Abstract
In this study we describe changes of gene expression that occur in the basolateral complex of the mouse amygdala (BLA) during the formation of fear memory. Through the combination of a behavioral training scheme with polymerase chain reaction-based expression analysis (subtractive hybridization and virtual Northern analysis) we were able to identify various gene products that are increased in expression after Pavlovian fear conditioning and are of potential significance for neural plasticity and information storage in the amygdala. In particular, a key enzyme of monoamine metabolism, aldehyde reductase, and the protein sorting and ubiquitination factor Praja1, showed pronounced and learning-specific induction six hours after fear conditioning training. Aldehyde reductase and Praja1, including a novel alternatively spliced isoform termed Praja1a, were induced in the BLA depending on the emotional stimulus presented and showed different expression levels in response to associative conditioning, training stress, and experience of conditioned fear. Stress and fear were further found to induce various signal transduction factors (transthyretin, phosphodiesterase1, protein kinase inhibitor-α) and structural reorganization factors (e.g., E2-ubiquitin conjugating enzyme, neuroligin1, actin, UDP-galactose transporter) during training. Our results show that the formation of Pavlovian fear memory is associated with changes of gene expression in the BLA, which may contribute to neural plasticity and the processing of information about both conditioned and unconditioned fear stimuli.[The Praja1a sequence has been deposited in GenBank data base under accession no. AF335250.]
The storage of long-term memories (LTMs) involves highly regulated and coordinated cellular and molecular changes in the central nervous system including the de novo synthesis of proteins, which are indispensable for the morphological and biochemical refinements that form a permanent memory trace (Stork and Welzl 1999). Fear memory is a particularly rapid LTM formed during an aversive experience, for example, during Pavlovian fear conditioning; it is thus highly amenable to neurochemical and electrophysiological investigation. Several studies have shown that the acquisition of Pavlovian fear memory depends at least in part on the functional integrity of the amygdala (Davis et al. 1994) and is prevented by disruption of neural plasticity in the amygdala through, for example, injection of NMDA receptor antagonists (Miserendino et al. 1990; Kim et al. 1992). Indeed, subsequent studies have confirmed that fear conditioning is associated with long-term potentiation and an increased synchronization of activity in projection neurons of the BLA (Quirk et al. 1995; Rogan et al. 1997; Paré and Collins 2000). Similar to other forms of LTM (Grecksch and Matthies 1980; Rose 1995; Tiunova et al. 1998), Pavlovian fear memory can be blocked with protein synthesis inhibitors applied shortly after training (Bourtchouladze et al. 1994; Schafe et al. 1999, 2000). If undisturbed, an induction of the transcription factors c-fos, c-jun, and NGFI-A occurs in the BLA during this initial phase of memory formation (e.g., Campeau et al. 1991; Beck and Fibiger 1995; Rosen et al. 1998). Studies in various behavioral paradigms (Grecksch and Matthies 1980; Tiunova et al. 1998) including fear conditioning (Stiedl et al. 1999) have furthermore suggested the existence of a second time window of protein synthesis several hours after training that is critical for memory consolidation. However, little is known about the molecular and cellular processes or the genes that are expressed during this time window.
In this study we sought to identify some of the effector genes that are expressed in the amygdala during this phase in the formation of Pavlovian fear memory. With a polymerase chain reaction (PCR)-based subtractive hybridization approach (Diatchenko et al. 1996) we have isolated several known and novel gene products with increased mRNA expression in the mouse BLA six hours after fear conditioning training. We focused on the analysis of identified gene products and distinguished putative learning-, stress- and fear-related changes of gene expression according to their induction following different training protocols.
RESULTS
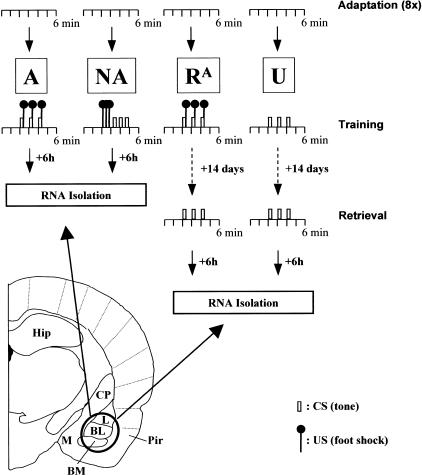
Genes that are differentially expressed in the BLA during the formation of fear memory were identified and characterized in two steps. First, we attempted to isolate a maximum number of fear conditioning-induced gene products by subtractive hybridization. Then, learning-specific footshock stress- and fear-related changes in expression were distinguished by virtual Northern analysis. To this end, RNA was isolated from the BLA of animals trained according to one of four different training protocols: A, the associative learning group; NA, the nonassociative learning group; RA, the retrieval group; and U, the unshocked control group.
Fear Conditioning Training
In a previous study, we have shown that the associative training protocol A (see Materials and Methods) induces robust Pavlovian fear memory in mice, whereas the nonassociative training protocol NA produces only weak and unspecific fear responses to the auditory cue (Stork et al. 2000). To confirm the responsiveness and successful conditioning of the animals in the present study, their behavior was observed by the experimenter during training and where possible also during the retrieval session. All animals in the learning group A, the stress control group NA, and the fear control group RA showed aversive behavioral responses including flight attempts, jumps, and vocalization when confronted with footshocks during training. They furthermore displayed an immediate fear response and pronounced freezing behavior after the footshock exposure. Animals of group RA also showed freezing behavior when re-exposed to the CS, confirming the formation of Pavlovian fear memory. Similar anxiety- and fear-related behaviors were not observed with animals of the unshocked control group U.
Subtractive Hybridization and Screening
As a first step in the isolation of fear conditioning-induced gene products, we enriched differentially expressed gene products in the associative learning group A through subtractive hybridization with the unshocked control group U. To control for subtraction efficiency, the abundance of the housekeeping gene G3PDH was estimated in both the unsubtracted sample A and the subtraction product. In both cases, PCR amplification produced the expected 450-bp cDNA fragment, however, five additonal cycles were needed for G3PDH detection after subtractive hybridization.
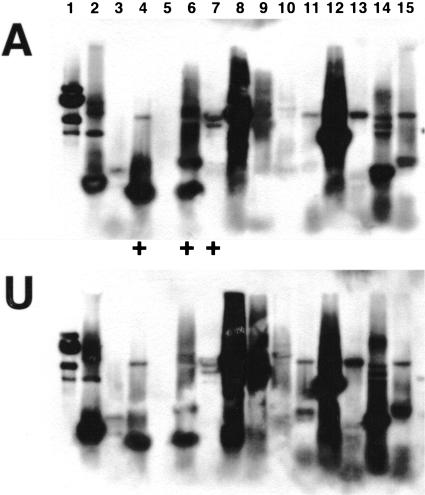
Following subcloning of subtraction products, 535 bacterial colonies were screened for their content of differentially expressed gene products. In a reverse Northern blot procedure, more than 95% of all cDNA fragments could be detected with probes from both group A and group U. Twenty-one of these products showed clearly increased labeling with probe A compared to probe U and were thus selected for further analysis (for an example see Fig. 1).
