Abstract
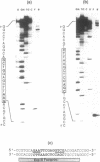
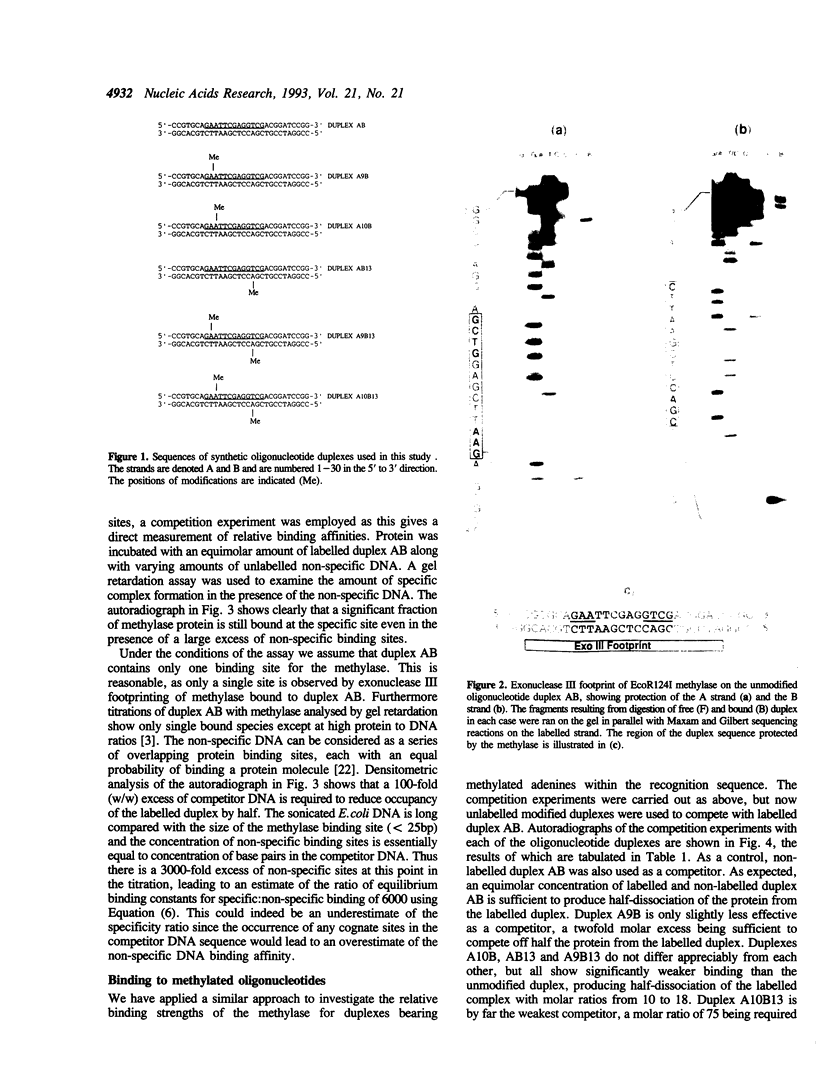
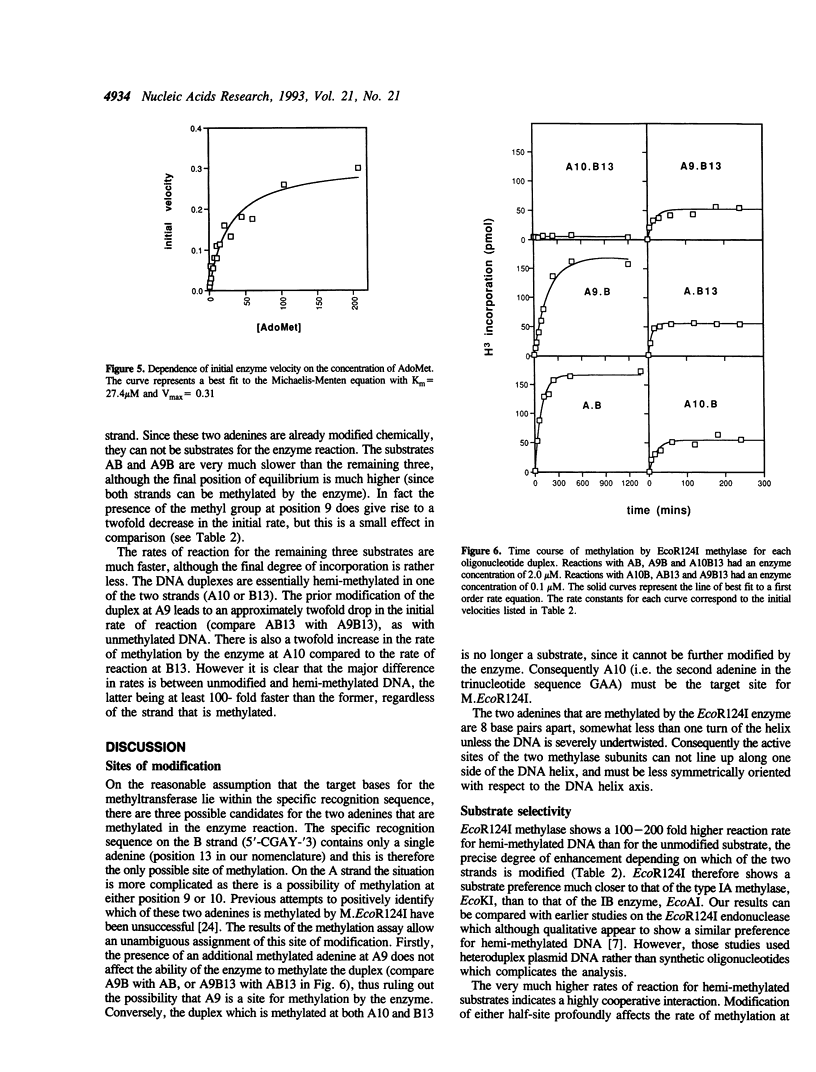
The type I DNA modification methylase M.EcoR124I binds sequence specifically to DNA and protects a 25bp fragment containing its cognate recognition sequence from digestion by exonuclease III. Using modified synthetic oligonucleotide duplexes we have investigated the catalytic properties of the methylase, and have established that a specific adenine on each strand of DNA is the site of methylation. We show that the rate of methylation of each adenine is increased at least 100 fold by prior methylation at the other site. However, this is accompanied by a significant decrease in the affinity of the methylase for these substrates according to competitive gel retardation assays. In contrast, methylation of an adenine in the recognition site which is not a target for the enzyme results in only a small decrease in both DNA binding affinity and rate of methylation by the enzyme.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cowan G. M., Gann A. A., Murray N. E. Conservation of complex DNA recognition domains between families of restriction enzymes. Cell. 1989 Jan 13;56(1):103–109. doi: 10.1016/0092-8674(89)90988-4. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Cowan G. M., Murray N. E. EcoA and EcoE: alternatives to the EcoK family of type I restriction and modification systems of Escherichia coli. J Mol Biol. 1985 Nov 5;186(1):65–75. doi: 10.1016/0022-2836(85)90257-8. [DOI] [PubMed] [Google Scholar]
- Gubler M., Braguglia D., Meyer J., Piekarowicz A., Bickle T. A. Recombination of constant and variable modules alters DNA sequence recognition by type IC restriction-modification enzymes. EMBO J. 1992 Jan;11(1):233–240. doi: 10.1002/j.1460-2075.1992.tb05046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubacek J., Glover S. W. Complementation analysis of temperature-sensitive host specificity mutations in Escherichia coli. J Mol Biol. 1970 May 28;50(1):111–127. doi: 10.1016/0022-2836(70)90108-7. [DOI] [PubMed] [Google Scholar]
- Kelleher J. E., Daniel A. S., Murray N. E. Mutations that confer de novo activity upon a maintenance methyltransferase. J Mol Biol. 1991 Sep 20;221(2):431–440. doi: 10.1016/0022-2836(91)80064-2. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., von Hippel P. H. