 2004). Homopolymeric alanine peptides of at least 11 residues exhibit toxicity (Giri et al., 2003), and <19 alanine residues are required for aggregation of green fluorescent protein (GFP) in mammalian cultured cells (Rankin et al., 2000). The inclusions are typically associated with proteasome subunits, ubiquitin, and chaperones (Calado et al., 2000; Abu-Baker et al., 2003; Berciano et al., 2004). Unlike the polyQ-expansion disorders, most polyA-expansion disorders can also be caused by point or frameshift mutations, duplications, deletions, or expansions (Amiel et al., 2004), suggesting that loss of function may be the cause of disease. Evidence has been growing, however, that shows that the polyA-expanded versions in patients confer dominant negative or gain-of-function effects (Amiel et al., 2004; Albrecht and Mundlos, 2005). In the case of OPMD, only polyA expansions in the poly(Ade)-binding protein PABPN1 have been identified in patients, and the disease is due to a dominant gain of function associated with its aggregation potential (Albrecht and Mundlos, 2005). A current model for PABPN1 polyA-expansion toxicity suggests that the expanded PABPN1 aggregates and sequesters RNA thus leading to transcriptional dysregulation (Kim et al., 2001; Brais, 2003). As in polyQ-expansions, however, the mechanism of cellular toxicity for polyA-expansions is not known.
Models of protein aggregation disorders in the budding yeast Saccharomyces cerevisiae have provided new insights into Parkinson's disease (Cooper et al., 2006; Gitler et al., 2009), amyotrophic lateral sclerosis (Johnson et al., 2008, 2009; Elden et al., 2010; Takahashi et al., 2010), and Huntington's disease (Meriin et al., 2002; Willingham et al., 2003; Giorgini et al., 2005; Duennwald et al., 2006a, 2006b; Duennwald and Lindquist, 2008). Studies using a yeast Parkinson's disease model found that α-synuclein (α-syn) disrupted endoplasmic reticulum-Golgi trafficking (Cooper et al., 2006), and discovered genetic modifiers that influence neuron loss in animals (Gitler et al., 2009). Yeast studies also led to the identification of a genetic link for amyotrophic lateral sclerosis susceptibility (Elden et al., 2010). The yeast Huntington's model has proven particularly useful. It led to the identification of flanking protein sequences that modulate huntingtin (Htt) polyQ-expansion toxicity (Duennwald et al., 2006a, 2006b), the discovery of proteins that enhance or suppress Htt polyQ-expansion toxicity (Willingham et al., 2003; Giorgini et al., 2005), and the discovery of a defect in endoplasmic reticulum stress systems in Huntington's disease (Duennwald and Lindquist, 2008); in addition, it helped decipher the roles of chaperones in Htt polyQ-expansion aggregate formation (Carmichael et al., 2000; Muchowski et al., 2000; Gokhale et al., 2005; Tam et al., 2006). Thus far, no model for polyA-expansion aggregation and cellular dysfunction has been constructed in yeast. Because the Htt polyQ-expansion model has been fruitful, we thought that development of a yeast polyA-expansion model could provide important new information about this class of homopolymeric tract expansion. Here, we describe our studies of polyA expansion in the yeast poly(Adenine)-binding protein Pab1.
RESULTS
Many yeast proteins possess native polyA tracts
Our first strategy in developing a yeast model of polyA expansion aggregation was to identify native yeast proteins that contain polyA tracts. To do this, we used the PatMatch search tool of the Saccharomyces Genome Database (http://www.yeastgenome.org/cgi-bin/PATMATCH/nph-patmatch) and identified 17 native yeast proteins with polyA tracts ≥6 alanines (Table 1). Interestingly, the longest polyA tract length was nine alanines, fewer than are typically associated with human polyA-expansion disorders. It is reasonable to suspect that if longer polyA tract lengths cause some form of cellular dysfunction, the existing tract lengths in the contemporary yeast proteome will have been kept below a toxic threshold length by natural selection. Similar to the collection of human polyA proteins that are associated with disease (Albrecht and Mundlos, 2005), most of the native yeast polyA proteins are nuclear and involved in transcription (Table 1).
TABLE 1:
PolyA proteins in S. cerevisiae.
| Protein | PolyA | Function |
|---|---|---|
| Ccp1 | 9A | Mitochondrial cytochrome c peroxidase |
| Def1* | 9A | RNA polymerase II degradation factor |
| Epl1* | 8A | Subunit of NuA4 histone acetyltransferase |
| Gdt1 | 6A | Unknown |
| Hem1 | 9A | 5-aminolevulinate synthase |
| Ino80* | 6A | ATPase with 3′ to 5′ DNA helicase activity |
| Ira2 | 9A | GTPase-activating protein |
| Ixr1* | 6A | Binds DNA with intrastrand cross-links |
| Mot3* | 6A | Transcription factor |
| Pab1 | 8A | Poly(Ade)-binding protein |
| Pdc2* | 6A | Transcription factor |
| Rap1* | 9A | DNA-binding protein |
| Reb1* | 7A, 8A | RNA polymerase I enhancer binding protein |
| Spt20* | 8A | Subunit of SAGA histone acetyltransferase |
| Ssn3* | 9A | Kinase component of RNA polymerase II |
| Sum1* | 6A | Transcriptional repressor |
| Yhr020w | 8A | Unknown |
| *nuclear |
Yeast Pab1 as a model for polyA length-dependent aggregation and toxicity
To understand the mechanisms of aggregation and cellular toxicity caused by expansion of polyA tracts in yeast, we chose Pab1 as an initial example to expand its native polyA tract. Pab1 is a nuclear-cytoplasmic poly(Ade)-binding protein that is a component of the 3′-end mRNA-processing complex (Amrani et al., 1997; Brune et al., 2005). Pab1 mediates interactions between the 5′ cap structure and the 3′ poly(Ade) tail of mRNA (Tarun and Sachs, 1996). We focused on Pab1 because it is similar to the human nuclear poly(Ade)-binding protein PABPN1, which possesses a polyA tract that when expanded results in OPMD (Brais et al., 1998), and is a homologue of PABPC1, which also possesses a polyA tract (Figure 1A). Expansion of PABPN1's polyA tract causes the protein to form aggregates in muscle cells and confers dominant-negative toxicity in the affected cells (Calado et al., 2000). Therefore we tested whether expansion of the yeast Pab1 polyA tract would cause aggregation and cellular dysfunction.
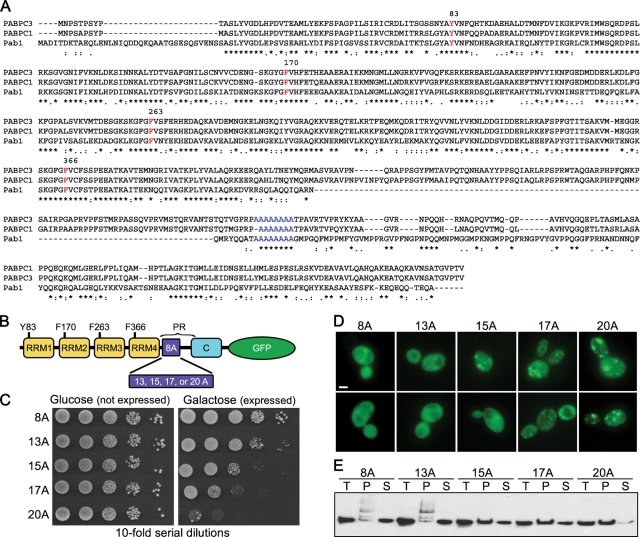
FIGURE 1:
Pab1 polyA-expansions are toxic and insoluble and aggregate in a polyA tract length–dependent manner. (A) Amino acid alignment of human PABPC1, PABPC3, and yeast Pab1. Residues required for RNA binding are indicated in red and the polyA tract in blue. (B) Schematic of Pab1 domain structure. C, C-terminal domain; PR, proline rich. (C) Tenfold serial dilutions of cells were spotted onto medium containing glucose (Pab1 expression repressed) or galactose (Pab1 expression induced) to measure spotting efficiency and toxicity of Pab1 polyA-expansions. (D) Pab1 expression (with indicated polyA tract length) was induced in wild-type cells for 16 h and imaged with epifluorescence microscopy for Pab1-GFP localization. Bar = 2 μm. (E) Solubility of Pab1 at 13,000 rcf. T, total; S, supernatant; P, pellet.