Figure 1.
A reverse Northern blot procedure was adapted to screen the subtracted cDNAs for differentially expressed molecules. Following colony-PCR, duplicate blots were prepared from different subtraction products and analyzed with probes from the associative learning group (A) and the unshocked control group (U). In the blot shown, clones #4, #6 (upper fragments), and #7 show increased labeling with probe A compared to group U, and were selected for further analysis.
Identification of Differentially Expressed Gene Products
All 21 selected cDNA clones were sequenced and compared to entries at the nonredundant NCBI gene bank. Eleven of the 21 clones could be identified this way (Table 1). They include the metabolic enzyme phosphofructokinase1c and various factors of signal transduction pathways (aldehyde reductase AKR1A4, membrane interacting protein of RGS16, neurocalcin, phosphodiesterase1, protein kinase inhibitor-α, and transthyretin) and structural reorganization processes (E2-ubiquitin conjugating enzyme, γ-actin, neuroligin1, proteasome subunit P31, and UDP-galactose transporter isozyme1). The remaining 10 clones showed no significant similarities to entries in the nonredundant database. However, clone #VIIG5 showed a match with two mouse expressed sequence tags (AA170872 and AA116522), which in turn aligned with the putative protein sorting factor Praja1 (U06944). To test whether the subtraction product indeed reflects part of a novel Praja1 isoform, 3′ rapid amplification of cDNA ends (RACE) and 5′ RACE procedures were performed using gene-specific primer oligonucleotides for clone #VIIG5. The 5′ RACE produced a single DNA fragment 640 bp in length, which aligned to both clone #VIIG5 and the known Praja1 sequence in the manner predicted by the EST-alignment. The 3′ RACE produced four fragments with sizes of 780 bp, 1.0 kb, 1.4 kb, and 1.8 kb. Each of these aligned precisely with clone #VIIG5 and with the previously reported Praja1 sequence at different positions of the coding region. They also contained the entire 3′ untranslated region of Praja1. Thus, the identity of clone #VIIG5 as part of a novel Praja1 isoform and the existence of at least four different Praja1 mRNAs were confirmed.
Table 1.
Identification and Expression Levels of Subtraction Products
| Clone #
|
Identity of subtraction product
|
Size of detected transcripts
|
Expression level (relative to U)
|
||
|---|---|---|---|---|---|
| A
|
NA
|
RA
|
|||
| IIIA1 | UDP-galactose transporter related isozyme 1 | 1.0 kb | 1.4 | 1.2 | 1.0 |
| IIID3, IIIE5 | Phosphodiesterase1 | 2.9 kb | 1.5 | 1.7 | 1.0 |
| 2.3 kb* | 1.5 | 1.4 | 1.0 | ||
| IIIF3 | Actin | 2.6 kb§ | 1.4 | 1.1 | 1.1 |
| 1.5 kb | 0.7 | 0.8 | 0.9 | ||
| IVB4 | Proteasome subunit P31 | 1.4 kb | A, NA > RA, U | ||
| 1.1 kb | |||||
| IVH4 | E2-ubiquitin conjugating enzyme | 1.5 kb§ | 1.2 | 1.3 | 0.7 |
| 0.7 kb** | 2.5 | 2.5 | 1.2 | ||
| IVH9 | Neuroligin1 | 5.0 kb | A, NA > RA, U | ||
| 4.8 kb | |||||
| 3.7 kb | A, NA > RA, U | ||||
| 3.5 kb | |||||
| IVH5.2 | Neurocalcin | 3.3 kb** | 1.3 | 1.1 | 1.1 |
| VIA8 | Phosphofructokinase1c | 2.8 kb | 1.3 | 1.2 | 1.0 |
| VIIG5 | Praja1-subtraction product | 2.7 kb*** | 6.5 | 4.9 | 1.6 |
| 2.3 kb*** | 12.1 | 6.5 | 3.2 | ||
| 2.0 kb§ | A, NA > RA, U | ||||
| Praja1-total | 2.7 kb** | 4.9 | 3.7 | 1.6 | |
| 2.3 kb** | 9.8 | 6.2 | 2.7 | ||
| 2.0 kb§ | A > NA > RA, U | ||||
| 1.8 kb* | 10.9 | 8.7 | 3.0 | ||
| 1.4 kb* | 4.8 | 5.2 | 2.2 | ||
| VIIH2 | Transthyretin | 1.1 kb§* | 1.7 | 1.7 | 1.0 |
| 0.8 kb | 1.4 | 1.4 | 1.0 | ||
| VIIIH6 | Aldehyde reductase | 1.5 kb*** | 7.2 | 4.3 | 2.2 |
| 0.7 kb§* | 2.5 | 1.8 | 1.2 | ||
| IXC1 | Protein kinase inhibitor α | 3.6 kb | A, NA > RA, U | ||
| Membrane interacting | 1.6 kb | A > NA, RA, U | |||
| protein of RGS16 | 1.3 kb | ||||
Of 21 clones selected during screening, the 12 listed here could be identified and were further investigated with virtual Northern analysis. The length of cDNA transcripts is largely in agreement with the mRNA sizes reported for the respective molecules; however, some molecules (phosphodiesterase1, neuroligin 1, proteasome subunit P31, and the membrane interacting protein of RGS16) showed doublet transcripts. Expression levels are given in relation to levels in the unshocked group U and are corrected for G3PDH. Proteasome subunit P31, neuroligin1, protein kinase inhibitor α, and the membrane interacting protein of RGS16, as well as some Praja1 transcripts, could not be quantified due to low expression levels; differences in their expression between behavioral groups were therefore judged by visual inspection.
Significant change of expression, P <0.05; **P <0.05; ***P <0.001; §minor band.
