Abstract
Control of Johne’s disease, caused by Mycobacterium avium subsp. paratuberculosis, has been difficult because of a lack of an effective vaccine. To address this problem we used targeted gene disruption to develop candidate mutants with impaired capacity to survive ex vivo and in vivo to test as a vaccine. We selected relA and pknG, genes known to be important virulence factors in Mycobacterium tuberculosis and Mycobacterium bovis, for initial studies. Deletion mutants were made in a wild type Map (K10) and its recombinant strain expressing the green fluorescent protein (K10-GFP). Comparison of survival in an ex vivo assay revealed deletion of either gene attenuated survival in monocyte-derived macrophages compared to survival of wild-type K10. In contrast, study in calves revealed survival in vivo was mainly affected by deletion of relA. Bacteria were detected in tissues from wild-type and the pknG mutant infected calves by bacterial culture and PCR at three months post infection. No bacteria were detected in tissues from calves infected with the relA mutant (P < 0.05). Flow cytometric analysis of the immune response to the wild-type K10-GFP and the mutant strains showed deletion of either gene did not affect their capacity to elicit a strong proliferative response to soluble antigen extract or live Map. Quantitative RT-PCR revealed genes encoding IFN-γ, IL-17, IL-22, T-bet, RORC, and granulysin were up-regulated in PBMC stimulated with live Map three months post infection compared to the response of PBMC pre-infection. A challenge study in kid goats showed deletion of pknG did not interfere with establishment of an infection. As in calves, deletion of relA attenuated survival in vivo. The mutant also elicited an immune response that limited colonization by challenge wild type Map. The findings show the relA mutant is a good candidate for development of a live attenuated vaccine for Johne’s disease.
Keywords: Mycobacterium avium subsp. paratuberculosis, Johne’s disease, paratuberculosis, Crohn’s disease, live vaccine
1. INTRODUCTION
Mycobacterium avium subsp. paratuberculosis (Map) is a pathogen with a broad-host range (reviewed in[1]). Although it can infect multiple species including humans [2, 3], it is primarily a disease problem in ruminants. It is the causative agent of Johne’s Disease (JD). The prevalence in U.S dairy herds is estimated to be 68% according to the most recent report from the National Animal Health Monitoring System [4]. A growing concern, however, is that Map from dairy cattle may also present a threat to individuals at risk for developing Crohn’s disease (CD), a form of inflammatory bowel disease [5]. Map has been identified in tissues from children [6, 7] and adults with CD [8]. The higher frequency of detection of Map [2, 3] and the presence of Map antigen (Ag) specific T cells [9] in ileal tissues from patients with CD supports the supposition that Map may be a contributing factor in CD pathogenesis. Additional evidence supporting the supposition may be the observation that childhood intestinal tuberculosis, caused by Mycobacterium tuberculosis (Mtb), has been occasionally misdiagnosed as CD [10]. These observations emphasize the importance of developing a strategy to control Johne’s disease (JD) for the livestock industry and human health.
JD has been difficult to control because of a limited understanding of the mechanisms of pathogenesis and the lack of a vaccine that prevents establishment of a persistent infection. When cattle are exposed to Map, they initially develop an immune response that controls but does not eliminate the pathogen. Efforts to overwhelm the response by exposing young calves to single and serial doses of up to 108 bacteria by the oral route or directly through an indwelling cannula to accelerate disease progression have shown the immune response is resilient and not readily suppressed [11, 12]. Similar results have been observed with calves inoculated with up to 1012 bacteria and treated with dexamethasone [13]. An immune response to Johnin purified protein derivative (jPPD) and soluble antigens (SAg), dominated by CD4 memory T cells, become consistently detectable two months post infection (PI). A less vigorous CD8 memory T cell response occurs during the same time frame becoming more pronounced one year PI. Both NK and γδ T cells become activated but activation appears to be nonspecific [12]. Over time, however, persistent infection triggers an event that dysregulates protective immunity and allows disease to progress in some animals, usually 2 or more years PI. This latent stage of infection is similar to latency in tuberculosis (TB). Studies conducted thus far in TB and JD have not identified the triggering event. However, investigations in Mycobacterium tuberculosis (Mtb) indicate dysregulation is associated with the capacity of pathogenic mycobacteria to establish a persistent infection in dendritic cells (DC) and macrophages [14-16] and modulate secretion of IL-12 and IL-23, cytokines that play a pivotal role in driving the differentiation of naïve CD4 and CD8 T cell subsets with effector activity (Th1, Th2, Th17 and Th22 cells) or regulatory activity (regulatory T cells, Tr cells) [17-21]. Th17 and Th22 cells are the source of newly identified proinflammatory cytokines IL-17 and IL-22, granulysin (a mycobactericidal peptide), and IFN-γ, the cytokine that primes DC for initiation of adaptive immunity [22-25]. Dysregulation of cytokine signaling from infected cells could be the triggering event. Studies in humans and mice suggest dysregulation of cytokine signaling leads to induction of regulatory T (Tr) cells that modulate the response to Mtb and contribute to the activation of Mtb in latently infected subjects [26, 27]. It is postulated that Tr play a similar role in modulating the immune response to Map [28]. Regardless of where the triggering event occurs it is clear that a vaccine must block the capacity of Map to establish an infection.
Killed vaccines have proven useful in limiting disease progression and more recently have shown potential as a therapeutic agent [29, 30]. However, they have not been effective in preventing infection [31]. Similar results have been obtained recently with subunit vaccines, emphasizing the potential and limitation of such vaccines for controlling JD [32]. The inability of killed and subunit vaccines to prevent establishment of infection leaves the potential for infected animals to break with disease if protective immunity wanes [31]. Further studies are clearly needed to determine if a vaccine can be developed that elicits an immune response that prevents establishment of a persistent infection. Because of the issues with killed and peptide based vaccines interest has increased in the potential of developing genetically attenuated mutants for evaluation as vaccines for JD. This strategy has been successful for other pathogens. Large libraries of mutants have been developed by transposon mutagenesis and are currently being screened to identify candidates for evaluation [33-35]. The sequencing of the Map genome, however, has provided an opportunity to take a more direct approach to developing mutants. To take advantage of this approach, we adapted the method of targeted allelic exchange mutagenesis for use with Map [36]. Three orthologues of genes associated with virulence or function in Mtb and M. bovis (Mbv), relA and pknG [37, 38], and lsr2 [39], were initially selected and successfully disrupted in the study.
The general procedure currently in use to evaluate deletion mutants of Map for potential as a vaccine has been to initially screen for attenuation in macrophages ex vivo on the premise that attenuation would be an indicator of how a mutant would survive in vivo. Based on the same premise, the mouse model has been selected for further evaluation of vaccine potential. The goat model has been selected for use in the final evaluation of candidate vaccines for efficacy (reviewed in [40]). Since we have been interested in determining the effect of mutation on the immune response to candidate vaccines as well as attenuation, we have focused on using the natural hosts of Map in our studies. We developed a flow cytometric assay and the monoclonal antibody reagents needed to study the immune response in cattle [11, 12]. We also developed a cannulated ileum model to provide continuous access to the ileum and means for studying the effect of vaccination on survival of challenge bacteria [11]. The objectives of the present study were to evaluate the effect of deletion of two of the candidate genes, relA and pknG, on survival ex vivo in macrophages and in vivo in calves and goats and also determine whether gene deletion affected the capacity of the mutant to elicit an immune response. We were especially interested in whether cells producing IL-17, IL-22, and granulysin were involved in the response to the mutant and wild type Map.
2. MATERIALS AND METHODS
2.1. Preparation of bacteria
A clinical isolate of wild-type Map strain, K10Wt, and its derivative expressing green fluorescence protein, K10Wt-GFP, were generously provided by Raul Barletta, University of Nebraska, Lincoln, NE [41]. The deletion mutants of K10Wt (K10pknG and K10relA) and K10Wt-GFP (K10pknG-GFP and K10relA-GFP) used in this study were previously generated in our laboratory [36]. Each strain was enriched from a single colony in Middlebrook 7H9 medium (Difco) supplemented with 6.7% para-JEM GS (Trek Diagnostic Systems, OH), 2 μg/ml of mycobactin J (Allied Monitor, MO), and 0.05 % Tween 80 (7H9 broth medium) [36]. When the broth culture reached an OD600 of 0.6 to 0.8, master stocks were prepared in 1.5 ml microcentrifuge screw-cap tubes, stored at -80° C, and used for each infection as previously described [11]. The bacterial numbers were estimated based on the OD600 values [36].
2.2. Calves
Fifteen newborn Holstein bull calves were obtained from the Washington State University research dairy. The dairy is a closed herd that is considered to be free of paratuberculosis. The dairy has no history of paratuberculosis for over 30 years and performs periodic whole-herd ELISA screening tests to verify disease free status [11]. All calves were fed 4 L of maternally derived colostrum within 6 hours of birth, and taken to a Biosafety Level 2 (BL-2) isolation unit within the first 24 hours of life. Calves were fed milk replacer, whey pellets, calf starter grain, and free choice alfalfa hay. All protocols and procedures were approved by the Washington State University Institutional Animal Care and Use Committee.