We created a series of increasing-length polyA tract expansions in Pab1, expanding the polyA tract from the endogenous 8 alanines to 13, 15, 17, and 20 alanines. To visualize the dynamics of the Pab1 polyA-expansion proteins in the cell, we fused GFP to the C-terminal domain of each protein. We placed the GFP-tagged versions under the control of the galactose-inducible GAL1 promoter to avoid constitutive expression of polyA-expansion proteins that might be toxic. All constructs were expressed concurrently with wild-type Pab1 expressed from its genomic promoter to assess if the polyA-expansion proteins had dominant effects. When the polyA tract of Pab1 was expanded from 8 to 13, 15, 17, and 20 alanines (Figure 1B), the polyA-expanded Pab1 became toxic to the cells with the severity of toxicity positively correlated with the increasing length of the polyA tract expansion (Figure 1C). Furthermore, the formation of visible inclusions within the cell became more apparent with the increasing length of the polyA tract (Figure 1D). Visible inclusion formation was also correlated with increasing insolubility for the polyA-expanded Pab1. The amount of Pab1 increased in the pellet fraction and decreased in the soluble fraction as the polyA tract was lengthened (Figure 1E). Thus as the polyA tract expansion became longer, cellular inclusion formation, insolubility, and toxicity of the expanded protein increased.
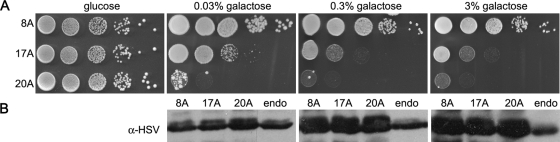
Most models of aggregation diseases often use high overexpression of the aggregating protein to induce a pathological phenotype. We were curious to see what level of expression over endogenous Pab1 was required to observe polyA-expanded toxicity. Therefore we variably induced the expression of Pab18A, Pab117A, and Pab120A with increasing concentrations of galactose (0.03%, 0.3%, or 3%) and compared the expression to endogenous Pab1 tagged with the identical 3xHSV-GFP at its C terminus. Even at the lowest induction conditions where polyA-expanded Pab1 levels were approximately equivalent to endogenous Pab1 levels, polyA-expanded Pab1 still exhibited tract length–dependent toxicity (Figure 2, A and B).
FIGURE 2:
Expression levels of endogenous and PolyA-expanded Pab1. (A) Tenfold serial dilutions of cells were spotted onto medium containing glucose (polyA-expanded Pab1 expression repressed) or varying concentrations of galactose (polyA-expanded Pab1 expression induced) to measure spotting efficiency and toxicity of Pab1 polyA-expansions at increasing Pab1 expression levels. (B) Immunoblot with anti-HSV to detect Pab1 tagged at its native locus (endo) and Pab1 polyA-expansion variants grown in increasing galactose concentrations. Equal OD equivalents were loaded on the polyacrylamide gel.
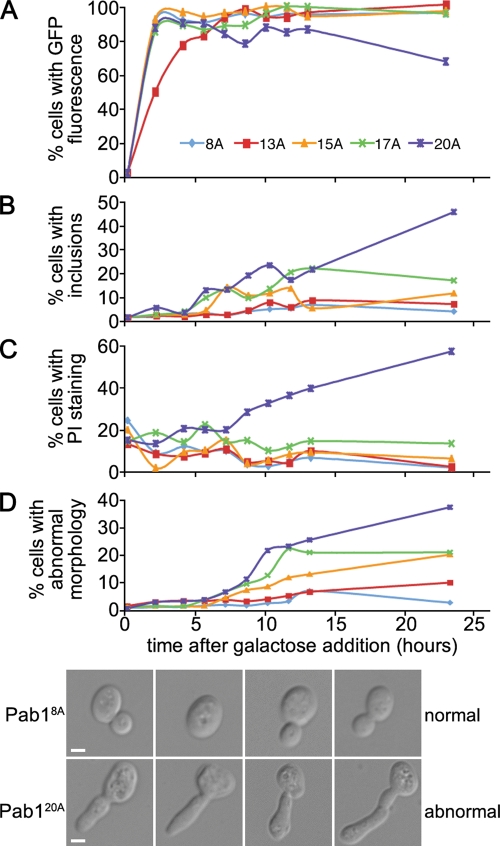
We next examined the dynamics of inclusion formation and toxicity in log-phase cultures. Within 2 h after addition of galactose to induce expression, each Pab1-GFP was maximally expressed in yeast cells (Figure 3A). Eight hours after galactose addition we observed increased formation of inclusions in cells expressing Pab117A-GFP and Pab120A-GFP (Figure 3B). In the case of Pab120A-GFP, this increase was accompanied by an increase in propidium iodide–positive cells, a marker for cell death (Figure 3C). Propidium iodide staining, however, was not always accompanied by visible inclusion formation in cells, which might indicate that the toxic moiety is not the large visible inclusions. In addition to toxicity and inclusion formation, we also observed an increased occurrence of cell morphology defects that correlated with polyA tract length (Figure 3D). The abnormal cell morphologies were marked by short and long protrusions from the cell body (Figure 3D), which are reminiscent of mutants that are missing components of the cytokinetic apparatus (Flescher et al., 1993; Fares et al., 1996).
FIGURE 3:
Time course of polyA-expanded Pab1 expression, toxicity, and aggregation. (A–D) Time course of Pab1–GFP expression (A), aggregate formation (B), cell death (C), and morphology defects (D). Cell death was measured by the accumulation of propidium iodide (PI) in cells. Reported as a percentage of total cells (A, C, and D) or as a percentage of GFP-positive cells (B). (D) Brightfield images of cells expressing Pab18A-GFP or Pab120A-GFP, as indicated. Bar = 2 μm.
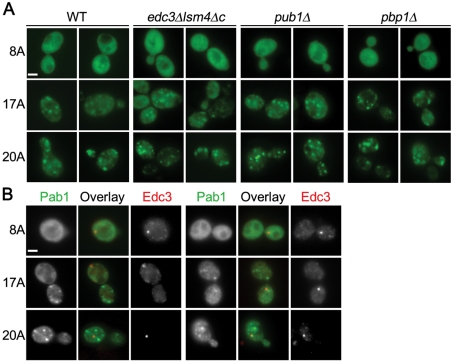
We wanted to determine whether the inclusions associated with the longer polyA tract expansions were new entities for Pab1 or whether they were previously defined structures in which Pab1 is known to be a component, such as P-bodies or stress granules (Buchan et al., 2008). P-bodies are dynamic inclusions that contain translationally silenced mRNAs and are thought to be involved in mRNA storage, translation repression, mRNA decapping, and nonsense-mediated decay (Parker and Sheth, 2007). Stress granules are also dynamic structures that contain untranslated mRNAs and form in response to translation initiation defects (Bond, 2006). To test whether Pab1 polyA-expansion inclusions are distinct from these defined cytoplasmic structures, Pab117A and Pab120A were expressed in yeast cells containing particular gene deletions (edc3Δlsm4ΔC, pub1Δ, or pbp1Δ) that render them defective in P-body or stress granule formation (Buchan et al., 2008). Pab117A-GFP and Pab120A-GFP formed inclusions in edc3Δ lsm4ΔC, pub1Δ, and pbp1Δ cells (Figure 4A), indicating that polyA-expansion–dependent inclusion formation still occurred in the absence of P-body or stress granule formation. Furthermore, the Pab117A- or Pab120A-positive inclusions did not colocalize with P-bodies marked by Edc3-GFP (Figure 4B). Thus Pab1 polyA-expansion inclusions are previously unknown cytoplasmic structures induced by the properties of the expanded polyA tract.
FIGURE 4:
Pab1 polyA-expansion inclusions are not P-bodies or stress granules. (A) Pab18A-GFP, Pab117A-GFP, or Pab120A-GFP expression was induced for 16 h in the indicated strains defective in P-body (edc3Δlsm4Δc) or stress granule formation (pub1Δ and pbp1Δ) and imaged with epifluorescence microscopy. (B) As in (A), except Pab18A-GFP, Pab117A-GFP, or Pab120A-GFP was coexpressed with Edc3-mCherry in wild-type cells. Bars = 2 μm.
Several polyA expansion-associated diseases result from a dominant gain of function for the polyA-expanded protein (Brais, 2003; Albrecht and Mundlos, 2005). In some cases, polyA-expanded inclusions also contain the native form of the protein in addition to the mutant expanded form (Albrecht et al., 2004; Klein et al., 2008). To determine whether polyA-expanded Pab1 is simply interfering with endogenous Pab1's essential functions by sequestering endogenous Pab1, we coexpressed Pab18A-mCherry with Pab117A-GFP or Pab120A-GFP to determine whether the wild type and expanded versions colocalize. In no instance did we find Pab18A associated with Pab117A or Pab120A inclusions (Figure 5A). Thus it is unlikely that the polyA-expanded forms of Pab1 are interfering with endogenous Pab1 function by sequestering endogenous Pab1.
