Abstract
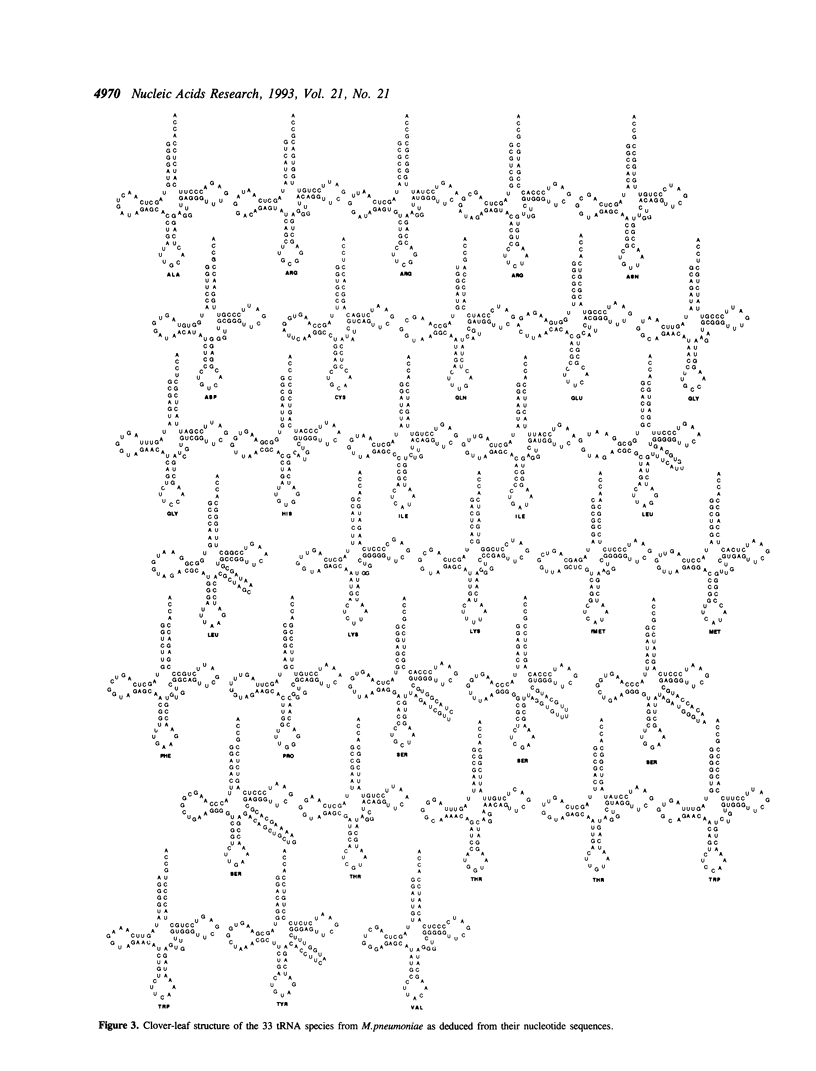
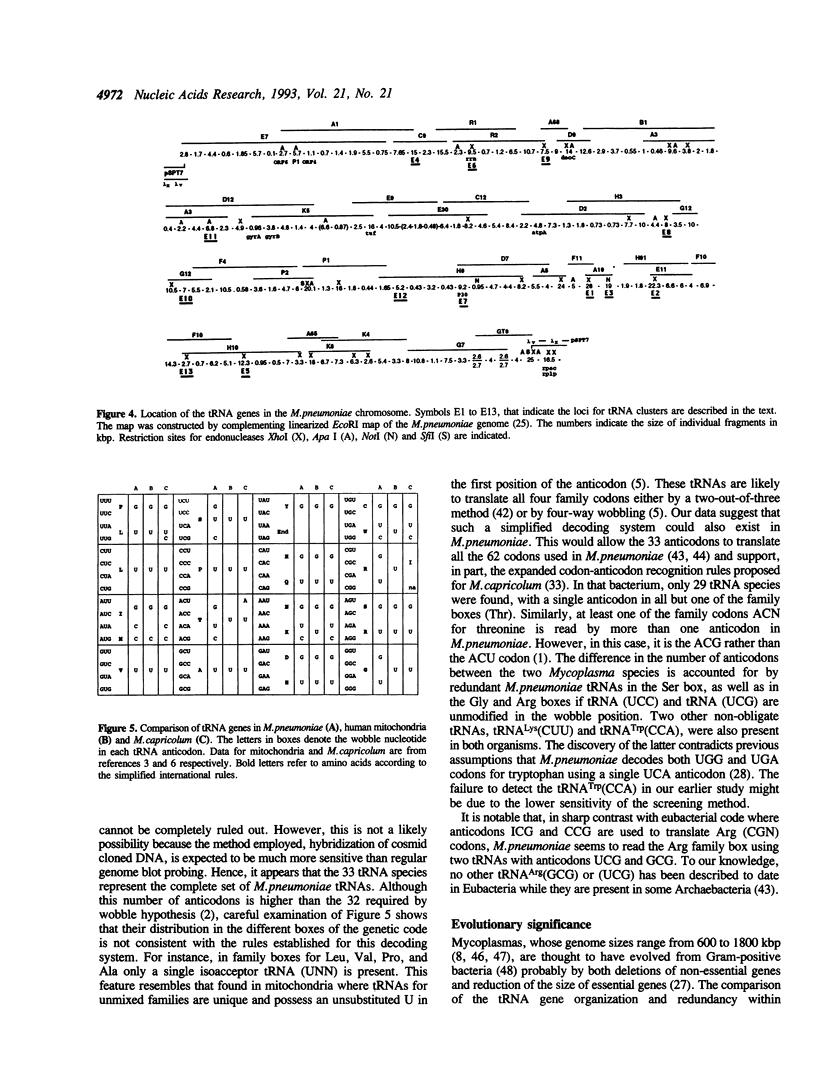
The 33 genes encoding the complete set of tRNA species in Mycoplasma pneumoniae have been cloned and sequenced. They are organized into 5 clusters in addition to 9 single genes. No redundant gene was found, indicating that 33 tRNAs correspond to 32 different anticodons and decode all 62 codons used in this organism. There is only one single tRNA for each of the Ala, Leu, Pro, and Val family boxes. Therefore, a simplified decoding system resembling that recently described for Mycoplasma capricolum (1) has to also exist in M.pneumoniae. However, analysis of the anticodon set and codon usage revealed features characteristic of the latter: (i) there is no obvious preference toward AT rich synonymous codons, (ii) CGG codons are assigned for arginine and are translated by tRNA Arg(UCG), and (iii) CNN or GNN anticodons are encountered in the Ser, Thr, Arg, and Gly family boxes. We thus propose that this codon-anticodon recognition pattern has emerged in the 'M.pneumoniae cluster' under a genomic economization strategy but without the influence of AT pressure.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amikam D., Glaser G., Razin S. Mycoplasmas (Mollicutes) have a low number of rRNA genes. J Bacteriol. 1984 Apr;158(1):376–378. doi: 10.1128/jb.158.1.376-378.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andachi Y., Yamao F., Iwami M., Muto A., Osawa S. Occurrence of unmodified adenine and uracil at the first position of anticodon in threonine tRNAs in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7398–7402. doi: 10.1073/pnas.84.21.7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andachi Y., Yamao F., Muto A., Osawa S. Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J Mol Biol. 1989 Sep 5;209(1):37–54. doi: 10.1016/0022-2836(89)90168-x. [DOI] [PubMed] [Google Scholar]
- Andersson S. G., Kurland C. G. Codon preferences in free-living microorganisms. Microbiol Rev. 1990 Jun;54(2):198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak A. L., Black F. T., Christiansen C., Freundt E. A. Genome size of mycoplasmal DNA. Nature. 1969 Dec 20;224(5225):1209–1210. doi: 10.1038/2241209a0. [DOI] [PubMed] [Google Scholar]