Expression Analysis of Identified Gene Products
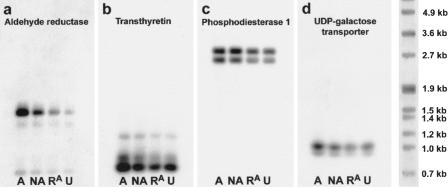
A detailed and quantitative expression analysis of identified gene products was performed with virtual Northern blots. Transcripts detected with this method were generally 0.2–0.4kb smaller than the respective full-length mRNA; this size shift is explained with a loss of polyA tails during reverse transcription. In addition, transcript doublets were observed for phosphodiesterase1, neuroligin1, proteasome subunit P31, and the membrane interacting protein of RGS16 and may be related to a premature incorporation of the SMART oligonucleotide during reverse transcription. To distinguish learning-, stress- and fear-related gene induction, we determined expression levels in all four behavioral groups (the associative learning group A, the nonassociative training group NA, the retrieval group RA, and the unshocked control group U). β-Tubulin and the polyadenylated mitochondrial 16S rRNA did not differ among the groups. For the 12 identified subtraction products including clone #VIIG5, however, an induction after fear conditioning training (i.e., in group A) was confirmed. Clone #IIIF3 (identified as γ-actin) showed a nonsignificant increase in the minor 2.6kb transcript (which likely reflects cross-hybridization with β-actin mRNA) and a mild decrease in the main signal at 1.5kb. Most other subtraction products showed a moderate (less than twofold) increase in expression in groups A and NA and little or no change in group RA, compared to group U. Such stress-related induction of gene expression was observed for transthyretin (ANOVA, F3,11 = 7.691, P <0.05; post-hoc comparison with Newman-Keul's test, A, NA vs. U: P <0.05), phosphodiesterase1 (F3,11 = 5.168, P <0.05; A, NA vs. U: not significant), and UDP-galactose transporter (not significant; shown in Fig. 2b-d), as well as E2-ubiquitin conjugating enzyme (F3,11 = 11.92, P <0.01; A, NA vs. U: P <0.05) and phosphofructokinase 1c (not significant). Exceptions are the induction of neurocalcin (F3,11 = 10.62, P <0.01; A vs. U: P <0.01) and membrane interacting protein of RGS16, which occurred only in group A. However, induction levels of these molecules were low (i.e., only + 30% for neurocalcin, Table 1).
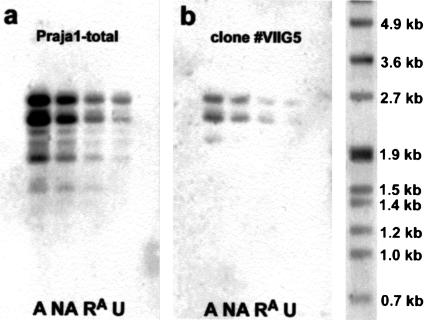
Only two of the identified factors showed a pronounced induction of gene expression after conditioning. Aldehyde reductase (F3,11 = 32.22; P <0.001), a key enzyme in monoamine metabolism, was induced sevenfold in the associative learning group A compared to the unshocked group U (P <0.001). In addition, this enzyme showed a graded increase across behavioral groups, with an approximately fourfold induction in the nonassociative training group NA (P <0.01) and a twofold induction in the retrieval group RA (not significant; Table 1, Fig. 2a). A similar observation was made for the putative protein sorting and ubiquitination factor Praja1. Four different Praja1 transcripts could be analyzed quantitatively, and all showed significant effects of training (F3,11 > 6.342; P <0.05). Two of them (2.3kb and 1.8kb) showed a greater than 10-fold increase and two (2.7kb and 1.4kb) showed a fivefold increase in group A, compared to group U (P <0.01). They were increased three- to eight-fold in group NA (P <0.05) and about two- to threefold in group RA (not significant). The larger Praja1 transcripts showed such a graded response across the different behavioral groups; however, the 1.4kb transcript showed similar expression levels in groups A and NA (Table 1, Fig. 3).
Figure 2.
Virtual Northern blot analysis of four selected clones obtained by subtractive hybridization: aldehyde reductase (a), transthyretin (b), phosphodiesterase1 (c), and UDP-galactose transporter (d). Aldehyde reductase showed a pronounced (sevenfold) and partly learning-specific increase in expression after fear conditioning (group A). Smaller increases in the nonassociative training group NA and the retrieval group RA reveal stress and fear effects that may also contribute to the increased expression of aldehyde reductase in group A. Transthyretin, phospodiesterase1, and the UDP-galactose transporter showed much weaker induction that was similar in groups A and NA, indicating a largely US-related response. Changes in group RA were not observed for these molecules. For quantification of the expression changes, refer to Table 1.
Figure 3.
Virtual Northern blot analysis of Praja1 expression using a 5′ Praja1 probe (a) and clone #VIIG5 probe (b). Both blots show strongly increased labeling in the associative learning group A and a graded response across behavioral groups with intermediate induction in the nonassociative training group NA and lowest (two– to threefold) increase of expression in the retrieval group RA. Praja1 showed four major and several minor bands between 1.4kb and 2.7kb. Clone #VIIG5 hybridized with the 2.7kb, 2.3kb, and in group A also with the 2.0kb Praja1 transcript. However, overall labeling was much weaker than with the 5′ Praja1 probe.
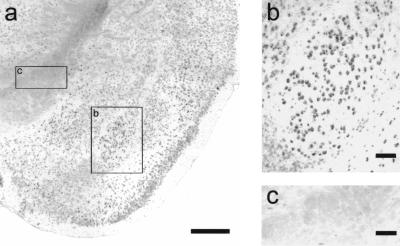
In situ hybridization revealed that Praja1 is widely expressed in cortical and subcortical brain areas. In the amygdala, the lateral, basal, medial, and cortical subnuclei could easily be distinguished according to their labeling intensity. For example, cells in the basolateral nucleus showed particularly strong labeling, whereas cells in the neighboring basomedial nucleus were only moderately stained. The lateral amygdala showed intermediate expression levels. The size and localization of Praja1-expressing cells in the BLA indicate that most of these cells are neuronal, although at present we cannot exclude Praja1 expression in glia. However, cell bodies along fiber tracts such as the internal capsule were generally not labeled, suggesting that most oligodendrocytes do not express Praja1 (Fig. 4). A predominantly neuronal expression of Praja1 was also apparent in the hippocampal formation, where we observed strong labeling in the pyramidal cell layer of areas CA1 and CA3 as well as the granule cell layer of the dentate gyrus (data not shown).
Figure 4.
Praja1 in situ hybridization. The Praja1 mRNA was found to be widely expressed in cortical and subcortical brain areas (a). Particularly strong labeling was observed in the basolateral nucleus of the amygdala; size and location of labeled cell bodies are indicative of neuronal cells (b). Fiber tracts, such as the internal capsule (c), showed no or only slight labeling. Sense controls were always negative (data not shown). Bars: 1 mm in (a), 100 μm in (b) and (c).
Sequence of PRAJA1a
The generation of various Praja1 mRNAs from a single Praja1 gene and the existence of a brain-specific Praja1 mRNA have been reported previously (Mishra et al. 1997). During subtractive hybridization, we isolated a piece (clone #VIIG5) of a novel Praja1 isoform with an insertion of 543 bp to the previously reported Praja1 sequence, as shown by 5′ RACE and 3′ RACE (Fig. 5). In fact, four different 3′ RACE products were obtained, which together with the detection of at least five different Praja1 transcripts in virtual Northern blots reveal the existence of various Praja1 isoforms in the brain. By virtual Northern analysis, clone #VIIG5 could be detected in transcripts with sizes of 2.7 kb, 2.3 kb, and 2.0 kb, corresponding to the three largest Praja1 mRNAs. It further showed increases of expression after behavioral stimulation that were comparable to those seen with a probe detecting the 5′ end of Praja1.