2.3. Ileal cannulation and inoculation of calves
Calves were raised in isolation facilities for 8-9 weeks until they were weaned from milk replacer and deemed to be large enough for surgery. Then animals were divided into three groups for different Map strain infection: K10relA-GFP (n=6), K10pknG-GFP (n=6), and K10Wt - GFP (n=3). The surgical procedure and post surgical care were performed as described by Allen et al. [11]. A calf in each mutant group was lost due to unexpected death.
Two weeks post surgery, the calves were inoculated with wild-type or mutant strains of Map (109 CFU in 20 ml of PBS) directly into the ileum. This was accomplished by inserting a French Foley catheter into the cannula and inflating it to block flow of ingesta. The catheter was left in place for 1 hour to allow the inoculum to remain in contact with ileal tissue. Each group of calves was housed in a separate isolation building to prevent cross infection. Fecal samples were collected at 1 wk before infection, and 24 h, 2, 6, and 12 wks PI to screen for bacterial shedding. The feces were stored at -20° C until processed. All fecal samples collected before infection were PCR negative. All infected animals were euthanized at three months PI as described [11].
2.4. Blood processing and isolation of peripheral blood mononuclear cells
Blood was collected from calves before inoculation, at 8 wks PI, and at the time of necropsy (three months PI). Peripheral blood mononuclear cells (PBMCs) were isolated as previously described [11]. Isolated PBMCs were re-suspended in RPMI-1640 medium supplemented with 10mM HEPES, 13 % bovine calf serum (BCS), 2-mercapto ethanol, 20 mM L-glutamine, and antibiotics (100 units/ml of penicillin G and 100 μg/ml of streptomycin sulfate), and then used as described.
2.5. Preparation and infection of bovine macrophages with Map
Bovine monocyte-derived macrophages (MΦ) were prepared in 24 well tissue culture plates as previously described [42]. The differentiated MΦ (approximately 105 cells/well) were infected with each Map strain at a multiplicity of infection of 10 for 3 hrs. Extracellular bacteria were removed by washing 3 times with warm RPMI-1640. Infected macrophages were lysed immediately (defined as day 0) or cultured in RPMI-1640 medium without antibiotics for 6 days (defined as day 6). Macrophages were lysed with 0.1 % saponin in PBS, and the lysates were plated in10 fold dilutions on 7H9 agar plates to determine the number of bacteria. The experiments were performed in triplicate and repeated 2 times.
To compare transcription of cytokines in infected cells, additional bovine MΦ were prepared in 60 mm-diameter tissue culture plates (Becton Dickinson), and infected as described above. At 6 h and 24 h PI, the culture medium was removed and MΦ were lysed in 1 ml of Trizol (Invitrogen). Total RNA was extracted from cells according to the manufacturer’s instructions.
2.6. Map culture and tissue sample processing
Nine different tissue sites of the intestine and lymph nodes were obtained at the time of necropsy. The method for culturing Map from tissues was slightly modified from a previous method [43]. In brief, 1.5 grams of tissue were homogenized in a 15 ml Closed Tissue Grinder System (Fisher Scientific) in 15 ml of distilled water. The homogenates were allowed to stand for 30 min. Supernatant (7 ml) was transferred into a new 15 ml tube. Two ml of the supernatant was taken and distributed into two 2 ml screw-cap tubes (1 ml for each). The tubes were centrifuged at 15,000 × g for 15 min. The pellets were stored at -20° C for identification of Map DNA by real-time PCR (RT-PCR). The remaining 5 ml of supernatant was decontaminated by mixing with the same volume of 1.5 % hexadecyl pyridinium chloride and incubated overnight. The tube was centrifuged at 3,800 × g for 30 min with no brake. The pellet was resuspended in 1 ml of 7H9 broth medium supplemented with 1.9 % para-JEM AS (antibiotic solution containing vancomycin, amphotericin B, nalidixic acid) (Trek Diagnostic Systems, OH). Three 7H9 agar plates containing 1.9 % para-JEM AS were inoculated with 0.2 ml of the re-suspended pellet. The cultures were incubated at 37° C for 14 to 16 wks.
2.7. Extraction and RT-PCR identification of Map DNA from tissue and fecal samples
The frozen tissue pellets were pre-treated in 360 μl of tissue lysis buffer (ATL buffer, Qiagen) and 40 μl of protein kinase (>600 mAU/ml, Qiagen) at 56° C for one hour. Tubes were then centrifuged at 15,000 xg for 15 min, and the supernatant carefully removed using a 1 ml pipette tip. The remaining steps for DNA extraction were performed with the DNeasy Blood and Tissue kit (Qiagen) following a protocol for Gram-positive bacteria. During the lysis steps in the protocol, the bacteria were mechanically disrupted with 0.1-mm zirconia-silica beads (BioSpec Products, Inc., OK) in FastPrep-24 instrument (MP biochemicals, CA) to facilitate DNA release. DNA was eluted in 200 μl of AE buffer.
Fecal DNA was extracted, in duplicate, using a method described by Schonenbruche et al. [44]. The lysing matrix D tube used in the original study was replaced with a 2 ml screw-cap tube containing 200 μl of 0.1-mm zirconia-silica beads.
RT-PCR detection of the IS900 sequence was performed using a StepOnePlus Real-Time PCR System machine (Applied Biosystems, CA). Five μl of eluted DNA sample were used in final 25 μl of reaction mixture. The RT-PCR conditions and sequences for primer and probe were the same as previously described [45]. Each sample was run in duplicate.
2.8. PBMC stimulation with live Map and Map antigens
PBMCs (107) were distributed into 6-well culture plates. Each well was prepared to contain 5 × 106 CFU of live Map K10, 20 μg of Map SAg, or none (“nil” treatment) in 5 ml of RPMI-1640 medium as previously described [11]. Identical plates were prepared for each of the experimental procedures. PBMCs stimulated with live Map, Map SAg, or medium alone were collected from three identical wells on day 6, and used for flow cytometric analysis (FC).
To compare the level of cytokine transcriptions between PBMCs isolated from calves before and after infection, three identical plates for live Map stimulation or un-stimulated (control) were set up in 24-well plates. After three days incubation, total RNA was extracted from the pooled PBMCs in triplicate.
2.9. Flow cytometric analysis of stimulated PBMCs
FC analysis was conducted as previously described [11]. The strategy used for FC analysis is shown in FIG. 1. PBMCs cultured for 6 days with/without Ag stimulation were labeled with a three color combination of monoclonal antibodies: CD4 (ILA11A, IgG2a), CD45R0 (ILA116A, IgG3), and CD25 (CACT116A, IgG1), or CD8 (7C2B, IgG2a), CD45R0, and CD25. The percent activated CD4 or CD8 memory T cells in total CD4 or CD8 T-cell pool was determined by using electronic gates to isolate CD4 and CD8 populations for analysis. Electronic gates, placed on small un-activated (resting) and large activated (proliferating) cells based on side-scatter (SSC) vs. forward-scatter (FSC), were used to track the proportion of activated cells in the culture as previously described [11]. Data were collected with Cell Quest software (Becton Dickinson Immunocytometry Systems, San Jose, CA) from a FACSort flow cytometer, and analyzed with FCS Express software (De Novo software, Thornton, Ontario, Ca).
FIG. 1.
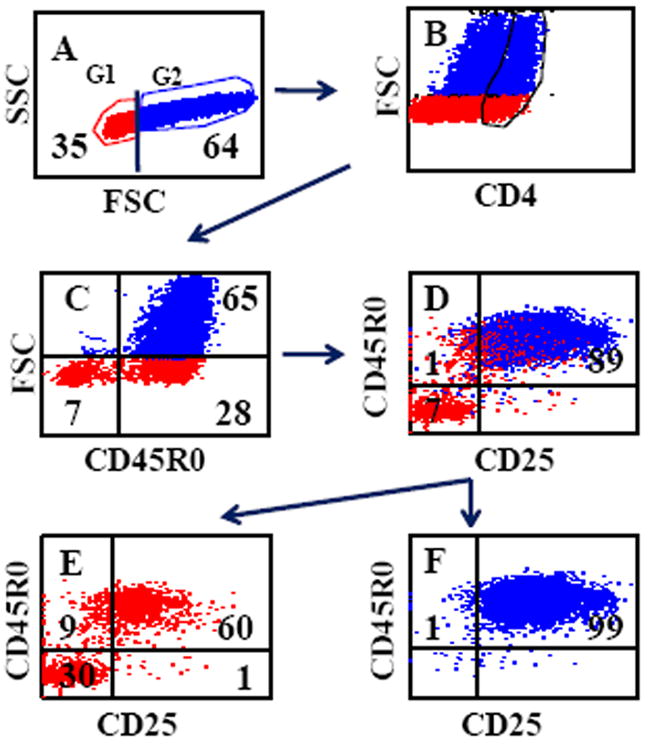
Gating strategy used to determine the frequency of activated CD4 memory T cells. Electronic gates 1 and 2 were placed on small unactivated and large activated cells (SSC vs FSC) and color coded to identify T cells proliferating in response to antigen stimulation (A). An additional electronic gate was used to isolate CD4 T cells to determine the frequency of CD45R0/CD25+ CD4 memory T cells proliferating in response to antigen stimulation (B). (C) Shows the frequency of activated CD4 memory T cells (FSC vs CD45R0). (D) Shows that resting and activated CD4 memory T cells express CD25. A combination of electronic gates placed on small unactivated or activated CD4 T cells, (E) and (F) respectively, were used to show that only unactivated and activated memory T cells express CD25. No naïve CD4 cells express CD25 (lower left quadrant in E). All activated memory CD4 T cells express CD25 (F). This observation has allowed us to use FSC vs CD45R0 to determine the frequency of activated CD4 memory T cells (C). The same gating strategy was used to determine the frequency of activated memory CD8 cells.