- Barrell B. G., Anderson S., Bankier A. T., de Bruijn M. H., Chen E., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A. Different pattern of codon recognition by mammalian mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3164–3166. doi: 10.1073/pnas.77.6.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonitz S. G., Berlani R., Coruzzi G., Li M., Macino G., Nobrega F. G., Nobrega M. P., Thalenfeld B. E., Tzagoloff A. Codon recognition rules in yeast mitochondria. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3167–3170. doi: 10.1073/pnas.77.6.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman S. D., Hu P. C., Litaker W., Bott K. F. A physical map of the Mycoplasma genitalium genome. Mol Microbiol. 1990 Apr;4(4):683–687. doi: 10.1111/j.1365-2958.1990.tb00638.x. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3' termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65(1):65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Green C. J., Vold B. S. Sequence analysis of a cluster of twenty-one tRNA genes in Bacillus subtilis. Nucleic Acids Res. 1983 Aug 25;11(16):5763–5774. doi: 10.1093/nar/11.16.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J Biol Chem. 1984 Aug 10;259(15):9461–9471. [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman J. E., Sarnoff J., Alzner-DeWeerd B., Yin S., RajBhandary U. L. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3159–3163. doi: 10.1073/pnas.77.6.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hu P. C., Collier A. M., Baseman J. B. Surface parasitism by Mycoplasma pneumoniae of respiratory epithelium. J Exp Med. 1977 May 1;145(5):1328–1343. doi: 10.1084/jem.145.5.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamine J. M., Ho K. C., Loechel S., Hu P. C. Evidence that UGA is read as a tryptophan codon rather than as a stop codon by Mycoplasma pneumoniae, Mycoplasma genitalium, and Mycoplasma gallisepticum. J Bacteriol. 1990 Jan;172(1):504–506. doi: 10.1128/jb.172.1.504-506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamine J. M., Loechel S., Hu P. C. Analysis of the nucleotide sequence of the P1 operon of Mycoplasma pneumoniae. Gene. 1988 Dec 15;73(1):175–183. doi: 10.1016/0378-1119(88)90323-x. [DOI] [PubMed] [Google Scholar]
- Lagerkvist U. Unorthodox codon reading and the evolution of the genetic code. Cell. 1981 Feb;23(2):305–306. doi: 10.1016/0092-8674(81)90124-0. [DOI] [PubMed] [Google Scholar]
- Loechel S., Inamine J. M., Hu P. C. Nucleotide sequence of the deoC gene of Mycoplasma pneumoniae. Nucleic Acids Res. 1989 Jan 25;17(2):801–801. doi: 10.1093/nar/17.2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Yokoyama S., Horie N., Matsuda A., Ueda T., Yamaizumi Z., Kuchino Y., Nishimura S., Miyazawa T. A novel lysine-substituted nucleoside in the first position of the anticodon of minor isoleucine tRNA from Escherichia coli. J Biol Chem. 1988 Jul 5;263(19):9261–9267. doi: 10.1351/pac198961030573. [DOI] [PubMed] [Google Scholar]