FIGURE 5:
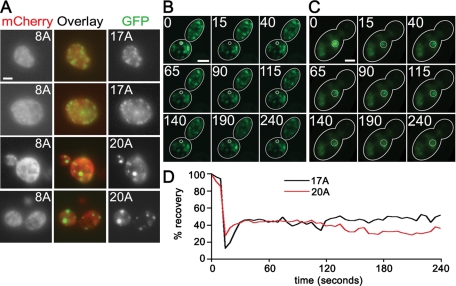
Pab117A and Pab120A inclusions do not contain Pab18A and do not exchange readily with the soluble pool. (A) Pab117A-GFP or Pab120A-GFP was coexpressed with Pab18A-mCherry and imaged with epifluorescence microscopy. (B and C) Images of FRAP time series, Pab117A-GFP (B) and Pab120A-GFP (C). Numbers represent time in seconds after start of imaging and correspond to graph in D. (D) Graph of FRAP experiments shown in B and C of Pab117A-GFP and Pab120A-GFP induced for 16 h. Bars = 2 μm.
Last, we examined the dynamics of polyA-expanded Pab1-GFP inclusions using fluorescence recovery after photobleaching (FRAP) to determine whether the inclusions are static once formed or whether they can exchange freely with Pab1-GFP not incorporated into inclusions. Pab117A-GFP and Pab120A-GFP inclusions were photobleached, and we monitored their fluorescence for up to 5 min. In no case (n = 11) did we observe full recovery of the GFP fluorescence for either Pab117A-GFP or Pab120A-GFP (example in Figure 5, B and C). The average fluorescence recovery after 2 min for Pab117A-GFP and Pab120A-GFP was 57 and 40%, respectively (Figure 5D), which represents the circulating cytoplasmic pool returning to the bleached area. The difference between the average recovery of Pab117A-GFP and Pab120A-GFP may indicate a slight increase in mobility of Pab117A with respect to Pab120A.
PolyA-expanded Pab1 inclusions and toxicity are different from polyQ-expanded inclusions and toxicity
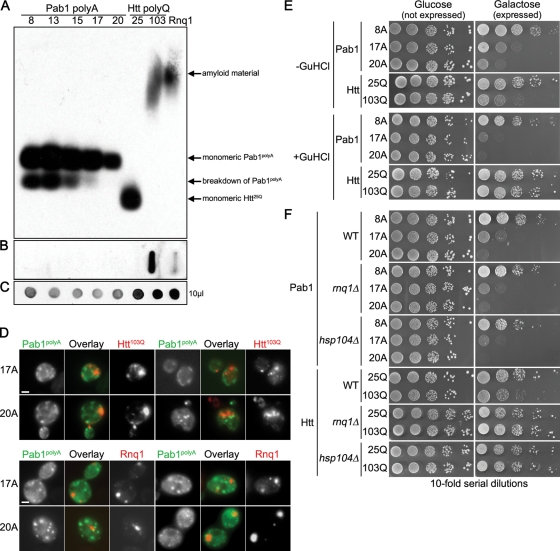
Because polyA-expanded Pab1 is most analogous to polyQ-expanded Htt in that both are homopolymeric amino acid tract expansion proteins that form cellular aggregates and cause toxicity, we examined whether the features of polyA-expanded Pab1 were similar to or different from those of polyQ-expanded Htt. For these studies, we used galactose-inducible Htt25Q, which represents the normal version of Htt, and Htt103Q, which represents the polyQ-expanded form (Krobitsch and Lindquist, 2000). We first tested the Pab1 polyA-expansion proteins for SDS-solubility and colocalization with Htt103Q inclusions. Expansion of Htt's polyQ tract from 25 to 103 glutamines results in the formation of SDS-insoluble, amyloid aggregates that can be visualized by their slower migration during semidenaturing agarose gel electrophoresis (Figure 6A). Yeast prions, like Rnq1, also form SDS-insoluble, amyloid aggregates (Douglas et al., 2008, and Figure 6A). By contrast, we found that all polyA-expanded Pab1 proteins were SDS-soluble (Figure 6A). The Htt103Q protein can also be trapped using a cellulose filter trap assay (Muchowski et al., 2000; Duennwald et al., 2006b). In no case could we similarly trap the polyA-expanded Pab1 proteins (Figure 6B). Furthermore, by fluorescence microscopy we observed that the inclusions formed by Pab117A or Pab120A were distinct and did not colocalize with inclusions formed by Htt103Q or the yeast prion Rnq1 (Figure 6D), which colocalizes with Htt103Q (Duennwald et al., 2006a).
FIGURE 6:
Pab1 polyA-expansion aggregates are distinct from polyQ-expansion and prion aggregates. (A–C) Immunoblotting with anti-GFP to detect Pab1-GFP polyA-expansion variants, Htt-GFP polyQ-expansion variants, or Rnq1-YFP. (A) Semidenaturing detergent agarose gel electrophoresis comparing SDS solubility of Pab1 polyA-expansions with Htt polyQ-expansions and Rnq1. (B) Filter retardation assay showing the ability of Pab1 polyA-expansion variants and Htt25Q but not Htt103Q to pass through 0.2-μm cellulose acetate filter. (C) A total of 10 μl of total protein extract was spotted onto nitrocellulose to determine relative protein concentration. (D) Cells coexpressing Pab117A-GFP or Pab120A-GFP with either Htt103Q-mRFP or Rnq1-mCherry were imaged by epifluorescence microscopy. Bars = 2 μm. (E) Tenfold serial dilutions of cells were spotted onto medium containing glucose (expression repressed) or galactose (expression induced) to measure spotting efficiency and toxicity of polyA-expansion Pab1 or polyQ-expansion Htt after three successive passages on guanidine-HCl (GuHCl) to cure yeast of prions. (F) Tenfold serial dilutions of cells were spotted onto medium containing glucose (expression repressed) or galactose (expression induced) to measure spotting efficiency and toxicity of polyA-expansion Pab1 or polyQ-expansion Htt in hsp104Δ or rnq1Δ cells.
Htt103Q toxicity in yeast depends upon the presence of the yeast prion Rnq1 and the molecular chaperone Hsp104 (Meriin et al., 2002). When yeast cells are cured of the Rnq1 prion by passage on guanidine-HCl, the toxicity of Htt103Q is ameliorated (Meriin et al., 2002, and Figure 6E). This was not the case for the toxicity of Pab117A and Pab120A (Figure 6E). Furthermore, whereas deletion of RNQ1 or HSP104 suppresses the toxicity of Htt103Q, these deletions had no effect on the toxicity of Pab117A and Pab120A (Figure 6F). Altogether, the evidence indicates that the aggregation and toxicity of Pab1 polyA expansions are distinct from polyQ expansions that have been studied in yeast.
PolyA-expanded Pab1 inclusions are distinct from TDP-43 and α-syn inclusions in yeast
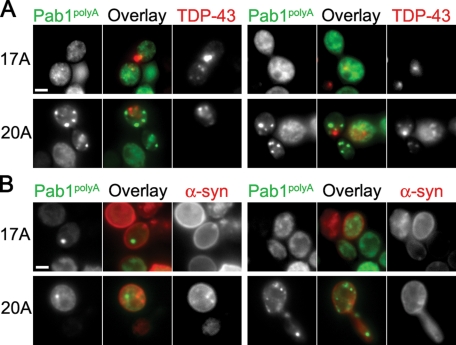
We also wanted to determine whether polyA-expanded Pab1 inclusions were different from inclusions of TAR DNA-binding protein 43 (TDP-43), which has been implicated in amyotrophic lateral sclerosis (Chen-Plotkin et al., 2010), and α-syn, which forms inclusions in Parkinson's disease (Lucking and Brice, 2000). In general, Pab117A-GFP and Pab120A-GFP inclusions and TDP-43–DsRed inclusions were distinct with little colocalization (Figure 7A), although we did observe limited colocalization with Pab1 polyA-expansion inclusions and TDP-43 at a low frequency. When we did observe colocalization, Pab120A-GFP and TDP-43–DsRed inclusions overlapped more than Pab117A-GFP and TDP-43–DsRed inclusions, which could be due to differences in the kinetics of inclusion formation. TDP-43 forms inclusions as early as 3 h after induction (Johnson et al., 2008), and Pab120A forms visible inclusions faster than Pab117A (Figure 3B). We did not detect any colocalization of Pab117A-GFP or Pab120A-GFP with α-syn–mCherry (Figure 7B), indicating that these inclusions are distinct.
FIGURE 7:
PolyA-expanded Pab1 inclusions are distinct from TDP-43 and α-syn inclusions. Cells coexpressing Pab117A-GFP or Pab120A-GFP with either TDP43-DsRed (A) or α-syn-mCherry (B) were imaged by epifluorescence microscopy. Bars = 2 μm.