Figure 5.
Sequence of the novel Praja1 fragment and its location within the Praja1a mRNA. Most of the fragment (bp316–808) was isolated in clone #VIIG5 after subtractive hybridization. NCBI gene bank analysis and RACE showed that the fragment represents part of a novel Praja1 isoform and expands the open reading frame by 543 bp. Putative protein kinase C (boxed) and casein kinase II (underlined) phosphorylation sites, as well as N-glycosylation sites (bold) and the D T DDDDDD motif (italic, bold) are indicated.
The novel fragment expands the open reading frame of Praja1 to 1818 bp, resulting in a 605-amino acid protein with a predicted molecular weight of 67 kD (Praja1a, AF335250). Mishra et al. (1997) reported a considerable deviation of the apparent from the predicted molecular weight, due to the high acidity of Praja1. Our novel fragment contains 32 strongly basic and 28 strongly acidic amino acids, thus only slightly reducing the acidity of Praja1a (pI = 4.88), compared to Praja1 without the insert (pI = 4.52). Transmembrane regions or obvious α-helical or β-sheet structures could not be found. However, the insert contains four protein kinase C and four casein kinase II phosphorylation sites, two N-glycosylation sites, and a conspicuous D S/T DDDDDD motif, which among other protein- and DNA-binding proteins is also found in the E2-ubiquitin conjugating enzyme and ubiquitin ligase (Fig. 5).
DISCUSSION
Transcriptional activation and protein synthesis in the amygdala have proven essential for the formation of fear memory (Bourtchouladze et al. 1994; Schafe et al. 1999, 2000). In addition, fear memory can be enhanced through localized overexpression in the BLA of the transcription factor CREB at the time of conditioning (Josselyn et al. 2001). In the present study we identified several signal transduction factors and mediators of structural organization processes that are induced in the mouse BLA during formation of Pavlovian fear memory. With behavioral control groups for learning-, stress- and fear-induced gene expression, we observed pronounced and learning-specific inductions of aldehyde reductase and the protein sorting and ubiquitination factor Praja1. These findings confirm and extend previous observations of fear conditioning-induced neural plasticity in the BLA (Quirk et al. 1995; Rogan et al. 1997; Paré and Collins 2000) and support the idea that the amygdala is a site of information storage during formation of fear memory. Moreover, a strong stress- and fear-related component in the expression of most genes including aldehyde reductase and Praja1 suggests that neural plasticity in the BLA is strongly involved in the processing of unconditioned stimulus-related information.
Fear Conditioning Training
Although the induction of neural plasticity in the BLA during Pavlovian fear conditioning is well documented, the precise role of the amygdala in aversive learning is still a matter of debate (Cahill et al. 2000; Wilensky et al. 2000). This is at least in part due to the fact that the amygdala is involved in not only the acquisition of conditioned fear during Pavlovian fear conditioning, but also in the modulation of aversive learning mediated by other brain areas and in the expression and coordination of both conditioned and unconditioned fear responses. Thus, not all of the gene products that are induced following fear conditioning training, which at the same time evokes acute foot-shock stress and unconditioned fear, may be indispensable for memory formation. To approach this problem, we studied gene expression in the BLA in two steps: First, mice conditioned with a conditioned stimulus/unconditioned stimulus (CS/US) pairing protocol that induces robust fear memory (the “associative learning group” A; Stork et al. 2000) were compared to an “unshocked control group” U. Using virtual Northern blot analysis we then tried to further distinguish learning-related from footshock stress- and fear-induced changes of gene expression with additional behavioral groups: a “nonassociative training group” NA was exposed to the same CS and US as group A but without temporal coincidence, and a “retrieval group” RA was established to study the effects of fear alone. This approach allowed us to identify learning-specific components of aldehyde reductase and Praja1 gene expression without disregarding stress- or fear- related aspects of gene expression in these and other gene products. However, it should be noted that the distinction between learning-related effects in group A and stress- or fear-related effects in groups A, NA, and RA may not be absolute, since a weak cued or contextual memory may still be formed in group NA and a reexperience of the CS during retrieval can destabilize fear memory in a protein synthesis-dependent manner (Nader et al. 2000). Accordingly, an activation of the transcription factors CREB and c-fos was recently observed following fear memory retrieval (Hall et al. 2001).
Expression of Signal Transduction and Structural Reorganization Factors
During subtractive hybridization, 21 clones with an increased expression after fear conditioning were isolated (i.e., 4% of the investigated 535 clones). Thirteen molecular factors in 12 clones could be identified; one of these is phosphofructokinase1c, the induction of which may reflect an increased activity and glucose metabolism in BLA neurons. The remaining 12 factors appear to fall into one of two major categories: factors that contribute to signal transduction pathways and factors that mediate morphological reorganization processes. This observation is in good agreement with our current knowledge of gene expression and cellular changes during the formation of LTM (Stork and Welzl 1999).
In particular, the prominent induction and graded increase across behavioral groups of aldehyde reductase, a key enzyme of monoamine metabolism, suggest that metabolic control of monoamine function plays an important role in the formation of fear memory and the stress- and fear-related modulation of amygdala function. This is in agreement with findings of enhanced fear conditioning after the targeted disruption of the gene for monoamine oxidase A (Kim et al. 1997). The increased expression in the present associative and nonassociative training groups A and NA of transthyretin, a transport protein of L-thyroxin in the brain, may further reflect a stress-induced activation of 3,3′,5-triiodo-L-thryronine (T3) in the amygdala (Baumgartner et al. 1998). T3 is known to modulate neurotransmitter release and GABAA receptor function and the expression of neuropeptides and neurotrophins, and to induce changes of synaptic density in the brain. It may thus be an important modulator of neural plasticity in the amygdala, which is characterized by particularly high levels of T3 receptors (Puymirat et al. 1991). In addition, the induction of neurocalcin gene expression in the present study's group A is indicative of a modification in Ca++-dependent mechanisms following conditioning. A role of Ca++ in LTM consolidation is less well established than its role during the induction of synaptic plasticity. However, a presynaptic increase of another Ca++-binding factor, calreticulin, was reported after sensitization of Aplysia californica sensory neurons (Kennedy et al. 1992). Similarly, an increase only in the present associative learning group A was seen for the membrane interacting protein of RGS16, involved in lipid metabolism and G-protein signaling (Zheng et al. 2000). While there is some indication for an involvement of another regulator of G-protein signaling (RGS2) in synaptic plasticity and emotional behavior (Ingi et al. 1998; Oliveira-Dos-Santos et al. 2000), our finding is the first evidence of a role for RGS16 in these processes. On the other hand, a great body of evidence indicates that changes in cAMP and protein kinase A activity are critical for LTM including the formation of fear memory (Bourtchouladze et al. 1994, Schafe et al. 1999, 2000). Increased expression of phosphodiesterase1 and protein kinase inhibitor-α may contribute to the regulation of this second messenger pathway following fear conditioning. Phosphodiesterase1 in particular, through its Ca++/calmodulin-dependent kinase responsiveness and synaptic localization, may play an important role in learning- and stress-related synaptic plasticity in the mammalian brain, much like its homolog dunce in Drosophila melanogaster (Preat 1998). In Aplysia californica neural plasticity on the other hand, elevated protein kinase A activity is maintained through degradation of its regulatory subunits in ubiquitin-mediated proteolysis (Hegde et al. 1997). One protein contributing to this proteolytic pathway is the E2-ubiquitin conjugating enzyme, which was increased in group A and group NA in our experiments.