- Muto A., Andachi Y., Yuzawa H., Yamao F., Osawa S. The organization and evolution of transfer RNA genes in Mycoplasma capricolum. Nucleic Acids Res. 1990 Sep 11;18(17):5037–5043. doi: 10.1093/nar/18.17.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Oba T., Andachi Y., Muto A., Osawa S. CGG: an unassigned or nonsense codon in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):921–925. doi: 10.1073/pnas.88.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara N., Moriya S., Yoshikawa H. Structure and organization of rRNA operons in the region of the replication origin of the Bacillus subtilis chromosome. Nucleic Acids Res. 1983 Sep 24;11(18):6301–6318. doi: 10.1093/nar/11.18.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo S., Muto A., Kawauchi Y., Yamao F., Osawa S. The ribosomal protein gene cluster of Mycoplasma capricolum. Mol Gen Genet. 1987 Dec;210(2):314–322. doi: 10.1007/BF00325700. [DOI] [PubMed] [Google Scholar]
- Osawa S., Jukes T. H. Codon reassignment (codon capture) in evolution. J Mol Evol. 1989 Apr;28(4):271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- Osawa S., Jukes T. H. Evolution of the genetic code as affected by anticodon content. Trends Genet. 1988 Jul;4(7):191–198. doi: 10.1016/0168-9525(88)90075-3. [DOI] [PubMed] [Google Scholar]
- Ozeki H., Inokuchi H. Organization of transfer RNA genes in prokaryotes. Adv Biophys. 1986;21:35–47. doi: 10.1016/0065-227x(86)90012-2. [DOI] [PubMed] [Google Scholar]
- Razin S. Molecular biology and genetics of mycoplasmas (Mollicutes). Microbiol Rev. 1985 Dec;49(4):419–455. doi: 10.1128/mr.49.4.419-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. J., Simmons J., Walker R. T., Weisburg W. G., Woese C. R., Tanner R. S., Robinson I. M., Stahl D. A., Olsen G., Leach R. H. Construction of the mycoplasma evolutionary tree from 5S rRNA sequence data. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1160–1164. doi: 10.1073/pnas.82.4.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson T., Axberg T., Borén T., Lagerkvist U. Unconventional reading of the glycine codons. J Biol Chem. 1983 Nov 10;258(21):13178–13184. [PubMed] [Google Scholar]
- Samuelsson T., Guindy Y. S., Lustig F., Borén T., Lagerkvist U. Apparent lack of discrimination in the reading of certain codons in Mycoplasma mycoides. Proc Natl Acad Sci U S A. 1987 May;84(10):3166–3170. doi: 10.1073/pnas.84.10.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong B. L., RajBhandary U. L. Mutants of Escherichia coli formylmethionine tRNA: a single base change enables initiator tRNA to act as an elongator in vitro. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8859–8863. doi: 10.1073/pnas.84.24.