The toxicity of Pab17A but not Pab20A depends on Pab1's RNA-binding domains
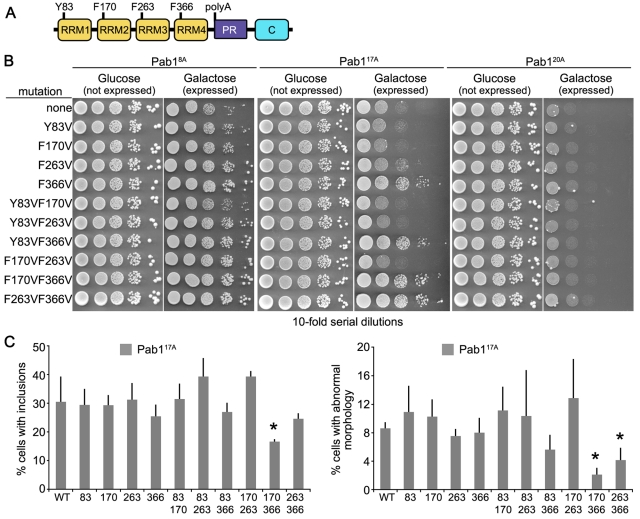
RNA binding has been shown to be a necessary feature of toxicity for several disease-associated mutations in RNA-binding proteins (Fan et al., 2001; Tavanez et al., 2005; Chartier et al., 2006; Johnson et al., 2008; Voigt et al., 2010). Therefore we examined whether this was also the case for polyA-expanded Pab1. Pab1 contains four RNA recognition motifs (RRMs): RRM1, RRM2, RRM3, and RRM4 (Sachs et al., 1986, and Figure 8A). RRM1 and RRM2 have poly(Ade)-binding capability (Burd et al., 1991), and are involved in 3′ mRNA poly(Ade)-tail binding, with RRM2 contributing the majority of affinity (Deardorff and Sachs, 1997). RRM3 and RRM4 possess nonspecific RNA-binding capability (Burd et al., 1991; Deardorff and Sachs, 1997), with RRM4 contributing most of the RNA-binding capability (Deardorff and Sachs, 1997). The RRM domain typically comprises two conserved motifs termed RNP1 and RNP2 (Maris et al., 2005). Within each motif, a single aromatic residue Phe/Tyr is necessary for RNA base-stacking interactions (Maris et al., 2005). Substitution of this residue with a nonaromatic residue (Leu or Val) reduces the ability of RRM-containing RNA-binding proteins to bind RNA (Deardorff and Sachs, 1997).
FIGURE 8:
Pab117A but not Pab120A requires RNA binding for toxicity and aggregation. (A) Schematic of Pab1 domain structure. C, C-terminal domain; PR, proline rich. (B) Tenfold serial dilutions of cells were spotted onto medium containing glucose (expression repressed) or galactose (expression induced) to measure spotting efficiency and toxicity of Pab117A and Pab120A RNA-binding mutants. (C) Cells expressing RNA-binding mutants of Pab117A were assayed for presence of GFP-positive inclusions and morphology defects after 16-h induction and reported as a percentage of GFP-positive cells. More than 200 cells in three independent cultures were counted for each. Bars are SD. *p < 0.05 by Student's t test when compared with wild-type Pab117A.
To determine whether RNA binding is required for Pab1 polyA-expansion toxicity and aggregation, we substituted the aromatic RNA-binding residues in the RNP1 sequence in each RRM (Y83 for RRM1, F170 for RRM2, F263 for RRM3, or F366 for RRM4) with Val individually or in combinations (Figure 8A). We examined the effect each substitution had on the toxicity of Pab117A-GFP or Pab120A-GFP (Figure 8B). Mutation of RRM1 (Y83V), RRM2 (F170V), or RRM3 (F263V) had no effect on Pab117A toxicity. Mutation of RRM4 (F366V) showed significant suppression of Pab117A toxicity. Most double mutations to the RRMs had no effect. Double mutations to RRM2 RRM4 (F170V, F366V) and RRM3 RRM4 (F263V, F366V), however, showed increased suppression of Pab117A toxicity over the single mutation to RRM4 (F366V). Interestingly, none of the mutations altered Pab120A toxicity, indicating that there is a difference between the longer 20A expansion and the slightly shorter 17A expansion in terms of the RNA-binding requirement for Pab1 polyA-expansion toxicity (Figure 8B).
We also examined the effects each mutation had on the ability of Pab117A to form inclusions in vivo. In general, all Pab117A constructs carrying the F366 mutation showed reduced inclusion formation, but only the Pab117A construct with simultaneous mutation of F170 and F366 resulted in a statistically significant reduction in inclusion formation (Figure 8C). The F170VF366V double mutant also nearly eliminated the appearance of cells with abnormal morphology (Figure 8C). Interestingly, none of the mutations that suppressed Pab117A inclusion formation altered Pab120A inclusion formation or morphology defects (unpublished data), which is consistent with their inability to suppress Pab120A toxicity (Figure 8B). Altogether, the mutational analysis demonstrated that altering RNA binding changes the toxicity of Pab1 polyA-expansions up to a certain length.
Mutations in the THO/TREX and TREX-2 mRNA export complexes suppress Pab117A toxicity and inclusion formation
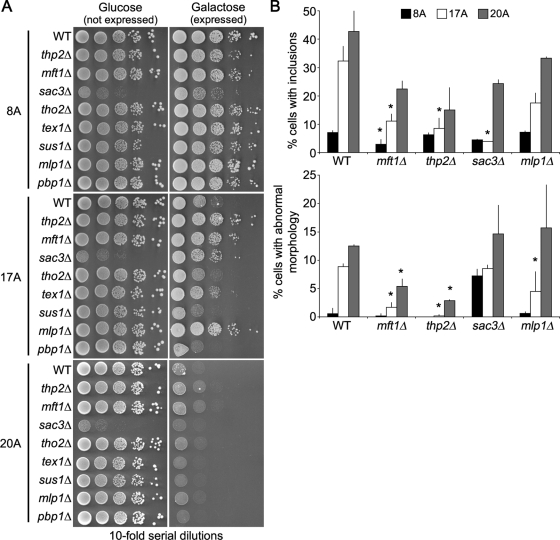
Last, we wanted to take advantage of yeast's powerful genetic methods to identify modifiers of Pab1 polyA-expansion toxicity. We were particularly interested in genetic mutations that could suppress toxicity. Therefore we used a strain in which Pab117A was expressed from a single genomically inserted copy of the PAB117A to screen the yeast MATa deletion collection for individual gene deletions that suppressed the growth defect caused by Pab117A expression. We independently conducted the screen three times and selected deletion strains that reproduced suppression of the Pab117A growth defect each time. From the independent screens, we identified 10 gene deletions that consistently suppressed the Pab117A growth defect (thp2Δ, mft1Δ, ynl140cΔ, sac3Δ, ade4Δ, ade8Δ, sse1Δ, sur4Δ, ptk2Δ, arg80Δ). Each putative suppressor that passed these criteria was recapitulated in an independent yeast strain and retested for suppression of Pab117A toxicity. Of these, only thp2Δ, mft1Δ, and sac3Δ suppressed the Pab117A growth defect (Figure 9A). Interestingly, none of these deletions suppressed the Pab120A growth defect (Figure 9A), once again indicating a profound difference between the toxicity of Pab117A compared with Pab120A.
FIGURE 9:
Pab117A but not Pab120A toxicity is suppressed in mRNA export mutants. (A) Tenfold serial dilutions of cells were spotted onto medium containing glucose (expression repressed) r galactose (expression induced) to measure spotting efficiency toxicity of Pab18A-GFP, Pab117A-GFP, or Pab120A-GFP in wild-type and deletion mutant cells. (B) Wild-type and deletion mutant cells expressing Pab18A-GFP, Pab117A-GFP, or Pab120A-GFP were assayed for presence of GFP-positive inclusions and morphology defects after 16-h induction and reported as a percentage of GFP-positive cells. More than 150 cells in three independent cultures were counted for each. Bars are SD. *p < 0.05 by Student's t test when polyA-expansion Pab1 mutant strains were compared with corresponding wild type.