E2-ubiquitin conjugating enzyme may also contribute to a structural reorganization associated with memory formation and synaptic plasticity. Indeed, the upregulation of the proteasome subunit P31 after fear conditioning training suggests the activation of these processes. Recent studies have further emphasized the importance of actin-mediated cytoskeletal rearrangements for pre- and postsynaptic plasticity (Hatada et al. 2000; Matus 2000). The differential expression of two actin transcripts after fear conditioning supports this idea. Neuroligin1 in turn, as a postsynaptic cell surface molecule and binding partner for Neurexins, can trigger presynaptic synaptogenesis (Scheiffele et al. 2000). Its increased expression in our training groups A and NA may reflect the morphological changes involved in fear memory formation and stress-induced functional changes in the BLA. Finally, the expression of the mRNA encoding a UDP-galactose transporter finally may reflect the production of cell-surface and extracellular glycoproteins, which are important mediators of synaptic reorganization and critical to memory formation (Rose 1995; Tiunova et al. 1998).
Praja1 — Involvement in Synaptic Tagging?
To achieve a synapse-specific neural plasticity, newly generated synaptic proteins must be incorporated and active at the very synapses that contribute to information storage. Frey and Morris (1998) suggested that during the induction of synaptic plasticity, molecular changes at the relevant synapses alter local protein constituents, which then can serve as a “synaptic tag” to guide newly synthesized proteins towards the site of stimulation. We suggest that the putative protein sorting factor Praja1 (Mishra et al. 1997) may be involved in this mechanism. Praja1 is expressed in many neuronal cells of the BLA and associated brain areas (Fig. 4). Praja1 shows some similarity to the neurodegeneration-associated protein neurodap1, which is found at the postsynaptic density of axosomatic synapses (Nakayama et al. 1995), and to RPM-1, an organizer of presynaptic terminals in Caenorhabditis elegans (Zhen et al. 2000). Recent studies have further suggested a role for Praja1 in substrate specification of E2-dependent ubiquitination (Lorick et al. 1999; Fang 2000), which was increased in our associative and nonassociative training groups A and NA. Following fear conditioning training, Praja1 was strongly (>10-fold) increased in expression. It showed a graded response across behavioral groups, indicating an expression in accordance with the type and intensity of emotionally relevant stimuli. Moreover, various Praja1 transcripts were detected that showed considerable difference in their induction following training. For example, the increases in the expression of the 2.3 kb and 1.8 kb transcripts was clearly stronger than that of the 2.7 kb and the 1.4 kb transcripts. A learning-specific component of induction could be observed for all except the 1.4 kb transcript. This suggests that in addition to transcriptional activation, a tissue-specific (Mishra et al. 1997) and activity-dependent generation of various Praja1 isoforms through alternative splicing may contribute to neural plasticity and information storage in the BLA. We identified a 543 bp region that codes for a stretch of 181 amino acids in the N-terminal third of a novel Praja1 isoform, Praja1a. A specific probe for this region labeled the larger three Praja1 transcripts and revealed an induction similar to that seen with the Praja1 total probe. Praja1a carries additional protein kinase C and casein kinase II phosphorylation sites and a D T/S DDDDDD motif found in several protein-binding and ubiquitination factors. Functional differences between Praja1 isoforms and their roles in the cellular processes underlying formation of fear memory remain to be investigated.
Conclusion
We have shown that an induction of gene expression occurs in the mouse BLA during a critical phase in the formation of fear memory, which is sensitive to protein synthesis inhibition. The genes that are induced reflect changes in signal transduction and structural reorganization processes during neural plasticity. Most of the observed effects appear to be related to the unconditioned stimulus presentation, indicating that many transcriptional processes may be commonly activated in the BLA during consolidation of fear memory and stress-induced changes of emotional state. Further, the contribution of stress-induced molecular changes in the BLA may be critical for the particular rapidity of the formation of long-term fear memory. It will be interesting to see whether comparable gene expression patterns are apparent in the BLA during other forms of aversive learning.
MATERIALS AND METHODS
Animals
Experiments were performed with 10-week-old male C57B/6 mice, which were obtained at an age of 6–7 wk (Japan SLC, Hamamatsu, Japan) and kept in our animal facility. Mice were housed in groups of 4–6 under standard laboratory conditions with a 12h day/night cycle (lights on at 0715) and food and water ad libitum. All animal experiments were performed in accordance with regulations under Japanese and German law and were approved by the Committee for Animal Research, Okazaki National Institutes (Nrs. A11–82–3 and A11–82–65) and the Regierungspräsidium Dessau (Nr. 2–375 UNI MD).
Fear Conditioning Protocol
Animals were conditioned in a Plexiglas compartment (25 × 25 × 20cm) that was equipped with a grid floor for delivery of the unconditioned stimulus (US) and an activity monitoring system with infrared photo beams (SCANET). The training compartment was enclosed in a soundproof isolation cubicle, which contained a fan providing fresh air and background noise and a tone generator for delivery of the conditioned stimulus (CS). Mice were separated for one week before being adapted to the training environment over a period of four days with two daily sessions, each of which was 6 min long. Preexperiment differences in anxiety level were estimated according to locomotor activity and the number of fecal boli produced during the last adaptation session. Mice were ranked according to their anxiety level and distributed equally into four different training groups: group A (“associative learning group”), group NA (“nonassociative training group”), group RA (“retrieval group”), and group U (“unshocked group”). On the 5th experimental day, after a habituation period of 2 min, the animals were trained according to the following protocols (Fig. 6): mice in groups A and RA received 3 CS (4 kHz tone, 75 dB for 10 sec) each coterminating with a US (0.7mA footshock, 1 sec) and separated by 50 sec of silence. Mice in group NA were confronted with 3 US and 1 min later with 3 CS, each 20 sec apart. Unshocked controls of group U received 3 CS, but no US during training. The behavioral response of the animals was inspected visually during the entire training session. Mice were returned to their home cages one minute after the last stimulus, and animals of groups A and NA were killed 6 h later. Mice in the groups RA and U were kept for 14 days, re-exposed to a set of 3 CS within a single 6-min retrieval session, and killed after 6 h.