2.10. Relative quantification of cytokine transcription
Quantitative reverse-transcription RT-PCR (qRT-PCR) was performed using extracted RNA (see above). The extracted RNA was treated with Turbo DNA-free kit (Applied Biosystems, TX) to remove genomic DNA contamination. RNA was reverse transcribed to cDNA using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, CA) according to the manufacturer’s instructions. The reaction mixture was prepared in a total of 20 μl containing 1 × Power SYBR Green PCR Master Mix (Applied Biosystems, CA), 200 nM of each primer, and cDNA. The qRT-PCR was performed in a StepOnePlus Real-Time PCR System machine (Applied Biosystems, CA) under the following conditions: 95° C for 10 min, 40 cycles of 95° C for 15 sec and 60° C for 1 min, followed by a dissociation analysis. The relative quantification (RQ) of gene transcription was analyzed using Step One software v2.1 program (Applied Biosystems, CA) by the 2-(ΔΔCt) method [46]. Briefly, ΔCt of each gene was calculated based on the mean Ct value of the two housekeeping genes (β-actin and GAPDH). Then, ΔΔCt was calculated using appropriate control groups as the calibrators (un-stimulated MΦ, un-stimulated PBMCs, or Map-stimulated PBMCs before infection). The results were expressed as relative quantification of transcription compared to those of control groups. The primer sequences used in this study are listed in Table 1. The primers developed in this study were designed using Primer Express 3.0 (Applied Biosystems, CA).
Table 1.
Primers used for qRT-PCR quantification for cytokine transcription.
| Gene | Forward primer | Reverse primer | Sourcea |
|---|---|---|---|
| β-actin | GGAATCCTGCGGCATTCAC | GGATGTCGACGTCACACTTCA | NM173979 |
| GAPDH | GCATCGTGGAGGGACTTATGA | GGGCCATCCACAGTCTTCTG | [59] |
| IL-10 | GAGCAAGGCGGTGGAGAAGG | GATGAAGATGTCAAACTCACTCATGG | NM174088 |
| IL-12p35 | CAGCAAGCCCAGGAAGGA | TGACAGCCCTCAGCAGGTTT | NM174355 |
| IL-17 | GGACTCTCCACCGCAATGAG | TGGCCTCCCAGATCACAGA | NM001008412 |
| IL-22 | GAGGTGCTGTTCCCCCAAT | GAAGGGCACCACCTTTTCC | NM001098379 |
| IL-23p19 | CACAAGGACTCAGGGACAACAG | AAAAACCAGGCCTCGATGAA | XM588269 |
| Granb | GACCTGCTGCTCCAAGGAGAA | CCAACTTGTCCATCAGATGTTGTATT | NM001075143 |
| Foxp3 | CCGGCATGCCTAGGAAAGA | ACGCCATTTGCTAGCAGTGAGT | DQ322170 |
| IFN-γ | CTTGAATGGCAGCTCTGAGAAA | TGCAGATCATCCACCGGAAT | [60] |
| T-bet | GAGGCGGCCTGCAACAC | CGGCGATGAACTGGGTTT | XM583748 |
| TGF-β | CATCTGGAGCCTGGATACACAGT | GAAGCGCCCGGGTTGT | [60] |
| TNF-α | TCTACCAGGGAGGAGTCTTCCA | GTCCGGCAGGTTGATCTCA | [61] |
| RORC | GAGTGCCTTGCGCTTTTCA | GATGAGGACGAGGGCTGTG | DQ667051 |
Gene accession no. is given for the primers developed in this study.
Granulysin.
2.11. Goat challenge study
Thirteen newborn goats were obtained from the goat herd maintained by the USDA-ARS unit at Washington State University. It has been maintained as a closed herd for 30 years. No animals have been diagnosed with JD during this time. All of the goats were maintained in a barn with concrete floor pens. All protocols and procedures were approved by the Washington State University Institutional Animal Care and Use Committee.
One goat (Y23) was euthanized, and tissue samples were collected. Fecal samples from all animals before inoculation were also collected. All tissue and fecal samples collected before inoculation were Map culture negative. The remaining goats (~8 months of age when used) were divided into 4 groups of 3 for the study and moved to separate BL-2 holding facilities for the duration of the studies. One group each was orally inoculated per os with 109 K10pknG, K10relA, or K10Wt. One group was not inoculated until the time of challenge. After 2 months, all groups were challenged with 109 K10Wt by oral route. All of the goats were euthanized and necropsied at 2 months post challenge. Tissues were collected and processed for bacterial culture as described above. Each 0.2 ml of the final re-suspension was inoculated on 7H9 agar plate with or without hygromycin (75 μg/ml), in duplicate, to distinguish the CFU originated from mutant or wild-type strains.
2.12. Statistical analysis
All statistical analyses were performed using MedCalc statistical software ver. 11. 2. 1 (Belgium). For comparison of intracellular survival of Map strains, the CFU data on day 6 were normalized by transforming to the percent survival, based on the CFU present at day 0. The results of survival rate of mutants and the relative quantification of gene transcription were analyzed using the Kruskal-Wallis test or one-way ANOVA. The numbers of Map culture positive animals between groups were compared by Fisher’s exact test. In all tests, a P value of less than 0.05 was considered significant.
3. RESULTS
3.1. Survival of Map mutants in bovine MΦ
The number of wild-type and mutant Map in MΦ was determined on days 0 and 6, and the survival rate of each strain on day 6 was calculated using the CFUs obtained on days 0 and 6. The survival rate of all Map strains decreased on day 6 compared to those at day 0, but the survival rates of K10pkng (5.1 ± 1.0 %; SD) and K10relA (12.5 ± 6.3 %) mutants on day 6 were significantly lower than that of the parental strain (29.4 ± 3.6 %) (P < 0.05). In addition, there was a statistically significant difference between K10pkng and K10relA (P < 0.05) (FIG. 2). These data suggest that the pknG mutant is more attenuated in bovine macrophages than relA in comparison to the wild-type strain.
FIG. 2.
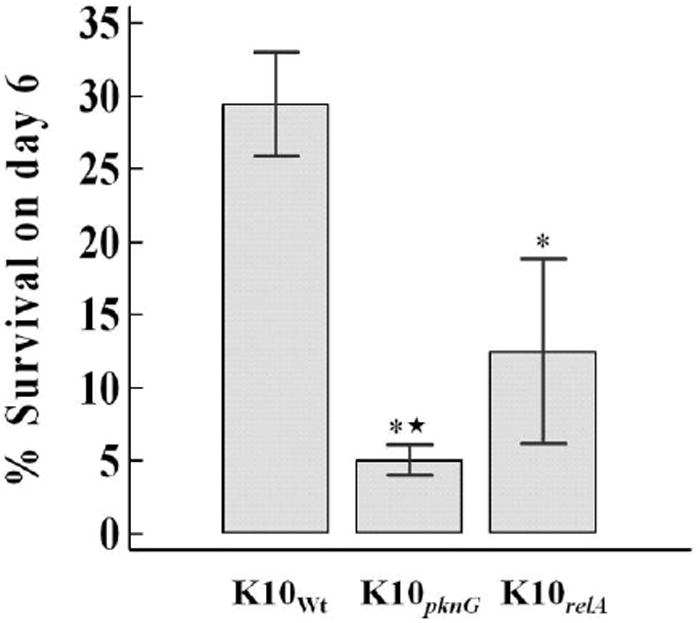
Survival of wild-type and mutant strains of Map in macrophages. Bovine macrophages were infected with wild-type or mutant Map strains. The bacterial numbers were counted on days 0 and 6. The percent survival of each strain on day 6 was calculated based on the CFU taken at day 0. The results are represented as the mean percent survival of each strain with error bar (SD) from two independent experiments (each in triplicate). * and ★, significant decrease compared to K10Wt and K10relA, respectively (P < 0.05).
3.2. Comparison of cytokine gene transcription in MΦ infected with wild-type and mutant strains of Map
The transcription levels of IL-10, TNF-α, IL-12p35, and IL-23p19 in bovine macrophages were measured by qRT-PCR at 6 and 24 h PI to determine the effect of infection on cytokine expression (FIG. 3). The expression of all 4 cytokines increased to some degree following infection, but significant differences were only observed for TNF-α and IL23 in all infected MΦ groups compared to those of uninfected MΦ at 24 h PI. While IL-23 transcription was strongly elevated in response to all the Map strains, the transcription of IL12p35 was only slightly changed. There were, however, no significant differences found between MΦ infected with wild-type and mutant strains for the cytokines measured in this study.
FIG. 3.