Thp2 and Mft1 are subunits of the THO complex, which mediates the export of mRNA from the nucleus (Strasser et al., 2002), and the TREX complex, which is recruited to activated genes and couples transcription to mRNA export (Kohler and Hurt, 2007). Other members of the THO complex include Hpr1 and Tho2/Rlr1 (Kohler and Hurt, 2007). We were unable to obtain viable hpr1Δ cells, but we were able to obtain viable tho2Δ cells. We examined Pab117A toxicity in a tho2Δ strain and found that this deletion did not suppress the Pab117A growth defect (Figure 9A). The THO complex associates with Tex1, Sub2, and Yra1 to form the TREX complex (Strasser et al., 2002). We were unable to obtain viable sub2Δ or yra1Δ cells, but we were able to obtain viable tex1Δ cells. The tex1Δ exhibited minimal suppression of the Pab117A growth defect (Figure 9A). Thus the suppression is specific for a subset of THO/TREX member deletions.
Sac3 is a member of the TREX-2 complex that is also involved in mRNA export from the nucleus (Fischer et al., 2002). The TREX-2 complex comprises Sac3, Thp1, Sus1, and Cdc31 (Kohler and Hurt, 2007). Cells with deletions of THP1 and CDC31 are inviable. We were, however, able to obtain viable sus1Δ cells, in which expression of Pab117A was still toxic. In addition, we identified the deletion of MLP1 in some rounds of the deletion screen as capable of suppressing the Pab117A growth defect. Mlp1 is a myosin-like protein associated with the nuclear envelope that interacts with Sac3 and is involved in the nuclear retention of unspliced mRNAs (Fischer et al., 2002; Fasken et al., 2008). We examined newly constructed mlp1Δ cells and found that the Pab117A growth defect was suppressed (Figure 9A). Thus some components of the TREX-2 complex, as well as components of the THO complex, may mediate Pab117A toxicity.
Last, Pab1 interacts with the yeast ataxin-2 homologue Pbp1 (Mangus et al., 1998). Deletion of PBP1 is known to suppress the toxicity of TDP-43 in yeast (Elden et al., 2010). Although we did not identify pbp1Δ as a suppressor of the Pab117A growth defect in the deletion screen, we tested whether loss of Pbp1 function could suppress Pab117A toxicity. pbp1Δ cells showed no suppression of the Pab117A growth defect (Figure 9A).
The thp2Δ, mft1Δ, sac3Δ, and mlp1Δ cells showed suppression of the Pab117A growth defect, so we examined Pab117A inclusion formation in these cells. In each case, Pab117A inclusion formation was reduced in thp2Δ, mft1Δ, sac3Δ, and mlp1Δ cells compared with wild-type cells (Figure 9B). The reduction in the toxicity of Pab117A in thp2Δ, mft1Δ, and mlp1Δ cells was also associated with a reduction in the frequency of cell morphology defects (Figure 9B). Because Thp2, Mft1, Sac3, and Mlp1 are commonly involved in the nuclear export of mRNA, these results suggest that regulation of mRNA levels in the cytoplasm can modulate Pab117A toxicity.
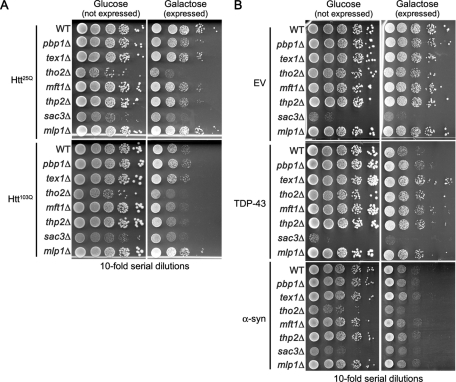
It could be that the mRNA nuclear export mutants are simply diminishing the amount of Pab117A mRNA in the cytoplasm and thus reducing Pab117A protein levels. The initial deletion screens were performed with the Pab117A gene integrated as one copy in the yeast genome. In our subsequent tests, we observed similar suppression of Pab117A when it was expressed from a multicopy plasmid (Figure 9A). Furthermore, none of the deletions suppressed Pab120A toxicity (Figure 9A), Htt103Q toxicity (Figure 10A), or α-syn toxicity (Figure 10B). Suppression of Pab120A, Htt103Q, or α-syn toxicity would have been expected if the effects of the deletions were simply to reduce mRNA levels to a point that disallowed sufficient expression.
FIGURE 10:
Htt103Q and α-syn toxicity is not suppressed in mRNA export mutants, whereas TDP-43 toxicity is suppressed. Tenfold serial dilutions of cells were spotted onto medium containing glucose (expression repressed) or galactose (expression induced) to measure spotting efficiency and toxicity of Htt25Q and Htt103Q (A), TDP-43 or α-syn (B) in wild-type and deletion mutant cells.
Next we wanted to determine whether Pab117A and the RNA-binding protein TDP-43 shared any common suppressors. TDP-43 toxicity in yeast can be suppressed by deletion of PBP1, an orthologue of the human polyQ protein ataxin-2 (Elden et al., 2010, and Figure 10C), whereas Pab117A toxicity was not altered by the pbp1Δ allele (Figure 9A). TDP-43 toxicity was suppressed by the mft1Δ, thp2Δ, and mlp1Δ alleles (Figure 10C), which also suppressed Pab117A toxicity (Figure 9A). TDP-43 was further suppressed, however, by the tex1Δ and tho2Δ alleles (Figure 10C), which was not the case for Pab117A toxicity (Figure 9A). Together, these results suggest that the two RNA-binding proteins share some similarities (as might be expected if RNA binding is requisite for the toxicity of each), but also have some differences in their cellular mechanisms of toxicity.
Mutations in RNA polymerase II suppress Pab117A toxicity and inclusion formation
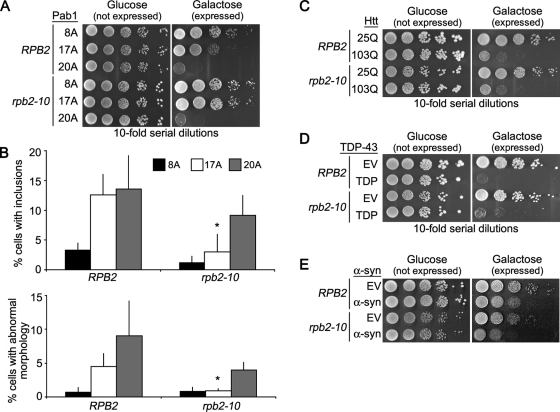
Because mRNA export mutants have lower cytoplasmic mRNA concentrations (Strasser et al., 2002), we hypothesized that the thp2Δ, mft1Δ, sac3Δ, and mlp1Δ alleles might suppress Pab117A toxicity by lowering global mRNA concentration in the cytoplasm. Another way to test this hypothesis is to use a hypomorphic RNA polymerase II mutant strain, rpb2–10, that also has modestly reduced global cellular mRNA levels (Lennon et al., 1998). Similar to the mRNA export mutants, rpb2–10 suppressed the Pab117A growth defect (Figure 11A), and this was accompanied by a lower frequency of cells with Pab117A inclusions and morphology defects (Figure 11B). The growth defects of Pab120A, Htt103Q, or α-syn were not rescued in rpb2–10 cells (Figure 11, A, C, and E), further supporting the idea that suppression of Pab117A was not simply due to reduced expression. TDP-43 toxicity was slightly suppressed in rpb2–10 cells (Figure 11D), again suggesting that TDP-43 and Pab117A toxicity might share a common mechanism, at least in part.
FIGURE 11:
Pab117A but not Pab120A toxicity is suppressed by globally lowering mRNA levels. (A) Tenfold serial dilutions of cells were spotted onto medium containing glucose (expression repressed) or galactose (expression induced) to measure spotting efficiency and toxicity of Pab117A-GFP or Pab120A-GFP in wild-type and RNA PolII mutant (rpb2–10) cells. (B) Wild-type and rpb2–10 cells expressing Pab18A-GFP, Pab117A-GFP, or Pab120A-GFP were assayed for presence of GFP-positive inclusions and morphology defects after 16-h induction and reported as a percentage of GFP-positive cells. More than 150 cells in three independent cultures were counted for each. Bars are SD. *p < 0.05 by Student's t test when Pab1polyA rpb2–10 strains were compared with corresponding RPB2 strain. (C–E) As in (A) except that cells expressing Htt25Q or Htt103Q (C), TDP-43-GFP (D), or α-syn (E) were assayed.
DISCUSSION
PolyA-expansion in a yeast protein causes aggregation and toxicity
Thus far the effects of polyA tract expansions have been primarily examined using human proteins wherein a polyA tract expansion has been identified as a cause for disease. No studies have yet been conducted to determine how general the phenomenon of polyA-expansion aggregation and toxicity might be for other eukaryotic proteins in different species. To pursue the possibility of generality, we analyzed the yeast proteome for native yeast proteins with endogenous polyA tracts. Of the 17 proteins that we identified with polyA tracts ≥6 alanines, all possessed tract lengths that were below the known toxic thresholds in human polyA proteins, supporting the idea that polyA tracts above a certain length in any of these yeast proteins might have deleterious consequences. Indeed, small expansions of the polyA tract in the yeast protein Pab1 resulted in aggregation and toxicity that was correlated with tract length, suggesting that expansion of polyA tracts will be a general phenomenon for eukaryotic proteins. Expansions of polyA tracts in other yeast proteins are underway so that we can begin investigations aimed at understanding the similarities and differences among polyA-expanded proteins from both yeast and humans.