Figure 6.
Outline of the behavioral training and RNA isolation protocol. Animals were divided into four groups (A, NA, RA, and U; N = 4 each) and habituated to the training environment over a period of four days with two daily sessions of 6 min each (“adaptation”). On the fifth day (“training”), the mice in groups A and RA were fear-conditioned with three CS/US pairings, while group NA received three CS and three US without temporal coincidence, and group U mice were exposed to three CS only. Groups RA and U were reexposed to the CS two weeks after training (“retrieval”). This design allowed us to distinguish learning-specific gene expression (in group A) from footshock stress- (groups A and NA) and fear-related (groups A, NA, and RA) effects. Six hours after their last stimulation, mice were killed for tissue collection and total RNA was isolated from tissue punches of the BLA (encircled area). L, lateral amygdala; BL, basolateral amygdala; BM, basomedial amygdala; M, medial amygdala; Pir, piriform cortex; CP, caudate putamen; Hip, hippocampus.
Preparation of cDNA
Animals were killed by cervical dislocation 6 h after the last stimulus. The brain was quickly removed from the skull and frozen with dry ice-cooled isopentan. Under binocular vision, the BLA was punched bilaterally from 180-μm-thick coronal sections. Tissue punches of each four animals were pooled and homogenized, and total RNA was isolated from the homogenate with RNA spin columns according to the manufacturer's protocol (QIAGEN, Leiden, The Netherlands). First-strand synthesis was done with MMLV reverse transcriptase (Superscript II, Promega) in the presence of modified oligo-dT cDNA synthesis primer and 5′ extension oligonucleotides (SMART II oligonucleotide; Clontech).
Subtractive Hybridization
Subtractive hybridization was done with the PCR-Select cDNA Subtraction system (Clontech) according to the manufacturer's protocol. In brief, first-strand cDNA of the associative learning group A and the unshocked control group U were amplified with 19 cycles of long-distance PCR, digested with Rsa1 and purified using QIAquick spin columns (QIAGEN). The tester cDNA (group A) was diluted and in two separate assays ligated to different adaptor oligonucleotides (PCR-Select). The products were separately hybridized with an excess of driver cDNA (group U) for 8 h at 68°C, then combined in the presence of additional driver cDNA and further hybridized overnight at 68°C. The enriched cDNAs were amplified from the hybridization product with a pair of PCR primer oligonucleotides that target the previously ligated adaptors (PCR-Select). The amplified subtraction products were subcloned into pBluescript SK+ using Eag1 sites in the adaptors, and grown in XL Blue competent cells (Stratagene).
Screening by Reverse Northern Blot Analysis
Subtracted amygdala cDNAs were amplified from 535 single bacterial colonies using standard T3 and T7 primer oligonucleotides (Stratagene). The PCR products were digested with Rsa1 to remove polylinker DNA and to separate hybridization products that had accidentally been connected during adaptor ligation or subcloning. Fragments were separated according to size by electrophoresis in agarose gels and transferred to nylon membranes (Hybond N; Amersham). For probe generation, Rsa1-digested cDNA from group A and group U were ligated with both adaptor nucleotides, amplified in the presence of digoxigenin-coupled dUTP (Roche Molecular Biochemicals) and again digested with Rsa1. After prehybridization with DIG-Easy-HYB (Roche Molecular Biochemicals), duplicate blots were hybridized with probe A or probe U at 65 °C and stringency washed with 0.1 × SSC, 0.1% SDS at the same temperature. Unspecific antibody binding was reduced by incubation in 1% blocking reagent in Tris-buffered saline (TBS) before signals were detected with alkaline phosphatase-coupled antidigoxigenin antibodies and CDPStar chemiluminescence substrate (Roche Molecular Biochemicals). After exposure to an autoradiographic film, staining intensity was evaluated visually and compared between group A and group U.
Sequence Analysis
The sequence of selected cDNA clones was determined using the Thermosequenase cycle sequencing system (Amersham) according to the manufacturer's protocols. In brief, 1μg of cDNA containing the insert of interest was amplified in the presence of fluorescence-labeled M13 forward or reverse primer oligonucleotide (Stratagene), and ddG, ddA, ddT, or ddC to terminate polymerization. Thirty cycles of 95°C for 30 sec, 50°C for 30 sec, and 70°C for 60 sec were applied. Products were loaded onto a polyacrylamide gel containing 7M urea, separated at 2000V and detected in a BDNA sequencer (Model 4000L, LI-COR). Sequences were analyzed using Lasergene99 software (DNASTAR) and the NCBI and PROSITE databanks.
Rapid Amplification of cDNA Ends
Rapid amplification of cDNA ends (RACE) was done for clone #VIIG5 using the SMART RACE system (Clontech) according to the manufacturer's protocol. Total RNA (group A) was reverse transcribed with Superscript II in the presence of a modified oligo-dT cDNA synthesis primer (for 3′ RACE) or oligo-dT primer and SMART II oligonucleotide (for 5′ RACE).
The 3′cDNA end was amplified using a nested 3′ RACE procedure: first, a clone #VIIG5-specific primer oligonucleotide (5′-TTC TGC CTC TCC CTC TCC AAG CTG TTG-3′) and the universal primer mix (SMART RACE) were used in a touchdown amplification procedure. The product was diluted and then reamplified with a nested VIIG5-specific primer (5′-CAA CAG CTT GGA GAG GGA GAG GCA GAA-3′) and the nested universal primer (SMART RACE). For the 5′ RACE, a VIIG5-specific primer (5′-GCG AAG GAA GAG TGT GGC AAG GTA GAA-3′) was used together with the universal primer mix for touchdown amplification with 18 cycles in the final step. The resulting fragments were isolated from agarose gels, cloned into pGEM-Teasy (Promega), and sequenced as described above.