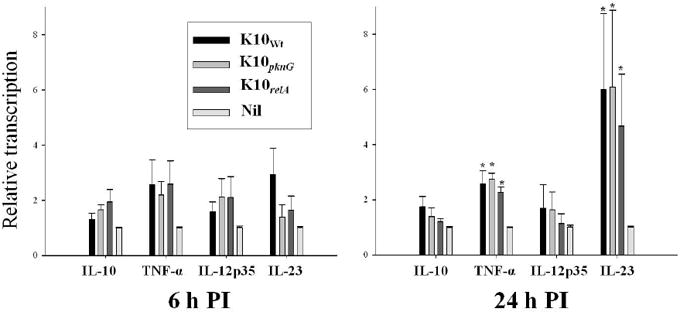
Transcription of cytokines in bovine macrophages infected with wild or mutant Map strains. Total RNA was extracted from macrophages after 6 and 24 h infection, and the relative quantification of cytokine transcription was measured by qRT-PCR. The relative quantification of gene transcription was calculated using uninfected macrophages (nil control) as calibrator by 2-(ΔΔCt) method. Two housekeeping genes (β-actin and GAPDH) were used as controls. Data are shown as the mean of two independent duplicated assays. Error bar indicates standard error. *, significant difference compared to the cytokine transcription in nil control (p < 0.05).
3.3. Map culture and RT-PCR identification in animal tissues and fecal samples
All three K10Wt-GFP infected calves were bacterial culture positive at necropsy, but only three of five K10pkng-GFP infected calves were positive at necropsy. All five K10relA-GFP infected calves were culture negative (Table 2), which was statistically significant (P < 0.05). The mean percentages of Map culture positive tissue were 48.1 % (28.7 – 68.1, 95 % CI) for K10Wt-GFP, 24.4 % (12.9 – 39.6) for K10pkng-GFP, and 0 % (0 – 7.87) for K10relA-GFP infected groups, respectively (Table 2). Screening for the presence of Map DNA by RT-PCR yielded results consistent with the culture results (kappa = 0.74). Seven tissues, defined as positive by Map culture, were negative by RT-PCR, but two negative tissues defined by Map culture were positive by RT-PCR. Six of the seven false negative tissues with RT-PCR had low colony counts (The sum of the CFUs from the triplicate plates was 5 or fewer). Importantly, none of the tissues from K10relA-GFP infected group were positive by RT-PCR.
Table 2.
Map culture results from 9 tissue sites from calves at necropsy.
| Strain | Animal | JP | JM | JD | IP | IM | ID | ICV | ML | ICL |
|---|---|---|---|---|---|---|---|---|---|---|
| 136 | -a | - | - | - | 5b | 285 | 46 | 3 | 35 | |
| K10Wt- | ||||||||||
| 139 | - | - | 11 | - | 125 | 3 | - | - | 9 | |
| GFP | ||||||||||
| 151 | - | - | - | - | 2 | - | 5 | 2 | 7 | |
| 134 | - | - | - | - | - | - | - | - | - | |
| 135 | - | - | - | - | - | - | - | - | - | |
| K10relA- | ||||||||||
| 137 | - | - | - | - | - | - | - | - | - | |
| GFP | ||||||||||
| 138 | - | - | - | - | - | - | - | - | - | |
| 140 | - | - | - | - | - | - | - | - | - | |
| 145 | - | - | - | - | - | - | - | - | - | |
| 146 | - | 1 | - | 690 | 410 | 13 | 18 | - | 1 | |
| K10pkng- | ||||||||||
| 147 | - | - | - | 1 | 5 | - | - | - | - | |
| GFP | ||||||||||
| 148 | - | - | - | - | 119 | 1 | 11 | - | - | |
| 149 | - | - | - | - | - | - | - | - | - |
JP, JM, JD: jejunum proximal, middle, distal; IP, IM, ID: ileum proximal, middle, distal; ICV: ileocecal valve; ML, ICL: mesenteric and ileocecal lymph nodes.
Negative.
Values are expressed as the total CFU obtained from each tissue.
Fecal Map-PCR results revealed that all infected animals had passive bacterial shedding at 24 h PI. One or more animals in each group was fecal Map-PCR positive at two wks PI. After two wks, only one of the K10pknG-GFP group and one of the K10Wt-GFP group were positive by fecal Map-PCR at 6 and 12 wks PI, respectively (Table 3).
Table 3.
Real-time PCR detection of IS900 in calve fecal samples.
| Group | 24 h | 2 wks | 6 wks | 12 wks |
|---|---|---|---|---|
| K10Wt-GFP | 3/3a | 1/3 | 0/3 | 1/3 |
| K10relA-GFP | 5/5 | 2/5 | 0/5 | 0/5 |
| K10pknG-GFP | 5/5 | 2/5 | 1/5 | 0/5 |
Values are expressed as the number of positive calves / the number of tested calves.
3.4. Flow cytometric analysis of PBMC from calves stimulated with live Map or Map SAg
The FC results with PBMC stimulated with live Map or Map SAg showed specific memory response against live Map and Map SAg after inoculation. The percentage of activated CD4 and CD8 memory T cells increased in response to stimulation with live Map and Map SAg until necropsy in the wild-type Map infected group (FIG. 4A). Remarkably, live Map elicited a more vigorous CD4 and CD8 T-cell proliferative response than Map SAg. In addition, the CD8 T-cell response was much higher for live Map than the CD4 T-cell response at 12 wks PI.
FIG. 4.
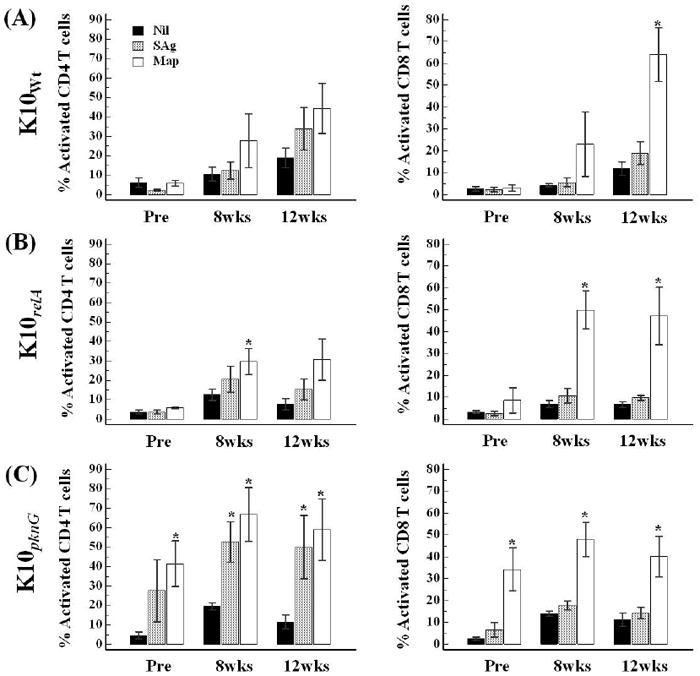
Comparison of memory T cell response in PBMCs isolated before and after infection by flow cytometric analysis. The data are presented as the mean percent (with SE) of activated CD4 or CD8 memory T cells in response to Map antigen stimulation or nil treatment at different time points. Pre, pre-infection; Nil, nil-stimulation; SAg, soluble anigen stimulation; Map, live Map stimulation; *, significant difference compared to the value of nil control (P < 0.05).
Overall, the memory T-cell response to the relA mutant infected group was similar to that of the wild-type Map infected group, although the peak was reached at 8 wks PI, and persisted until necropsy. Significant differences compared to un-stimulated PBMC were observed only when stimulated with live Map (both 8 and 12 wks PI). The CD8 T-cell response against live Map stimulation was also more vigorous than the CD4 T-cell response (FIG. 4B). The pattern of memory T-cell response of the pknG mutant infected group was similar to that of the relA mutant infected group. However, as noted in a previous study, there was a high response to live Map and Map SAg at pre-infection (FIG. 4C) [11].
3.5. Cytokine transcription in PBMCs stimulated with live Map
Relative changes of cytokine transcription in PBMCs stimulated for three days with live Map were compared between two time points (pre-infection vs necropsy) by qRT-PCR. The K10pknG infected group was excluded from this analysis due to the unexpected high responsiveness to Map SAg at pre-infection. The data have been grouped to compare the cytokine profiles considered to define Th1, Th17, and regulatory T cells (Treg).
The general profiles of transcriptional change of cytokine expression between pre-infection and three months PI were similar in the K10Wt and K10relA groups. The transcription of IFN-γ, an indicator of a Th1 response, was elevated significantly in both K10Wt and K10relA infected groups (mean fold changes: 896 and 106, respectively). The transcription factor, T-bet, was also increased (FIG. 5A). Cytokines secreted by Th17 cells, IL-17 and IL-22, were also up-regulated in both infected groups at three months PI. While the transcriptional changes of the two cytokines were significant in the K10relA infected group (P < 0.05), they were not significant in the K10Wt infected group. A transcription factor for Th17 cells, RORC, was simultaneously up-regulated in both groups (FIG. 5B). Regulatory cytokines secreted by Treg, IL-10 and TGF-β, and a transcription factor, Foxp3, were not changed or were slightly decreased in the level of transcription at three months PI compared to those at pre-infection (no significant changes in both experimental groups, P > 0.05) (FIG. 5C).
FIG. 5.