The role of mRNA in polyA-expanded Pab1 toxicity
A number of human RNA-binding proteins that aggregate have been shown to require the intact function of their RNA-binding domains to elicit their toxic effects. These include PABPN1, TDP-43, and FUS/TLS (Tavanez et al., 2005; Voigt et al., 2010; Sun et al., 2011). Therefore we chose to expand the polyA tract in Pab1 as an initial example in large part because of Pab1's RNA-binding properties. Similar to the aforementioned proteins, we found that the mRNA-binding function conferred by Pab1's RRM4 domain was primarily responsible for Pab117A toxicity, with some contribution from the RRM3 domain. Thus Pab117A is an additional member of the class of aggregation-prone proteins that requires RNA binding for toxicity.
There are two potential models of how RNA binding might contribute to toxicity. The first is that Pab1 polyA-expansion aggregates “trap” mRNA molecules leading to translational dysregulation (Calado et al., 2000; Chartier et al., 2006). The second is that the mRNA itself is required to form the toxic Pab1 polyA-expansion species. The results from our genetic suppression analyses do not support an mRNA trapping model for the Pab1 polyA-expansions. If the mRNA trapping model were operative, it would be expected that decreasing cytoplasmic mRNA levels would enhance the toxicity of Pab1 polyA-expansions. Decreasing mRNA nuclear export (in mft1Δ, thp2Δ, and sac3Δ cells; Figure 9A) or globally down-regulating mRNA expression (in rbp2–10 cells; Figure 11A) alleviated Pab117A toxicity, however. Thus we think that the suppression of toxicity by decreased cytoplasmic mRNA levels points to mRNA playing a promotional role in the aggregation and toxicity of Pab117A. Additionally, Pab120A aggregation and toxicity were unaffected after mutating the RRM domains, indicating that this longer form of polyA-expanded Pab1 does not require RNA binding to elicit its toxic effects. If RNA trapping were the mechanism of toxicity, we would expect that RNA binding would still be required.
We propose the following model for mRNA-dependent Pab117A toxicity: Pab1 binds to both polyadenylated mRNA (Deardorff and Sachs, 1997) and other Pab1 monomers (Yao et al., 2007). Because the polyA tract is adjacent to Pab1's oligomerization domain (Yao et al., 2007), an expanded polyA tract is poised to strengthen Pab1's self-association by formation of stabilized β-sheet conformations seen with lengthened polyA tracts (Shinchuk et al., 2005). In the case of polyA tract expansions ≤17, this strengthening is not sufficient to generate a toxic aggregation-prone species; mRNA binding of the oligomeric Pab117A is also required to strengthen the interactions and generate the toxic species. Thus decreasing the mRNA concentration in the cytoplasm would reduce toxic oligomer formation. Because Pab120A toxicity did not require RNA binding, we propose that mRNA facilitation is not critical for the formation of the toxic oligomeric form when the tract is expanded beyond a certain length because the polyA tract length is now sufficiently long to stabilize the toxic oligomeric form on its own.
Our data hint at a structural role for mRNA in Pab117A toxicity, but this model requires further validation. To support this model, we attempted to visualize mRNA in polyA-expanded Pab1 inclusions using in situ hybridization, but we were unsuccessful (unpublished data). We also attempted to disrupt polyA-expanded Pab1 inclusions in situ by permeabilizing cells and treating with RNase, and in vitro by incubating cell lysates with RNase. In neither case were we successful (unpublished data). Our lack of success was not altogether surprising, however, because from our FRAP data it appears that both Pab117A and Pab120A cellular inclusions are relatively stable once formed (Figure 5C); thus any mRNA in the inclusions or aggregates is likely to be inaccessible to in situ probes or RNase molecules. We therefore cannot confirm Pab117A-mRNA interactions in the inclusions at this time. mRNA interactions in inclusions have yet to be confirmed with other RNA-binding proteins that aggregate, and thus remains one of the primary challenges for understanding the role that RNA binding plays in aggregation and toxicity.
Differences between 17A- and 20A-expanded Pab1
Both cell and animal models of polyQ- and polyA-expansion diseases have advanced our understanding of their cellular pathogenesis. A large majority, however, of the polyQ- and polyA-expansion models use long expansions to generate extremely toxic forms of the proteins that produce a robust phenotype in the shortest period of time. Our polyA-expansion model with Pab1 indicates that, by exploring the mechanistic details of only the longest expansions, we may be missing important features of shorter disease-associated expansions. None of the suppressors identified for Pab117A were able to rescue Pab120A toxicity, indicating that these two proteins differ in either the formation of the toxic moiety or the mechanism of their toxicity. Because there may be different mechanisms of toxicity for shorter and longer expansions, we think it is important to validate findings using expansions of varying length. It is intriguing that a difference of only three alanine residues can dramatically alter the determinants of polyA-expansion Pab1 toxicity. Whether this difference comes from the propensity of Pab120A to gain a different toxic form than Pab117A or alter its downstream actions will be an area of future investigation.
MATERIALS AND METHODS
Yeast strains, media, and plasmids
Yeast strains used in this study were BY4741 (met15Δ0, his3Δ1, ura3Δ0, leu2Δ0) (Brachmann et al., 1998), RGY3265 (BY4741 pbp1Δ), RGY3266 (BY4741 pub1Δ), RGY3275 (BY4741 thp2Δ), RGY3276 (BY4741 mft1Δ), RGY3277 (BY4741 sac3Δ), RGY3278 (BY4741 mlp1Δ), RGY3280 (BY4741 tex1Δ), RGY4176 (BY4741 tho2Δ), RGY4203 (BY4741 sus1Δ), yRP2338 (Decker et al., 2007), and DY105 (Lennon et al., 1998). Standard yeast media and yeast genetic methods were used (Guthrie and Fink, 1991). Unless otherwise noted, yeasts were grown in synthetic media containing 2% glucose (repressing), 3% raffinose (nonrepressing), or 0.03–3% galactose (inducing) with the appropriate amino acids.
Standard molecular biology techniques were used to construct all Pab1 polyA-expansions. pRG1361, pRG1363, pRG1364, pRG1365, and pRG1362 are Pab18A, Pab113A, Pab115A, Pab117A, and Pab120A, respectively, tagged C-terminally with GFP (yeast codon adapted S65T) behind the GAL1 promoter in pRS416 (Brachmann et al., 1998). pRG1494, pRG1496, pRG1497, pRG1498, and pRG1495 are Pab18A, Pab113A, Pab115A, Pab117A, and Pab120A, respectively, tagged C-terminally with GFP behind the GAL1 promoter in pRS406 (Brachmann et al., 1998). pRG2881 is α-syn-mCherry behind the GAL1 promoter in pRS426. pAG426Gal-TDP-43-DsRed is TDP-43-DsRed behind the GAL1 promoter in pRS426. Untagged α-syn (Willingham et al., 2003) and TDP-43-GFP (Johnson et al., 2008) were described previously. Edc3-mCherry (pRP1574) was a gift from Roy Parker (Buchan et al., 2008). Htt25Q-GFP, Htt103Q-GFP, Htt25Q-mRFP, and Htt103Q-mRFP were gifts from Michael Sherman (Meriin et al., 2002, 2007). Rnq1-mCherry was a gift from Judith Frydman (Kaganovich et al., 2008). Details of all plasmids and oligos used will be provided upon request.
Spotting assays
Cells were picked from a fresh plate and resuspended in sterile water. Five 10-fold serial dilutions in sterile water were subsequently made from the initial resuspension. Seven microliters of each dilution was plated onto solid medium containing either 2% glucose, 0.5% galactose = 2.5% raffinose, or 3% galactose. Cells were grown at 30ºC for 2–6 d.