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974 Jun 25;86(2):469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Gough J. A., Suri B., Bickle T. A. Structural homologies among type I restriction-modification systems. EMBO J. 1982;1(5):535–539. doi: 10.1002/j.1460-2075.1982.tb01205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J., Taylor I., Dutta C. F., Kneale G., Firman K. High-level expression of the cloned genes encoding the subunits of and intact DNA methyltransferase, M.EcoR124. Gene. 1992 Mar 1;112(1):21–27. doi: 10.1016/0378-1119(92)90298-4. [DOI] [PubMed] [Google Scholar]
- Price C., Lingner J., Bickle T. A., Firman K., Glover S. W. Basis for changes in DNA recognition by the EcoR124 and EcoR124/3 type I DNA restriction and modification enzymes. J Mol Biol. 1989 Jan 5;205(1):115–125. doi: 10.1016/0022-2836(89)90369-0. [DOI] [PubMed] [Google Scholar]
- Price C., Pripfl T., Bickle T. A. EcoR124 and EcoR124/3: the first members of a new family of type I restriction and modification systems. Eur J Biochem. 1987 Aug 17;167(1):111–115. doi: 10.1111/j.1432-1033.1987.tb13310.x. [DOI] [PubMed] [Google Scholar]
- Price C., Shepherd J. C., Bickle T. A. DNA recognition by a new family of type I restriction enzymes: a unique relationship between two different DNA specificities. EMBO J. 1987 May;6(5):1493–1497. doi: 10.1002/j.1460-2075.1987.tb02391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich N. O., Olsen C., Osti F., Murphy J. In vitro specificity of EcoRI DNA methyltransferase. J Biol Chem. 1992 Aug 5;267(22):15802–15807. [PubMed] [Google Scholar]
- Sain B., Murray N. E. The hsd (host specificity) genes of E. coli K 12. Mol Gen Genet. 1980;180(1):35–46. doi: 10.1007/BF00267350. [DOI] [PubMed] [Google Scholar]
- Suri B., Bickle T. A. EcoA: the first member of a new family of type I restriction modification systems. Gene organization and enzymatic activities. J Mol Biol. 1985 Nov 5;186(1):77–85. doi: 10.1016/0022-2836(85)90258-x. [DOI] [PubMed] [Google Scholar]
- Suri B., Nagaraja V., Bickle T. A. Bacterial DNA modification. Curr Top Microbiol Immunol. 1984;108:1–9. doi: 10.1007/978-3-642-69370-0_1. [DOI] [PubMed] [Google Scholar]
- Suri B., Shepherd J. C., Bickle T. A. The EcoA restriction and modification system of Escherichia coli 15T-: enzyme structure and DNA recognition sequence. EMBO J. 1984 Mar;3(3):575–579. doi: 10.1002/j.1460-2075.1984.tb01850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I., Patel J., Firman K., Kneale G. Purification and biochemical characterisation of the EcoR124 type I modification methylase. Nucleic Acids Res. 1992 Jan 25;20(2):179–186. doi: 10.1093/nar/20.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]