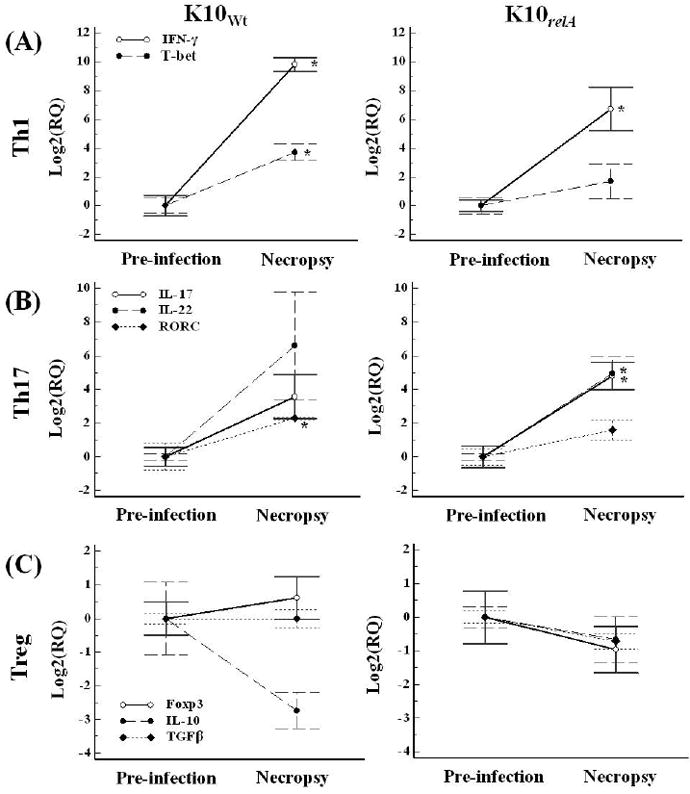
Relative transcription of cytokines in PBMCs in response to live Map stimulation measured by qRT-PCR. PBMCs isolated at pre-infection and necropsy were simulated with live Map for 3 days. The relative transcription was calculated using the value at pre-infection as the calibrator with two housekeeping genes (β-actin and GAPDH). Data are presented as the mean value of each group with error bar (SD). RQ, relative quantification; *, significant difference compared to the value at pre-infection (P < 0.05).
Stimulation with live Map induced expression of granulysin mRNA in PBMCs from calves before infection (mean, 4.4 – 6.6-fold change). Nevertheless, larger differences in expression of granulysin mRNA were observed when compared to the same treated PBMCs from naïve (before infection) and infected calves (at necropsy). Un-stimulated PBMCs isolated at necropsy produced significantly higher amounts of granulysin mRNA compared to expression in PBMCs isolated before infection (P < 0.05 for both infected groups). The increased level was higher in the K10Wt infected group (mean, 186.2-fold change) than in K10relA mutant infected group (mean, 18.2-fold change). This increase in transcription of granulysin mRNA in PBMCs from infected calves was further increased when stimulated with live Map. The mean fold change (Map stimulation compared to un-stimulation) in PBMCs from infected calves was 3.7-fold for the wild-type infected group (not significant, P > 0.05) and 15.5-fold for the mutant infected group (significant, P < 0.05), respectively (FIG. 6).
FIG. 6.
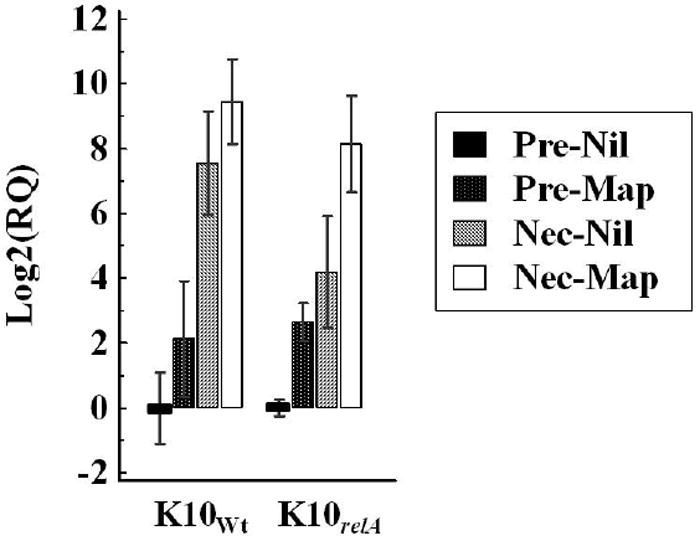
Relative transcription of granulysin mRNA. PBMCs isolated at pre-infection on and necropsy were cultured with live Map or medium alone (nil treatment) for 3 days. The level of mRNA transcription was measured by qRT-PCR. The relative quantification of mRNA transcription was calculated using a calibrator (nil-treated PBMCs at pre-infection) and 2 housekeeping genes (β-actin and GAPDH). Data are shown as the mean fold changes with error bars (SD). RQ, relative quantification; Pre, pre-infection; Nec, necropsy; Nil, nil-stimulation; Map, live Map stimulation.
3.6. Effect of exposure to K10pknG and K10relA mutants on survival of K10Wt in goats
The findings with calves suggested one of the mutants might be a candidate for further evaluation as a vaccine. To pursue this possibility, a pilot study was conducted with a smaller ruminant animal, goats, to determine if the immune response elicited by either mutant blocks or diminishes the capacity of K10Wt to establish an infection. As shown in Table 4, the goats exposed two times with K10Wt at two month interval had more culture positive tissues and higher bacterial numbers than those observed from animals only exposed once at the time of challenge. Interestingly, inoculation with the K10pknG mutant before challenge resulted in an increase in the number of culture positive tissues and bacterial burden in comparison with tissues from goats exposed once or twice to K10Wt. The frequency of colonies growing in the presence and absence of hygromycin indicate most of the colonies obtained from doubly-infected goats were K10pknG mutants. This finding differs from results obtained in calves that were only exposed to the K10pknG mutant. In contrast, no K10relA mutants were cultured from any of the tissues from goats inoculated with the K10relA mutant and challenged with K10Wt. In addition, inoculation with K10relA attenuated colonization with K10Wt. K10Wt were only cultured from a single tissue from each goat pre-exposed to K10relA (Table 4).
Table 4.
Map culture results from 9 tissue sites in challenged goats.
| Pre-exposure | Challenge | Goat | JP | JM | JD | IP | IM | ID | ICV | ML | ICL |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 19 | -a | 2/1b | 0/1 | C/Cc | 157/151 | TN/TNd | 192/168 | 4/3 | 47/32 | ||
| K10pknG | K10Wt | 21 | 26/22 | - | - | 2/1 | 6/1 | 64/68 | 122/123 | 1/1 | 18/25 |
| 33 | 0/1 | 1/0 | 1/0 | 2/2 | 58/49 | TN/TN | 194/173 | 1/3 | 40/28 | ||
| 26 | - | - | - | - | - | 10/0 | - | - | - | ||
| K10relA | K10Wt | 27 | - | - | - | - | - | 8/0 | - | - | - |
| 30 | - | - | - | - | - | - | 2/0 | - | - | ||
| 22 | - | - | - | - | 20/0 | 1/0 | 1/0 | - | 30/0 | ||
| K10Wt | K10Wt | 25 | - | - | - | - | 15/0 | 22/0 | - | 3/0 | 61/0 |
| 20 | - | - | - | - | - | 8/0 | 9/0 | 5/0 | 110/0 | ||
| 16 | - | - | - | 6/0 | - | 12/0 | - | - | 3/0 | ||
| NPe | K10Wt | 24 | - | - | - | - | - | - | 5/0 | - | 2/0 |
| 31 | - | - | - | - | 7/0 | 1/0 | 2/0 | 1/0 | - |
JP, JM, JD: jejunum proximal, middle, distal; IP, IM, ID: ileum proximal, middle, distal; ICV: ileocecal valve; ML, ICL: mesenteric and ileocecal lymph nodes.
Negative.
Values are expressed as the total CFU obtained from normal 7H9 agar plates / the total CFU from 7H9 agar plates containing hygromycin.
Contamination happened during incubation.
Too numerous to count.
No pre-exposure.
4. DISCUSSION
The present study was designed to determine the effect of gene disruption on 1) ex vivo and in vivo survival of Map and 2) the capacity of mutants to elicit an immune response as determined by flow cytometry and gene expression profiles. The results revealed that deletion of relA, but not pknG, altered the capacity of the mutants to establish an infection without altering its capacity to elicit an immune response. In addition, gene expression profiles of T-cell secreting cytokines and granulysin demonstrated similar profiles regardless of infection with relA or wild-type Map. To our knowledge, this is the first demonstration of a mutation in Map that affects in vivo survival while retaining the capacity to elicit a strong response to challenge exposure to wild type Map.
Improvement in the use of targeted gene disruption in Map has allowed us to take a more direct approach to developing a vaccine that elicits an immune response while blocking the capacity of the pathogen to evade immune elimination and establish a persistent infection. The improvement has permitted us to select genes for disruption that might alter intracellular survival without affecting immunogenicity [36]. Orthologues of two virulence genes in Mtb and Mbv were selected for initial evaluation. The relA gene is a global regulator of the stringent response in many bacteria, including pathogenic mycobacteria. Deletion of relA in Mtb was shown to decrease the capacity of the mutant to survive in vivo in a mouse model [37]. Protein kinase G (encoded by pknG) is secreted by Mtb and BCG-Mbv within the phagosome. pknG inhibits phagosome-lysosome fusion. Deletion of the gene in BCG-Mbv was shown to significantly decrease survival in MΦ suggesting deletion would also impair survival in vivo [38]. However, examination of survival in calves and goats, as noted in the present study, showed survival in macrophages didn’t predict survival in vivo.