Sedimentation assay
Cells were grown at 30ºC in 3% raffinose medium to ∼0.25 × 107 cells/ml. Galactose was added to 3% and the cells were incubated for 16 h at 30ºC. Cells were harvested and lysed in lysis buffer (100 μM Tris, pH 7.5, 200 mM NaCl, 1 mM EDTA, 5% glycerol, 1 mM dithiothreitol, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 8 mM phenylmethylsulfonyl fluoride) by glass bead disruption. The lysate was clarified by centrifugation at 13,000 rpm. The supernatant (S) was removed, and an equal volume of SUMEB (8 M urea, 1% SDS, 10 mM MOPS, pH 6.8, 10 mM EDTA, bromophenol blue) was added. The pellet (P) was resuspended in equal cellular equivalents of SUMEB. All fractions were heated at 65ºC for 10 min, and insoluble debris was removed by high-speed centrifugation (13,000 × g for 5 min). Proteins were resolved on an 8–16% gradient SDS–PAGE gel (Pierce, Rockford, IL), transferred to nitrocellulose, and immunoblotted with anti-GFP antibodies (Sigma, St. Louis, MO).
Semidenaturing detergent agarose gel electrophoresis and filter retardation assay
Cells were grown at 30ºC in 3% raffinose medium to ∼0.25 × 107 cells/ml. Galactose was added to 3%, and the cells were incubated for 16 h at 30ºC. Cells were harvested, washed once with water, and lysed in (500 × g for 2 min). Loading buffer was added to each sample to 1× (0.5× TAE, 5% glycerol, 2% SDS, bromophenol blue). Semidenaturing detergent agarose gel electrophoresis was performed essentially as described (Halfmann and Lindquist, 2008). Briefly, 100 μl of protein extracts was subjected to electrophoresis (1.8% agarose in 1× TAE with 0.1% SDS). Proteins were transferred to nitrocellulose membrane overnight via wicking using Tris-buffered saline as the transfer medium. Filter retardation assays were performed as previously described (Muchowski et al., 2000). Briefly, 100 μl of protein extracts was filtered through cellulose acetate (0.2-μm pore size). Proteins were detected by immunoblotting with anti-GFP (Sigma) antibodies.
Fluorescence microscopy
Cells were grown at 30ºC in 3% raffinose medium to ∼0.25 × 107 cells/ml. Galactose was added to 3%, and the cells were incubated for 16 h at 30ºC. For live cell imaging, cells in growth medium were placed onto a 1% agarose pad, covered with a coverslip, at imaged at room temperature. For fixed cells, 1 ml of cells was harvested, fixed in 4% paraformaldehyde in 0.1 M sucrose for 15 min, washed in wash buffer (1.2 M sorbitol, 0.4 M KPO4), stained with DAPI for 5 min in wash buffer plus 2% Triton X-100, and washed two times in wash buffer. Cells were imaged on either 1) a Nikon Diaphot 200, with a 100× objective (PlanApo N.A. 1.4), filters for green (ex492/18, em535/40) or red (ex572/23, em630/60) fluorescence, and a Princeton Instruments MicroMax cooled CCD camera with MetaMorph acquisition software (Molecular Devices, Sunnyvale, CA) or 2) a Nikon Eclipse 90i with a 100× objective (DIC N2 N.A. 1.4), filters for green (ET470/40x, T495LP, ET525/50m) or red (ET560/40x, T585LP, ET630/75m) fluorescence, and a Photometrics Cool Snap HQ2 cooled CCD camera with NIS-Elements acquisition software. FRAP experiments were performed on a Zeiss 510 Meta confocal microscope equipped with a 100× NA 1.45 Plan-fluor TIRF objective and Zeiss LSM acquisition software. Circular areas were photobleached using 50 iterations of the 488-nm line from a 30-mW argon laser operating at 100% laser power. Fluorescence recovery was monitored at 5-s intervals. All images were processed using Photoshop (Adobe Systems).
Yeast deletion screen
This screen was performed as described previously (Tong et al., 2001, 2004, 2006), with some modifications, using a Singer RoToR HDA (Singer Instruments, Somerset, UK). A galactose-inducible PAB17A expression construct was integrated at the GAL1 locus in the MATα strain Y7092 (gift from C. Boone, University of Toronto) to generate the query strain. This query strain was mated to the yeast haploid deletion collection of nonessential genes (MATa, each gene deleted with KanMX cassette [confers resistance to G418]). Haploid mutants harboring the PAB17A-expression plasmid were grown in the presence of glucose (Pab117A expression “off”) or galactose (Pab117A expression “on”). Following growth at 30ºC for 2 d, plates were photographed and colony sizes measured by ImageJ image analysis software (Collins et al., 2006). The entire screen was repeated three times, and only hits that reproduced all three times were selected for further validation by regenerating the deletions in a fresh PAB117A strain.
Acknowledgments
We thank Roy Parker, Michael Sherman, Charlie Boone, and Judith Frydman for strains and plasmids. R.G.G. thanks Sandy Bajjalieh for insights on expansions, polyalanine and otherwise. This work was supported by a Genetic Approaches to Aging National Institutes of Health (NIH)/National Institute on Aging (NIA) training grant (5T32AG000057 to C.A.K.), a Training in Molecular Neurobiology NIH/National Institute of Neurological Disorders and Stroke (NINDS) training grant (5T32NS007332 to C.A.K.), an NIH Director's New Innovator Award (1DP2OD004417 to A.D.G.), an NIH/NINDS grant (R01NS065317 to A.D.G.), a Pew Scholar in the Biomedical Sciences grant supported by The Pew Charitable Trusts (to A.D.G.), an Ellison Medical Foundation New Scholar Award in Aging (to R.G.G.), a University of Washington Research Royalty Foundation grant (to R.G.G.), and a Marian E. Smith Junior Faculty Award (to R.G.G.).
Abbreviations used:
- α-syn
α-synuclein
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- Htt
huntingtin
- OPMD
oculopharyngeal muscular dystrophy
- polyA
polyalanine
- poly(Ade)
poly(Adenine)
- polyQ
polyglutamine
- RRM
RNA recognition motif
- TDP-43
TAR DNA-binding protein 43
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-01-0037) on April 20, 2011.
REFERENCES
- Abu-Baker A, Messaed C, Laganiere J, Gaspar C, Brais B, Rouleau GA. Involvement of the ubiquitin-proteasome pathway and molecular chaperones in oculopharyngeal muscular dystrophy. Hum Mol Genet. 2003;12:2609–2623. doi: 10.1093/hmg/ddg293. [DOI] [PubMed] [Google Scholar]
- Albrecht A, Mundlos S. The other trinucleotide repeat: polyalanine expansion disorders. Curr Opin Genet Dev. 2005;15:285–293. doi: 10.1016/j.gde.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Albrecht AN, Kornak U, Boddrich A, Suring K, Robinson PN, Stiege AC, Lurz R, Stricker S, Wanker EE, Mundlos S. A molecular pathogenesis for transcription factor associated poly-alanine tract expansions. Hum Mol Genet. 2004;13:2351–2359. doi: 10.1093/hmg/ddh277. [DOI] [PubMed] [Google Scholar]
- Amiel J, Trochet D, Clement-Ziza M, Munnich A, Lyonnet S. Polyalanine expansions in human. Hum Mol Genet. 2004;13 Spec No 2:R235–R243. doi: 10.1093/hmg/ddh251. [DOI] [PubMed] [Google Scholar]
- Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol Cell Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berciano MT, Villagra NT, Ojeda JL, Navascues J, Gomes A, Lafarga M, Carmo-Fonseca M. Oculopharyngeal muscular dystrophy-like nuclear inclusions are present in normal magnocellular neurosecretory neurons of the hypothalamus. Hum Mol Genet. 2004;13:829–838. doi: 10.1093/hmg/ddh101. [DOI] [PubMed] [Google Scholar]
- Bond U. Stressed out! Effects of environmental stress on mRNA metabolism. FEMS Yeast Res. 2006;6:160–170. doi: 10.1111/j.1567-1364.2006.00032.x. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brais B. Oculopharyngeal muscular dystrophy: a late-onset polyalanine disease. Cytogenet Genome Res. 2003;100:252–260. doi: 10.1159/000072861. [DOI] [PubMed] [Google Scholar]
- Brais B, et al. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat Genet. 1998;18:164–167. doi: 10.1038/ng0298-164. [DOI] [PubMed] [Google Scholar]
- Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA. 2005;11:517–531. doi: 10.1261/rna.7291205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Matunis EL, Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol. 1991;11:3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caburet S, Demarez A, Moumne L, Fellous M, De Baere E, Veitia RA. A recurrent polyalanine expansion in the transcription factor FOXL2 induces extensive nuclear and cytoplasmic protein aggregation. J Med Genet. 2004;41:932–936. doi: 10.1136/jmg.2004.024356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A, Tome FM, Brais B, Rouleau GA, Kuhn U, Wahle E, Carmo-Fonseca M. Nuclear inclusions in oculopharyngeal muscular dystrophy consist of poly(A) binding protein 2 aggregates which sequester poly(A) RNA. Hum Mol Genet. 2000;9:2321–2328. doi: 10.1093/oxfordjournals.hmg.a018924. [DOI] [PubMed] [Google Scholar]