Expression Analysis with Virtual Northern Blots
For virtual Northern blot analysis, first-strand cDNA of all four behavioral groups (the associative learning group A, the nonassociative training group NA, the retrieval group RA and the unshocked group U) was amplified with long-distance PCR. Quantification of the total PCR product in the range of 0.5–10 kb revealed that samples were still exponentially amplified after 19 cycles, which were therefore applied in all experiments. Each 1μg of the product was loaded on an agarose gel, separated by gel-electrophoresis and blotted to nylon membranes (Hybond N). Digoxigenin-labeled probes were generated from selected clones through amplification with standard T3 and T7 primers in the presence of digoxigenin-coupled dUTP, followed by digestion with Rsa1 and purification with QIAquick spin columns. The following cDNA fragments were used as probes (subtraction products are underlined; other fragments were obtained through direct amplification with gene specific primer pairs): UDP-galactose transporter isozyme1 (D87990): bp89–790, bp144–414; phosphodiesterase1 (NM015744): bp525–1516, bp1719–2008; γ-actin (NM009609): bp435–948; proteasome subunit P31 (NM002812): bp191–842, bp255–544; E2-ubiquitin conjugating enzyme (AF159230): bp1–649, bp73–331; neuroligin1 (U22952): bp235–436; bp3038–3572; neurocalcin (D10884): bp12–563; phosphofructokinase1c (AF123533): bp610–1156; Praja1a (AF335250, Fig. 5): bp30–642; bp316–808; transthyretin (NM013697): bp87–282, bp90–416; aldehyde reductase AKR1A4 (NM021473): bp180–996; protein kinase inhibitor-α (NM008862): bp1089–1720; membrane interacting protein of RGS16 (AF212861): bp839–1203; glycerinaldehyde-3-phosphate dehydrogenase (G3PDH; NM008084): bp576–1017; β-tubulin (BC005547): bp888–1238; mitochondrial 16S rRNA (AF089815): bp1345–1595. Hybridization, stringency washing, and signal detection with CDPStar chemiluminescence substrate were done as described above, followed by a quantitative analysis of signal intensity with a chemiluminescence detection system (ChemiDoc, BIO-RAD) using G3PDH as a standard. Based on three independent behavioral experiments (with N = 4 for each of the four behavioral groups), three virtual Northern blot analyses were performed. The results of these three experiments were evaluated statistically with repeated measures ANOVA, and groups were compared post-hoc with Newman-Keul's test.
In Situ Hybridization
In a cryostat, coronal sections of 14 μm thickness were cut at the level of the amygdala and thaw-mounted onto silane-coated slide glasses. Digoxigenin-labeled Praja1 sense and antisense cRNA were generated by in vitro transcription from a vector containing bp30–642 of the Praja1a cDNA. In situ hybridization was performed with these probes as described previously (Stork et al. 2000). Briefly, after fixation in 4% paraformaldehyde in phosphate-buffered saline, sections were acetylated, dehydrated, and prehybridized for 2 h at room temperature. After hybridization overnight and washing at 55°C with 0.2 × SSC containing 50% formamide, labeled cells were detected with alkaline phosphatase-coupled antidigoxigenin antibody and subsequently stained using 4-nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate as substrate (Roche Molecular Biochemicals).
Acknowledgments
We thank Drs. H. Welzl and P. Landgraf for helpful discussions and comments on the manuscript. This work was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan (grant nos. 07279107 and 10480229), the Leibniz Programm of the Deutsche Forschungsgemeinschaft (to H.C.P.), and a fellowship of the Science & Technology Program of the European Union (1998/1999) to O.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL oliver.stork@medizin.uni-magdeburg.de; FAX 49-391-671-5819.
Article and publication are at http://www.learnmem.org/cgi/doi/10.1101/lm.39401.
REFERENCES
- Baumgartner A, Hiedra L, Pinna G, Eravci M, Prengel H, Meinhold H. Rat brain type II 5′-iodothyronine deiodinase activity is extremely sensitive to stress. J Neurochem. 1998;71:817–826. doi: 10.1046/j.1471-4159.1998.71020817.x. [DOI] [PubMed] [Google Scholar]
- Beck CHM, Fibiger HC. Conditioned fear-induced changes in behavior and in the expression of the immediate early gene c-fos: With and without diazepam pretreatment. J Neurosci. 1995;15:709–720. doi: 10.1523/JNEUROSCI.15-01-00709.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtchouladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva A. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- Campeau S, Hayward MD, Hope BT, Rosen JB, Nestler EJ, Davis M. Induction of the c-fos proto-oncogene in rat amygdala during unconditioned and conditioned fear. Brain Res. 1991;565:349–352. doi: 10.1016/0006-8993(91)91669-r. [DOI] [PubMed] [Google Scholar]
- Cahill L, Vazdarjanova A, Setlow B. The basolateral amygdala complex is involved with, but is not necessary for, rapid acquisition of Pavlovian ‘fear conditioning’. Eur J Neurosci. 2000;12:3044–3050. doi: 10.1046/j.1460-9568.2000.00187.x. [DOI] [PubMed] [Google Scholar]
- Davis M, Rainnie D, Cassell M. Neurotransmission in the rat amygdala related to fear and anxiety. Trends Neurosci. 1994;17:208–214. doi: 10.1016/0166-2236(94)90106-6. [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, et al. Suppression subtractive hybridization: A method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- Frey U, Morris RGM. Synaptic tagging: Implications for late maintenance of hippocampal long-term potentiation. Trends Neurosci. 1998;21:181–188. doi: 10.1016/s0166-2236(97)01189-2. [DOI] [PubMed] [Google Scholar]
- Grecksch G, Matthies H. Two sensitive periods for the amnesic effect of anisomycin. Pharmacol Biochem Behav. 1980;12:663–665. doi: 10.1016/0091-3057(80)90145-8. [DOI] [PubMed] [Google Scholar]
- Hall J, Thomas KL, Everitt BJ. Fear memory retrieval induces CREB phosphorylation and Fos expression within the amygdala. Eur J Neurosci. 2001;13:1453–1458. doi: 10.1046/j.0953-816x.2001.01531.x. [DOI] [PubMed] [Google Scholar]
- Hatada Y, Wu F, Sun ZY, Schacher S, Goldberg DJ. Presynaptic morphological changes associated with long-term synaptic facilitation are triggered by actin polymerization at preexisting varicositis. J Neurosci. 2000;20:RC82. doi: 10.1523/JNEUROSCI.20-13-j0001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde AN, Inokuchi K, Pei W, Casadio A, Ghirardi M, Chain DG, Martin KC, Kandel ER, Schwartz JH. Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell. 1997;89:115–126. doi: 10.1016/s0092-8674(00)80188-9. [DOI] [PubMed] [Google Scholar]