Initial screening of the mutants in an ex vivo MΦ assay revealed deletion of either gene reduced the capacity of Map to survive in comparison to wild-type Map at 6 days PI. Between the two mutants, the decrease in survival rate of K10pknG was more significant than that of K10relA (FIG. 2). Although the results from the pknG deletion mutant were expected [38], the results obtained with the relA mutant differed from a previous study of the Mtb relA mutant [47]. In the Mtb study, no difference was observed in growth rate between the relA mutant and wild-type Mtb in a human MΦ cell line. The authors suggested that it is because the primary carbon source for intracellular growth of Mtb depends on lipid catabolism. Recent studies provide a possible explanation for our finding whereby mycobacteria may encounter an inorganic phosphate limited condition in MΦ, which could induce a stringent response [48-50]. In addition, the decrease in survival in MΦ has also been observed with relA deletion in other intracellular pathogens [51]. In contrast to the ex vivo studies, the in vivo experiments revealed deletion of relA had a greater effect than deletion of pknG. All tissues from the K10relA infected calves were negative as assessed by culture and RT-PCR (Table 2) whereas some tissues from the K10pknG infected group were still positive at three months PI in some animals. Based on the ex vivo findings in MΦ with BCG-Mbv [38] and Map (in this study) this finding was unexpected. Nevertheless, the lower frequency of tissue positive sites in K10pknG infected group compared with the K10Wt infected group indicates deletion may have had some effect. This difference between the ex vivo and in vivo results emphasizes the importance of an in vivo assay to identify mutants for further analysis.
Further analysis showed the deletion of either gene did not alter the capacity of mutant to induce effector memory T-cell responses. The FC results revealed that a strong proliferative T-cell response is elicited against live Map and Map SAg following infection with wild-type and mutant strains of Map. As noted in previous studies, the CD8 T-cell proliferative response was less vigorous than the CD4 T-cell response to Map SAg and jPPD during the first 3 months PI [11, 12]. This was clearly evident with PBMCs from animals infected with wild-type and the relA mutant. An additional observation obtained from this study was the difference in response to live Map. In contrast to the CD8 T-cell response to Map SAg, the CD8 T-cell response to live Map was much more vigorous and accounted for a higher proportion of proliferating cells in the cultures (FIG. 4). The finding indicates that only examining the proliferative response to Map antigens (such as jPPD and SAg) does not fully reflect the immune status of memory CD4 and CD8 T cells and their potential effector activity against Map.
The proliferative response of PBMC from calves inoculated with the pknG mutant is more difficult to interpret. The response was already elevated in most of the calves at the initiation of the study. The explanation for the elevated response is not clear. The WSU dairy has not had any cases of JD during the past 30 years and we have used the dairy as a source of our calves for JD research for 10 years. We have considered two possibilities that might account for the increase in activity. In spite of efforts to minimize opportunity for exposure, it is possible that this group of calves was exposed to environmental mycobacteria. Another possibility, however, is that activated NK cells could have increased the proliferative activity. We have noted in some of our studies that the proliferative activity of CD4 and CD8 T cells is increased in cultures containing activated NK cells ([12] and unpublished observations). We did note that NK cells were activated and proliferating in cultures obtained from this group of animals. Although elevated, the proliferative response noted with the K10pknG treated group was consistent with the response obtained with K10Wt and K10relA treated groups. The findings point to the need to assess the activation status of all subsets when conducting studies in cattle.
Analysis of the cytokine profile in infected MΦ also revealed that deletion of either gene did not alter the capacity of the mutants to stimulate increased expression of genes encoding cytokines that drive differentiation of naïve T cells to effector T cells that differentially express IFN-γ, IL-17, or IL-22 [18, 19]. Of interest, infection with K10Wt Map or the mutants increased expression of IL-23 in macrophages, the cytokine associated with driving differentiation of Th17 cells [20, 52], but not IL-12, the cytokine associated with driving differentiation of Th1 cells. This observation is consistent with the finding in MΦ stimulated with Mtb Ags [53].
Comparison of the cytokine profiles obtained from PBMCs from K10Wt and K10relA treated groups cultured with live Map revealed disruption of relA did not alter the capacity of the mutant to elicit a response comparable to the response elicited by K10Wt. Expression of IFN-γ, IL-17, and IL-22 and their transcription factors were elevated while the expression of IL-10, Foxp3, and TGF-β were decreased. This difference in expression is consistent with the presence of effector T cells with immunoprotective activity. The finding that IFN-γ, IL-17, and IL-22 are all elevated indicates the proliferating population was comprised of two or more subsets expressing the respective cytokine genes. No information has been obtained thus far that demonstrate how many subsets exist in ruminants that express one or more of the cytokines as noted in other species [18, 19]. Further studies are needed to determine how many of these new CD4 and CD8 T-cell subsets exist in cattle and the relative contribution of subsets to the transcription profiles observed in this study. The findings show the response to Map is more complex and that understanding the immune response to Map will require development of additional reagents to identify and characterize the function of T-cell subsets.
Although granulysin is under intensive investigation as a mycobactericidal peptide that may play a role in control of tuberculosis, it has not been examined for a potential role in control of JD. Both innate and adaptive immune cells have been shown to secrete the peptide [22, 54-57]. In addition to its broad cytotoxic activity towards bacteria and some types of cancer cells it is a chemotactic attractant that promotes recruitment and activation of DC and potentiation of antigen-specific immune responses. Granulysin gene expression was highly up regulated in live Map treated cultures of PBMC from animals infected with wild-type and the relA mutant. A similar result has been reported from BCG-Mbv vaccinated animals. Due to the increased level of granulysin gene transcription after vaccination, the authors suggested granulyisn could be a biomarker for the presence of protective immunity against Mbv [58]. Similarly, the findings obtained here suggest granulysin could play an important role in host defense against Map.
Attenuation of survival in macrophage ex vivo has been used as a criterion for selection of mutants for further testing in vivo. Although the mouse is not the natural host of Map it has been considered the animal model of choice for further selection of mutants for evaluation in goats or cattle. However, a recent report comparing attenuation in the different models has raised a question as to whether this is the best approach for identifying candidates for testing in vivo [33]. The findings indicate survival in macrophages and the mouse model may not predict survival of mutants in the natural host or their capacity to elicit a protective immune response. Their conclusion is that even though screening in macrophage cultures ex vivo and mice in vivo appears to be an economical approach to narrowing down the strains to be tested for attenuation, the potential of strains as vaccine candidates can only be assessed in ruminants, the natural hosts of Map. We have taken this possibility into consideration in our efforts to develop a live attenuated vaccine. The challenge trial with goats was a follow-up pilot study to determine if further studies with either mutant were warranted for evaluation as a candidate vaccine in ruminants such as cattle. We were particularly interested in determining whether the pattern of in vivo survival of the mutants would be similar in another species and also whether the immune response elicited by the mutants would have any effect on establishment of an infection with K10Wt, In contrast to the calf-infection study, the pknG mutant was detected in more tissue sites with higher concentration of bacteria in the tissues than detected in tissues from goats inoculated once or twice with K10Wt, Outgrowth of bacteria was comparable in the presence and absence of hygromycin indicating the majority of the bacteria were from the hygromycin marked mutant, K10pknG. This was an unexpected result based on the previous results in the current study and others [38]. In contrast, the relA mutant was not able to establish an infection but was able to elicit an immune response that attenuated the capacity of K10Wt, to establish an infection. A few K10Wt, were cultured from a single tissue from each animal inoculated with the relA mutant followed by challenge with wild-type Map. The significance of this finding is that it shows the relA mutant elicits an immune response that differs from the response elicited by the pknG mutant and K10Wt. To our knowledge this is the first report showing such a difference. Further studies are needed to elucidate the basis for the difference in the immune response to the relA mutant and wild-type Map.
In conclusion, the strategies used to identify and evaluate Map mutants for potential use as a vaccine have yielded a mutant that meets the criteria for further testing. Disruption of relA attenuated survival ex vivo in macrophages without altering its capacity to activate cytokine genes comparable to wild-type Map. Disruption also attenuated survival in vivo in cattle as well as goats. The findings suggest deletion of relA blocked the capacity of Map to establish a persistent infection in DC and macrophages, the cells used by Map to establish persistent infection and dysregulate protective immunity.