- Carmichael J, Chatellier J, Woolfson A, Milstein C, Fersht AR, Rubinsztein DC. Bacterial and yeast chaperones reduce both aggregate formation and cell death in mammalian cell models of Huntington's disease. Proc Natl Acad Sci USA. 2000;97:9701–9705. doi: 10.1073/pnas.170280697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier A, Benoit B, Simonelig M. A Drosophila model of oculopharyngeal muscular dystrophy reveals intrinsic toxicity of PABPN1. EMBO J. 2006;25:2253–2262. doi: 10.1038/sj.emboj.7601117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Lee VM, Trojanowski JQ. TAR DNA-binding protein 43 in neurodegenerative disease. Nat Rev Neurol. 2010;6:211–220. doi: 10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff JA, Sachs AB. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J Mol Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas PM, Treusch S, Ren HY, Halfmann R, Duennwald ML, Lindquist S, Cyr DM. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci USA. 2008;105:7206–7211. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald ML, Jagadish S, Giorgini F, Muchowski PJ, Lindquist S. A network of protein interactions determines polyglutamine toxicity. Proc Natl Acad Sci USA. 2006a;103:11051–11056. doi: 10.1073/pnas.0604548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci USA. 2006b;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald ML, Lindquist S. Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 2008;22:3308–3319. doi: 10.1101/gad.1673408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Dion P, Laganiere J, Brais B, Rouleau GA. Oligomerization of polyalanine expanded PABPN1 facilitates nuclear protein aggregation that is associated with cell death. Hum Mol Genet. 2001;10:2341–2351. doi: 10.1093/hmg/10.21.2341. [DOI] [PubMed] [Google Scholar]
- Fares H, Goetsch L, Pringle JR. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J Cell Biol. 1996;132:399–411. doi: 10.1083/jcb.132.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasken MB, Stewart M, Corbett AH. Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated protein, Mlp1, in mRNA export. J Biol Chem. 2008;283:27130–27143. doi: 10.1074/jbc.M803649200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faux NG, Bottomley SP, Lesk AM, Irving JA, Morrison JR, de la Banda MG, Whisstock JC. Functional insights from the distribution and role of homopeptide repeat-containing proteins. Genome Res. 2005;15:537–551. doi: 10.1101/gr.3096505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Strasser K, Racz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, Lechner J, Hurt E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flescher EG, Madden K, Snyder M. Components required for cytokinesis are important for bud site selection in yeast. J Cell Biol. 1993;122:373–386. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgini F, Guidetti P, Nguyen Q, Bennett SC, Muchowski PJ. A genomic screen in yeast implicates kynurenine 3-monooxygenase as a therapeutic target for Huntington disease. Nat Genet. 2005;37:526–531. doi: 10.1038/ng1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri K, Ghosh U, Bhattacharyya NP, Basak S. Caspase 8 mediated apoptotic cell death induced by beta-sheet forming polyalanine peptides. FEBS Lett. 2003;555:380–384. doi: 10.1016/s0014-5793(03)01294-8. [DOI] [PubMed] [Google Scholar]
- Gitler AD, et al. Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat Genet. 2009;41:308–315. doi: 10.1038/ng.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale KC, Newnam GP, Sherman MY, Chernoff YO. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J Biol Chem. 2005;280:22809–22818. doi: 10.1074/jbc.M500390200. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–863. [PubMed] [Google Scholar]
- Halfmann R, Lindquist S. Screening for amyloid aggregation by semi-denaturing detergent-agarose gel electrophoresis. J Vis Exp. 2008;17(pii):838. doi: 10.3791/838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana NR, Nukina N. Recent advances in understanding the pathogenesis of polyglutamine diseases: involvement of molecular chaperones and ubiquitin-proteasome pathway. J Chem Neuroanat. 2003;26:95–101. doi: 10.1016/s0891-0618(03)00029-2. [DOI] [PubMed] [Google Scholar]
- Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci USA. 2008;105:6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Noguchi S, Hayashi YK, Tsukahara T, Shimizu T, Arahata K. The product of an oculopharyngeal muscular dystrophy gene, poly(A)-binding protein 2, interacts with SKIP and stimulates muscle-specific gene expression. Hum Mol Genet. 2001;10:1129–1139. doi: 10.1093/hmg/10.11.1129. [DOI] [PubMed] [Google Scholar]
- Klein AF, Ebihara M, Alexander C, Dicaire MJ, Sasseville AM, Langelier Y, Rouleau GA, Brais B. PABPN1 polyalanine tract deletion and long expansions modify its aggregation pattern and expression. Exp Cell Res. 2008;314:1652–1666. doi: 10.1016/j.yexcr.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon JC 3rd, Wind M, Saunders L, Hock MB, Reines D. Mutations in RNA polymerase II and elongation factor SII severely reduce mRNA levels in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:5771–5779. doi: 10.1128/mcb.18.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucking CB, Brice A. Alpha-synuclein and Parkinson's disease. Cell Mol Life Sci. 2000;57:1894–1908. doi: 10.1007/PL00000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Amrani N, Jacobson A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol Cell Biol. 1998;18:7383–7396. doi: 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Meriin AB, Zhang X, Alexandrov IM, Salnikova AB, Ter-Avanesian MD, Chernoff YO, Sherman MY. Endocytosis machinery is involved in aggregation of proteins with expanded polyglutamine domains. FASEB J. 2007;21:1915–1925. doi: 10.1096/fj.06-6878com. [DOI] [PubMed] [Google Scholar]
- Meriin AB, Zhang X, He X, Newnam GP, Chernoff YO, Sherman MY. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Rankin J, Wyttenbach A, Rubinsztein DC. Intracellular green fluorescent protein-polyalanine aggregates are associated with cell death. Biochem J. 2000;348 Pt 1:15–19. [PMC free article] [PubMed] [Google Scholar]
- Riley BE, Orr HT. Polyglutamine neurodegenerative diseases and regulation of transcription: assembling the puzzle. Genes Dev. 2006;20:2183–2192. doi: 10.1101/gad.1436506. [DOI] [PubMed] [Google Scholar]
- Ross CA. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Sachs AB, Bond MW, Kornberg RD. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986;45:827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- Shinchuk LM, Sharma D, Blondelle SE, Reixach N, Inouye H, Kirschner DA. Poly-(L-alanine) expansions form core beta-sheets that nucleate amyloid assembly. Proteins. 2005;61:579–589. doi: 10.1002/prot.20536. [DOI] [PubMed] [Google Scholar]
- Strasser K, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Sun Z, Zamia D, Xiaodon F, Hart MP, Chesi A, Shorter J, Gitler AD. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Katada S, Onodera O. Polyglutamine diseases: where does toxicity come from? What is toxicity? Where are we going? J Mol Cell Biol. 2010;2:180–191. doi: 10.1093/jmcb/mjq005. [DOI] [PubMed] [Google Scholar]
- Tam S, Geller R, Spiess C, Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol. 2006;8:1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ, Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Tavanez JP, Calado P, Braga J, Lafarga M, Carmo-Fonseca M. In vivo aggregation properties of the nuclear poly(A)-binding protein PABPN1. RNA. 2005;11:752–762. doi: 10.1261/rna.7217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Tong XS, Xu S, Zheng S, Pivnichny JV, Martin J, Dufresne C. High throughput metabolic stability screen for lead optimization in drug discovery. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;833:165–173. doi: 10.1016/j.jchromb.2006.01.037. [DOI] [PubMed] [Google Scholar]
- van Ham TJ, Breitling R, Swertz MA, Nollen EA. Neurodegenerative diseases: lessons from genome-wide screens in small model organisms. EMBO Mol Med. 2009;1:360–370. doi: 10.1002/emmm.200900051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A, Herholz D, Fiesel FC, Kaur K, Muller D, Karsten P, Weber SS, Kahle PJ, Marquardt T, Schulz JB. TDP-43-mediated neuron loss in vivo requires RNA-binding activity. PLoS One. 2010;5:e12247. doi: 10.1371/journal.pone.0012247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham S, Outeiro TF, DeVit MJ, Lindquist SL, Muchowski PJ. Yeast genes that enhance the toxicity of a mutant huntingtin fragment or alpha-synuclein. Science. 2003;302:1769–1772. doi: 10.1126/science.1090389. [DOI] [PubMed] [Google Scholar]
- Yao G, Chiang YC, Zhang C, Lee DJ, Laue TM, Denis CL. PAB1 self-association precludes its binding to poly(A), thereby accelerating CCR4 deadenylation in vivo. Mol Cell Biol. 2007;27:6243–6253. doi: 10.1128/MCB.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]