- Ingi T, Krumins AM, Chidiac P, Brothers GM, Chung S, Snow BE, Barnes CA, Lanahan AA, Siderovski DP, Ross EM, et al. Dynamic regulation of RGS2 suggests a novel mechanism in G-protein signaling and neuronal plasticity. J Neurosci. 1998;18:7178–7188. doi: 10.1523/JNEUROSCI.18-18-07178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josselyn SA, Shi C, Carlezon WAJ, Neve RL, Nestler EJ, Davis M. Long-term memory is facilitated by cAMP response element-binding protein overexpression in the amygdala. J Neurosci. 2001;21:2404–2412. doi: 10.1523/JNEUROSCI.21-07-02404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy TE, Kuhl D, Barzilai A, Sweatt JD, Kandel ER. Long-term sensitization training in Aplysia leads to an increase in calreticulin, a major presynaptic calcium-binding protein. Neuron. 1992;9:1013–1024. doi: 10.1016/0896-6273(92)90062-i. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Fanselow MS, DeCola JP, Landeira-Fernandez J. Selective impairment of long-term but not short-term conditional fear by the N-methyl-D-aspartate antagonist APV. Behav Neurosci. 1992;106:591–596. doi: 10.1037//0735-7044.106.4.591. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Shih JC, Chen K, Chen L, Bao S, Maren S, Anagnostaras SG, Fanselow MS, De Maeyer E, Seif I, et al. Selective enhancement of emotional, but not motor, learning in monoamine oxidase A-deficient mice. Proc Natl Acad Sci. 1997;94:5929–5933. doi: 10.1073/pnas.94.11.5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A. Actin-based plasticity in dendritic spines. Science. 2000;290:754–758. doi: 10.1126/science.290.5492.754. [DOI] [PubMed] [Google Scholar]
- Miserendino MJ, Sananes CB, Melia KR, Davis M. Blocking of acquisition but not expression of conditioned fear-potentiated startle by NMDA antagonists in the amygdala. Nature. 1990;345:716–718. doi: 10.1038/345716a0. [DOI] [PubMed] [Google Scholar]
- Mishra L, Tully RE, Monga SP, Yu P, Cai T, Makalowski W, Mezey E, Pavan WJ, Mishra B. Praja1, a novel gene encoding a RING-H2 motif in mouse development. Oncogene. 1997;15:2361–2368. doi: 10.1038/sj.onc.1201405. [DOI] [PubMed] [Google Scholar]
- Nader K, Schafe GE, LeDoux JE. Fear memories require protein synthesis in the amygdala for reconsolidation after retrieval. Nature. 2000;406:722–726. doi: 10.1038/35021052. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Miyake T, Gahara Y, Ohara O, Kitamura T. A novel RING-H2 motif protein downregulated by axotomy: Its characteristic localization at the postsynaptic density of axosomatic synapse. J Neurosci. 1995;15:5238–5248. doi: 10.1523/JNEUROSCI.15-07-05238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Dos-Santos AJ, Matsumoto G, Snow BE, Bai D, Houston FP, Whishaw IQ, Mariathasan S, Sasaki T, Wakeham A, Ohashi PS, et al. Regulation of T cell activation, anxiety, and male aggression by RGS2. Proc Natl Acad Sci. 2000;97:12272–12277. doi: 10.1073/pnas.220414397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parè C, Collins DR. Neuronal correlates of fear in the lateral amygdala: Multiple extracellular recordings in conscious cats. J Neurosci. 2000;20:2701–2710. doi: 10.1523/JNEUROSCI.20-07-02701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preat T. Decreased odor avoidance after electric shock in Drosophila mutants biases learning and memory tests. J Neurosci. 1998;18:8534–8538. doi: 10.1523/JNEUROSCI.18-20-08534.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puymirat J, Miehe M, Marchand R, Sarlieve L, Dussault JH. Immunocytochemical localization of thyroid hormone receptors in the adult rat brain. Thyroid. 1991;1:173–184. doi: 10.1089/thy.1991.1.173. [DOI] [PubMed] [Google Scholar]
- Quirk GJ, Repa JC, LeDoux JE. Fear conditioning enhances short-latency auditory responses of lateral amygdala neurons: Parallel recordings in the freely behaving rat. Neuron. 1995;15:1029–1039. doi: 10.1016/0896-6273(95)90092-6. [DOI] [PubMed] [Google Scholar]
- Rogan MT, Staubli UV, LeDoux JE. Fear conditioning induces associative long-term potentiation in the amygdala. Nature. 1997;390:604–607. doi: 10.1038/37601. [DOI] [PubMed] [Google Scholar]
- Rose SPR. Glycoproteins and memory formation. Behav Brain Res. 1995;66:73–78. doi: 10.1016/0166-4328(94)00127-2. [DOI] [PubMed] [Google Scholar]
- Rosen JB, Fanselow MS, Young SL, Sitcoske M, Maren S. Immediate-early gene expression in the amygdala following footshock stress and contextual fear conditioning. Brain Res. 1998;796:132–142. doi: 10.1016/s0006-8993(98)00294-7. [DOI] [PubMed] [Google Scholar]
- Schafe GE, Nadel NV, Sullivan GM, Harris A, LeDoux JE. Memory consolidation for contextual and auditory fear conditioning is dependent on protein synthesis, PKA, and MAP kinase. Learn Mem. 1999;6:97–110. [PMC free article] [PubMed] [Google Scholar]
- Schafe GE, LeDoux JE. Memory consolidation of auditory Pavlovian fear conditioning requires protein synthesis and protein kinase A in the amygdala. J Neurosci. 2000;20:RC96. doi: 10.1523/JNEUROSCI.20-18-j0003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Stiedl O, Palve M, Radulovic J, Birkenfeld K, Spiess J. Differential impairment of auditory and contextual fear conditioning by protein synthesis inhibition in C57BL/6N mice. Behav Neurosci. 1999;113:496–506. doi: 10.1037//0735-7044.113.3.496. [DOI] [PubMed] [Google Scholar]
- Stork O, Welzl H. Memory formation and the regulation of gene expression. Cell Mol Life Sci. 1999;55:575–592. doi: 10.1007/s000180050316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork O, Welzl H, Cremer H, Schachner M. Increased intermale aggression in mice deficient for the neural cell adhesion molecule (NCAM) Eur J Neurosci. 1997;9:1117–1125. doi: 10.1111/j.1460-9568.1997.tb01464.x. [DOI] [PubMed] [Google Scholar]
- Stork O, Welzl H, Wolfer D, Schuster T, Mantei N, Stork S, Hoyer D, Lipp H-P, Obata K, Schachner M. Recovery of emotional behaviour in neural cell adhesion molecule (NCAM) null mutant mice through transgenic expression of NCAM180. Eur J Neurosci. 2000;12:3291–3306. doi: 10.1046/j.1460-9568.2000.00197.x. [DOI] [PubMed] [Google Scholar]
- Tiunova A, Anokhin KV, Rose SPR. Two critical periods of protein and glycoprotein synthesis in memory consolidation for visual categorization learning in chicks. Learn Mem. 1998;4:401–410. doi: 10.1101/lm.4.5.401. [DOI] [PubMed] [Google Scholar]
- Wilensky AE, Schafe GE, LeDoux JE. The amygdala modulates memory consolidation of fear-motivated inhibitory avoidance learning but not classical fear conditioning. J Neurosci. 2000;20:7059–7066. doi: 10.1523/JNEUROSCI.20-18-07059.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen M, Huang X, Bamber B, Jin Y. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron. 2000;26:331–343. doi: 10.1016/s0896-6273(00)81167-8. [DOI] [PubMed] [Google Scholar]
- Zheng B, Chen D, Farquhar MG. MIR16, a putative membrane glycerophosphodiester phosphodiesterase, interacts with RGS16. Proc Natl Acad Sci. 2000;97:3999–4004. doi: 10.1073/pnas.97.8.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]