Acknowledgments
This project was funded in part with funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. N01-AI-30055, the JDIP program (USDA-CSREES-NRI-CAP Award No.2007-01019) awarded to WCD, USDA-APHIS 03-9100-0788-GR and 03-9100-07-GR, and an intramural grant USDA Animal Health WNV-00150 and the WSU Monoclonal Antibody Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.National Research Council (U.S.). Committee on Diagnosis and Control of Johne’s Disease. Diagnosis and control of Johne’s Disease. Washington, D.C.: National Academies Press; 2003. [PubMed] [Google Scholar]
- 2.Juste RA, Elguezabal N, Pavon A, Garrido JM, Geijo M, Sevilla I, et al. Association between Mycobacterium avium subsp. paratuberculosis DNA in blood and cellular and humoral immune response in inflammatory bowel disease patients and controls. Int J Infect Dis. 2009 Mar;13(2):247–54. doi: 10.1016/j.ijid.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 3.Singh AV, Singh SV, Makharia GK, Singh PK, Sohal JS. Presence and characterization of Mycobacterium avium subspecies paratuberculosis from clinical and suspected cases of Crohn’s disease and in the healthy human population in India. Int J Infect Dis. 2008 Mar;12(2):190–7. doi: 10.1016/j.ijid.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 4.USDA. Animal and Plant Health Inspection Service. Johne’s disease on U.S. dairies. 1991-2007. Report. 2008 [Google Scholar]
- 5.Sibartie S, Scully P, Keohane J, O’Neill S, O’Mahony J, O’Hanlon D, et al. Mycobacterium avium subsp. paratuberculosis (MAP) as a modifying factor in Crohn’s disease. Inflamm Bowel Dis. 2010 Feb;16(2):296–304. doi: 10.1002/ibd.21052. [DOI] [PubMed] [Google Scholar]
- 6.Kirkwood CD, Wagner J, Boniface K, Vaughan J, Michalski WP, Catto-Smith AG, et al. Mycobacterium avium subspecies paratuberculosis in children with early-onset Crohn’s disease. Inflamm Bowel Dis. 2009 Nov;15(11):1643–55. doi: 10.1002/ibd.20967. [DOI] [PubMed] [Google Scholar]
- 7.Lee A, Griffiths TA, Parab RS, King RK, Dubinsky MC, Urbanski SJ, et al. Association of Mycobacterium avium subspecies paratuberculosis With Crohn Disease in pediatric patients. J Pediatr Gastroenterol Nutr. 2011 Feb;52(2):170–4. doi: 10.1097/MPG.0b013e3181ef37ba. [DOI] [PubMed] [Google Scholar]
- 8.Naser SA, Ghobrial G, Romero C, Valentine JF. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn’s disease. Lancet. 2004 Sep 18-24;364(9439):1039–44. doi: 10.1016/S0140-6736(04)17058-X. [DOI] [PubMed] [Google Scholar]
- 9.Olsen I, Tollefsen S, Aagaard C, Reitan LJ, Bannantine JP, Andersen P, et al. Isolation of Mycobacterium avium subspecies paratuberculosis reactive CD4 T cells from intestinal biopsies of Crohn’s disease patients. PLoS One. 2009;4(5):e5641. doi: 10.1371/journal.pone.0005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiappini E, de Martino M, Mangiantini F, Lionetti P. Crohn disease and mycobacterial infection in children: an intriguing relationship. J Pediatr Gastroenterol Nutr. 2009 Nov;49(5):550–8. doi: 10.1097/MPG.0b013e3181b0f908. [DOI] [PubMed] [Google Scholar]
- 11.Allen AJ, Park KT, Barrington GM, Lahmers KK, Hamilton MJ, Davis WC. Development of a bovine ileal cannulation model to study the immune response and mechanisms of pathogenesis of paratuberculosis. Clin Vaccine Immunol. 2009 Apr;16(4):453–63. doi: 10.1128/CVI.00347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koo HC, Park YH, Hamilton MJ, Barrington GM, Davies CJ, Kim JB, et al. Analysis of the immune response to Mycobacterium avium subsp. paratuberculosis in experimentally infected calves. Infect Immun. 2004 Dec;72(12):6870–83. doi: 10.1128/IAI.72.12.6870-6883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stabel JR, Palmer MV, Harris B, Plattner B, Hostetter J, Robbe-Austerman S. Pathogenesis of Mycobacterium avium subsp. paratuberculosis in neonatal calves after oral or intraperitoneal experimental infection. Vet Microbiol. 2009 May 12;136(3-4):306–13. doi: 10.1016/j.vetmic.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 14.Demangel C, Britton WJ. Interaction of dendritic cells with mycobacteria: where the action starts. Immunol Cell Biol. 2000 Aug;78(4):318–24. doi: 10.1046/j.1440-1711.2000.00935.x. [DOI] [PubMed] [Google Scholar]
- 15.Martino A. Mycobacteria and innate cells: critical encounter for immunogenicity. J Biosci. 2008 Mar;33(1):137–44. doi: 10.1007/s12038-008-0029-4. [DOI] [PubMed] [Google Scholar]
- 16.Niedergang F, Didierlaurent A, Kraehenbuhl JP, Sirard JC. Dendritic cells: the host Achille’s heel for mucosal pathogens? Trends Microbiol. 2004 Feb;12(2):79–88. doi: 10.1016/j.tim.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Awasthi A, Murugaiyan G, Kuchroo VK. Interplay between effector Th17 and regulatory T cells. J Clin Immunol. 2008 Nov;28(6):660–70. doi: 10.1007/s10875-008-9239-7. [DOI] [PubMed] [Google Scholar]
- 18.Brand S. Crohn’s disease: Th1, Th17 or both? The change of a paradigm: new immunological and genetic insights implicate Th17 cells in the pathogenesis of Crohn’s disease. Gut. 2009 Aug;58(8):1152–67. doi: 10.1136/gut.2008.163667. [DOI] [PubMed] [Google Scholar]
- 19.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010 Feb 26;327(5969):1098–102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoeve MA, Savage ND, de Boer T, Langenberg DM, de Waal Malefyt R, Ottenhoff TH, et al. Divergent effects of IL-12 and IL-23 on the production of IL-17 by human T cells. Eur J Immunol. 2006 Mar;36(3):661–70. doi: 10.1002/eji.200535239. [DOI] [PubMed] [Google Scholar]
- 21.Langrish CL, McKenzie BS, Wilson NJ, de Waal Malefyt R, Kastelein RA, Cua DJ. IL-12 and IL-23: master regulators of innate and adaptive immunity. Immunol Rev. 2004 Dec;202:96–105. doi: 10.1111/j.0105-2896.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- 22.Dhiman R, Indramohan M, Barnes PF, Nayak RC, Paidipally P, Rao LV, et al. IL-22 produced by human NK cells inhibits growth of Mycobacterium tuberculosis by enhancing phagolysosomal fusion. J Immunol. 2009 Nov 15;183(10):6639–45. doi: 10.4049/jimmunol.0902587. [DOI] [PubMed] [Google Scholar]
- 23.Dieli F, Troye-Blomberg M, Ivanyi J, Fournie JJ, Krensky AM, Bonneville M, et al. Granulysin-dependent killing of intracellular and extracellular Mycobacterium tuberculosis by Vgamma9/Vdelta2 T lymphocytes. J Infect Dis. 2001 Oct 15;184(8):1082–5. doi: 10.1086/323600. [DOI] [PubMed] [Google Scholar]
- 24.Lieberman J. The ABCs of granule-mediated cytotoxicity: new weapons in the arsenal. Nat Rev Immunol. 2003 May;3(5):361–70. doi: 10.1038/nri1083. [DOI] [PubMed] [Google Scholar]
- 25.Lockhart E, Green AM, Flynn JL. IL-17 production is dominated by gammadelta T cells rather than CD4 T cells during Mycobacterium tuberculosis infection. J Immunol. 2006 Oct 1;177(7):4662–9. doi: 10.4049/jimmunol.177.7.4662. [DOI] [PubMed] [Google Scholar]
- 26.Kursar M, Koch M, Mittrucker HW, Nouailles G, Bonhagen K, Kamradt T, et al. Cutting Edge: Regulatory T cells prevent efficient clearance of Mycobacterium tuberculosis. J Immunol. 2007 Mar 1;178(5):2661–5. doi: 10.4049/jimmunol.178.5.2661. [DOI] [PubMed] [Google Scholar]
- 27.Sharma PK, Saha PK, Singh A, Sharma SK, Ghosh B, Mitra DK. FoxP3+ regulatory T cells suppress effector T-cell function at pathologic site in miliary tuberculosis. Am J Respir Crit Care Med. 2009 Jun 1;179(11):1061–70. doi: 10.1164/rccm.200804-529OC. [DOI] [PubMed] [Google Scholar]