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoneau P., Hu P. C. The gene for a 4.5S RNA homolog from Mycoplasma pneumoniae: genetic selection, sequence, and transcription analysis. J Bacteriol. 1992 Jan;174(2):627–629. doi: 10.1128/jb.174.2.627-629.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoneau P., Wenzel R., Herrmann R., Hu P. C. Nucleotide sequence of a tRNA cluster from Mycoplasma pneumoniae. Nucleic Acids Res. 1990 May 11;18(9):2814–2814. doi: 10.1093/nar/18.9.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vold B. S. Structure and organization of genes for transfer ribonucleic acid in Bacillus subtilis. Microbiol Rev. 1985 Mar;49(1):71–80. doi: 10.1128/mr.49.1.71-80.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrousek E. F., Hansen J. N. Structure and organization of a cluster of sic tRNA genes in the space between tandem ribosomal RNA gene sets in Bacillus subtilis. J Biol Chem. 1983 Jan 10;258(1):291–298. [PubMed] [Google Scholar]
- Weisburg W. G., Tully J. G., Rose D. L., Petzel J. P., Oyaizu H., Yang D., Mandelco L., Sechrest J., Lawrence T. G., Van Etten J. A phylogenetic analysis of the mycoplasmas: basis for their classification. J Bacteriol. 1989 Dec;171(12):6455–6467. doi: 10.1128/jb.171.12.6455-6467.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel R., Herrmann R. Cloning of the complete Mycoplasma pneumoniae genome. Nucleic Acids Res. 1989 Sep 12;17(17):7029–7043. doi: 10.1093/nar/17.17.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel R., Pirkl E., Herrmann R. Construction of an EcoRI restriction map of Mycoplasma pneumoniae and localization of selected genes. J Bacteriol. 1992 Nov;174(22):7289–7296. doi: 10.1128/jb.174.22.7289-7296.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley J. C., Muto A., Finch L. R. A physical map for Mycoplasma capricolum Cal. kid with loci for all known tRNA species. Nucleic Acids Res. 1991 Jan 25;19(2):399–400. doi: 10.1093/nar/19.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamao F., Iwagami S., Azumi Y., Muto A., Osawa S., Fujita N., Ishihama A. Evolutionary dynamics of tryptophan tRNAs in Mycoplasma capricolum. Mol Gen Genet. 1988 May;212(2):364–369. doi: 10.1007/BF00334708. [DOI] [PubMed] [Google Scholar]
- Yamao F., Muto A., Kawauchi Y., Iwami M., Iwagami S., Azumi Y., Osawa S. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F., Laigret F., Whitley J. C., Citti C., Finch L. R., Carle P., Renaudin J., Bové J. M. A physical and genetic map of the Spiroplasma citri genome. Nucleic Acids Res. 1992 Apr 11;20(7):1559–1565. doi: 10.1093/nar/20.7.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]