- 28.de Almeida DE, Colvin CJ, Coussens PM. Antigen-specific regulatory T cells in bovine paratuberculosis. Vet Immunol Immunopathol. 2008 Oct 15;125(3-4):234–45. doi: 10.1016/j.vetimm.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 29.Singh SV, Singh PK, Singh AV, Sohal JS, Sharma MC. Therapeutic Effects of a New “Indigenous Vaccine” Developed Using Novel Native “Indian Bison Type” Genotype of Mycobacterium avium Subspecies paratuberculosis for the Control of Clinical Johne’s Disease in Naturally Infected Goatherds in India. Vet Med Int. 2010;2010:351846. doi: 10.4061/2010/351846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juste RA, Alonso-Hearn M, Molina E, Geijo M, Vazquez P, Sevilla IA, et al. Significant reduction in bacterial shedding and improvement in milk production in dairy farms after the use of a new inactivated paratuberculosis vaccine in a field trial. BMC Res Notes. 2009;2:233. doi: 10.1186/1756-0500-2-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddacliff L, Eppleston J, Windsor P, Whittington R, Jones S. Efficacy of a killed vaccine for the control of paratuberculosis in Australian sheep flocks. Vet Microbiol. 2006 Jun 15;115(1-3):77–90. doi: 10.1016/j.vetmic.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 32.Kathaperumal K, Kumanan V, McDonough S, Chen LH, Park SU, Moreira MA, et al. Evaluation of immune responses and protective efficacy in a goat model following immunization with a coctail of recombinant antigens and a Mycobacterium avium subsp. paratuberculosis. Vaccine. 2009 Jan 1;27(1):123–35. doi: 10.1016/j.vaccine.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 33.Scandurra GM, de Lisle GW, Cavaignac SM, Young M, Kawakami RP, Collins DM. Assessment of live candidate vaccines for paratuberculosis in animal models and macrophages. Infect Immun. 2010 Mar;78(3):1383–9. doi: 10.1128/IAI.01020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin SJ, Wu CW, Steinberg H, Talaat AM. Identification of novel virulence determinants in Mycobacterium paratuberculosis by screening a library of insertional mutants. Infect Immun. 2006 Jul;74(7):3825–33. doi: 10.1128/IAI.01742-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris NB, Feng Z, Liu X, Cirillo SL, Cirillo JD, Barletta RG. Development of a transposon mutagenesis system for Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol Lett. 1999 Jun 1;175(1):21–6. doi: 10.1111/j.1574-6968.1999.tb13597.x. [DOI] [PubMed] [Google Scholar]
- 36.Park KT, Dahl JL, Bannantine JP, Barletta RG, Ahn J, Allen AJ, et al. Demonstration of allelic exchange in the slow-growing bacterium Mycobacterium avium subsp. paratuberculosis, and generation of mutants with deletions at the pknG, relA, and lsr2 loci. Appl Environ Microbiol. 2008 Mar;74(6):1687–95. doi: 10.1128/AEM.01208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dahl JL, Kraus CN, Boshoff HI, Doan B, Foley K, Avarbock D, et al. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci U S A. 2003 Aug 19;100(17):10026–31. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, Huygen K, et al. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science. 2004 Jun 18;304(5678):1800–4. doi: 10.1126/science.1099384. [DOI] [PubMed] [Google Scholar]
- 39.Colangeli R, Helb D, Vilcheze C, Hazbon MH, Lee CG, Safi H, et al. Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr2 in M. tuberculosis. PLoS Pathog. 2007 Jun;3(6):e87. doi: 10.1371/journal.ppat.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hines ME, 2nd, Stabel JR, Sweeney RW, Griffin F, Talaat AM, Bakker D, et al. Experimental challenge models for Johne’s disease: a review and proposed international guidelines. Vet Microbiol. 2007 Jun 21;122(3-4):197–222. doi: 10.1016/j.vetmic.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 41.Harris NB, Zinniel DK, Hsieh MK, Cirillo JD, Barletta RG. Cell sorting of formalin-treated pathogenic Mycobacterium paratuberculosis expressing GFP. Biotechniques. 2002 Mar;32(3):522–4. 6–7. doi: 10.2144/02323st03. [DOI] [PubMed] [Google Scholar]
- 42.Woo SR, Sotos J, Hart AP, Barletta RG, Czuprynski CJ. Bovine monocytes and a macrophage cell line differ in their ability to phagocytose and support the intracellular survival of Mycobacterium avium subsp. paratuberculosis. Vet Immunol Immunopathol. 2006 Mar 15;110(1-2):109–20. doi: 10.1016/j.vetimm.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 43.Greig A, Stevenson K, Perez V, Pirie AA, Grant JM, Sharp JM. Paratuberculosis in wild rabbits (Oryctolagus cuniculus) Vet Rec. 1997 Feb 8;140(6):141–3. doi: 10.1136/vr.140.6.141. [DOI] [PubMed] [Google Scholar]
- 44.Schonenbrucher H, Abdulmawjood A, Failing K, Bulte M. New triplex real-time PCR assay for detection of Mycobacterium avium subsp. paratuberculosis in bovine feces. Appl Environ Microbiol. 2008 May;74(9):2751–8. doi: 10.1128/AEM.02534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irenge LM, Walravens K, Govaerts M, Godfroid J, Rosseels V, Huygen K, et al. Development and validation of a triplex real-time PCR for rapid detection and specific identification of M. avium subsp. paratuberculosis in faecal samples. Vet Microbiol. 2009 Apr 14;136(1-2):166–72. doi: 10.1016/j.vetmic.2008.09.087. [DOI] [PubMed] [Google Scholar]
- 46.Coussens PM, Verman N, Coussens MA, Elftman MD, McNulty AM. Cytokine gene expression in peripheral blood mononuclear cells and tissues of cattle infected with Mycobacterium avium subsp. paratuberculosis: evidence for an inherent proinflammatory gene expression pattern. Infect Immun. 2004 Mar;72(3):1409–22. doi: 10.1128/IAI.72.3.1409-1422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Primm TP, Andersen SJ, Mizrahi V, Avarbock D, Rubin H, Barry CE., 3rd The stringent response of Mycobacterium tuberculosis is required for long-term survival. J Bacteriol. 2000 Sep;182(17):4889–98. doi: 10.1128/jb.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rifat D, Bishai WR, Karakousis PC. Phosphate depletion: a novel trigger for Mycobacterium tuberculosis persistence. J Infect Dis. 2009 Oct 1;200(7):1126–35. doi: 10.1086/605700. [DOI] [PubMed] [Google Scholar]
- 49.Ulrichs T, Kaufmann SH. Mycobacterial persistence and immunity. Front Biosci. 2002 Feb 1;7:d458–69. doi: 10.2741/A788. [DOI] [PubMed] [Google Scholar]
- 50.Spira B, Silberstein N, Yagil E. Guanosine 3’,5’-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi. J Bacteriol. 1995 Jul;177(14):4053–8. doi: 10.1128/jb.177.14.4053-4058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dean RE, Ireland PM, Jordan JE, Titball RW, Oyston PC. RelA regulates virulence and intracellular survival of Francisella novicida. Microbiology. 2009 Dec;155(Pt 12):4104–13. doi: 10.1099/mic.0.031021-0. [DOI] [PubMed] [Google Scholar]
- 52.Curtis MM, Way SS, Wilson CB. IL-23 promotes the production of IL-17 by antigen-specific CD8 T cells in the absence of IL-12 and type-I interferons. J Immunol. 2009 Jul 1;183(1):381–7. doi: 10.4049/jimmunol.0900939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verreck FA, de Boer T, Langenberg DM, Hoeve MA, Kramer M, Vaisberg E, et al. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proc Natl Acad Sci U S A. 2004 Mar 30;101(13):4560–5. doi: 10.1073/pnas.0400983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gansert JL, Kiessler V, Engele M, Wittke F, Rollinghoff M, Krensky AM, et al. Human NKT cells express granulysin and exhibit antimycobacterial activity. J Immunol. 2003 Mar 15;170(6):3154–61. doi: 10.4049/jimmunol.170.6.3154. [DOI] [PubMed] [Google Scholar]
- 55.Olsen I, Boysen P, Kulberg S, Hope JC, Jungersen G, Storset AK. Bovine NK cells can produce gamma interferon in response to the secreted mycobacterial proteins ESAT-6 and MPP14 but not in response to MPB70. Infect Immun. 2005 Sep;73(9):5628–35. doi: 10.1128/IAI.73.9.5628-5635.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sada-Ovalle I, Chiba A, Gonzales A, Brenner MB, Behar SM. Innate invariant NKT cells recognize Mycobacterium tuberculosis-infected macrophages, produce interferon-gamma, and kill intracellular bacteria. PLoS Pathog. 2008 Dec;4(12):e1000239. doi: 10.1371/journal.ppat.1000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tewary P, Yang D, de la Rosa G, Li Y, Finn MW, Krensky AM, et al. Granulysin activates antigen-presenting cells through TLR4 and acts as an immune alarmin. Blood. 2010 Nov 4;116(18):3465–74. doi: 10.1182/blood-2010-03-273953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Capinos Scherer CF, Endsley JJ, de Aguiar JB, Jacobs WR, Jr, Larsen MH, Palmer MV, et al. Evaluation of granulysin and perforin as candidate biomarkers for protection following vaccination with Mycobacterium bovis BCG or M. bovisDeltaRD1. Transbound Emerg Dis. 2009 Aug;56(6-7):228–39. doi: 10.1111/j.1865-1682.2008.01058.x. [DOI] [PubMed] [Google Scholar]
- 59.Chan PS, Caron JP, Rosa GJ, Orth MW. Glucosamine and chondroitin sulfate regulate gene expression and synthesis of nitric oxide and prostaglandin E(2) in articular cartilage explants. Osteoarthritis Cartilage. 2005 May;13(5):387–94. doi: 10.1016/j.joca.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 60.Seo KS, Lee SU, Park YH, Davis WC, Fox LK, Bohach GA. Long-term staphylococcal enterotoxin C1 exposure induces soluble factor-mediated immunosuppression by bovine CD4+ and CD8+ T cells. Infect Immun. 2007 Jan;75(1):260–9. doi: 10.1128/IAI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Evans AC, Ireland JL, Winn ME, Lonergan P, Smith GW, Coussens PM, et al. Identification of genes involved in apoptosis and dominant follicle development during follicular waves in cattle. Biol Reprod. 2004 May;70(5):1475–84. doi: 10.1095/biolreprod.103.025114. [DOI] [PubMed] [Google Scholar]


