Abstract
The Notch signalling pathway is an evolutionarily conserved intercellular signalling mechanism that is essential for cell fate specification and proper embryonic development. We have analysed the expression, regulation and function of the jagged 2 (Jag2) gene, which encodes a ligand for the Notch family of receptors, in developing mouse teeth. Jag2 is expressed in epithelial cells that give rise to the enamel-producing ameloblasts from the earliest stages of tooth development. Tissue recombination experiments showed that its expression in epithelium is regulated by mesenchyme-derived signals. In dental explants cultured in vitro, the local application of fibroblast growth factors upregulated Jag2 expression, whereas bone morphogenetic proteins provoked the opposite effect. Mice homozygous for a deletion in the Notch-interaction domain of Jag2 presented a variety of severe dental abnormalities. In molars, the crown morphology was misshapen, with additional cusps being formed. This was due to alterations in the enamel knot, an epithelial signalling structure involved in molar crown morphogenesis, in which Bmp4 expression and apoptosis were altered. In incisors, cytodifferentiation and enamel matrix deposition were inhibited. The expression of Tbx1 in ameloblast progenitors, which is a hallmark for ameloblast differentiation and enamel formation, was dramatically reduced in Jag2−/− teeth. Together, these results demonstrate that Notch signalling mediated by Jag2 is indispensable for normal tooth development.
Keywords: Jagged 2, Notch signalling, Tooth, Embryonic development, Ameloblast, Enamel, Cell fate, Tissue interactions, Tbx1, Bmp4, FGF, Barx1, Pax9, Pitx2, Mouse
INTRODUCTION
The Notch signalling pathway is an evolutionarily conserved mechanism that enables adjacent cells to adopt different fates (Artavanis-Tsakonas et al., 1995; Gridley, 1997; Robey, 1997; Weinmaster, 1997). Four isoforms of the Notch receptors (Notch1, Notch2, Notch3 and Notch4) have been identified in vertebrates, whereas only one isoform is found in Drosophila. The Notch receptor is a transmembrane protein with a large extracellular domain carrying multiple epidermal growth factor (EGF)-like repeats and a cytoplasmic domain required for signal transduction. Notch activation is achieved through direct interaction with membrane-bound ligands that contain, in their extracellular domain, multiple EGF-like motifs and the Delta/Serrate/Lag-2 (DSL) domain (Henderson et al., 1994; Muskavitch, 1994). Five ligands [jagged 1 (Jag1), Jag2, delta-like 1 (Dll1), Dll3 and Dll4] have been identified in vertebrates (D'Souza et al., 2008; Radtke et al., 2005). All of these ligands are transmembrane proteins. The signal induced by ligand binding is transmitted by the intracellular part of the receptor in a process involving proteolysis and interactions with cytoplasmic and nuclear proteins (Fortini, 2009; Fortini and Bilder, 2009; Jarriault et al., 1995; Kopan and Ilagan, 2009; Kopan et al., 1996).
Signals exchanged between neighbouring cells through the Notch receptors influence proliferation, differentiation and apoptotic events at all stages of development, controlling organ formation and morphogenesis (Artavanis-Tsakonas et al., 1995; Artavanis-Tsakonas et al., 1999; Cornell and Eisen, 2005; Lewis, 2008; Robey, 1997). Notch malfunction has been shown to disrupt aspects of neurogenesis, somite formation, angiogenesis, and kidney and lymphoid development (Conlon et al., 1995; Hrabe de Angelis et al., 1997; Limbourg et al., 2005; Louvi and Artavanis-Tsakonas, 2006; McCright et al., 2001; Nye et al., 1994; Radtke et al., 2005; Swiatek et al., 1994; Wilson and Radtke, 2006). In humans, mutations in the NOTCH1, NOTCH3 and JAG1 genes are associated, respectively, with a lymphoblastic leukaemia, a neurological disease known as CADASIL, and an inherited malformative disorder known as Alagille syndrome that affects the liver, heart, vertebrae, eyes and face (Ellisen et al., 1991; Gridley, 2003; Joutel et al., 1996; Li et al., 1997; Louvi et al., 2006; Oda et al., 1997).
The tooth represents a powerful model for elucidating the molecular mechanisms involved in cell fate determination and differentiation of various cell lineages during embryonic development (Mitsiadis and Graf, 2009). Teeth arise from reciprocal inductive interactions between the oral epithelium and the underlying neural crest-derived mesenchyme (Bluteau et al., 2008; Cobourne and Mitsiadis, 2006; Thesleff and Hurmerinta, 1981). These interactions progressively transform the tooth primordia into complex mineralised structures of various cell types. In mice at embryonic day (E) 10, factors derived from the oral epithelium, such as fibroblast growth factors (FGFs), bone morphogenetic proteins (BMPs), Wnt factors and sonic hedgehog (Shh), signal to the mesenchyme and initiate tooth development (Aberg et al., 1997; Dassule et al., 2000; Dassule and McMahon, 1998; Kettunen and Thesleff, 1998; Mitsiadis, 2001; Tummers and Thesleff, 2009). These molecular events are followed by cellular activities that are visualised as local epithelial thickenings at the sites of the future teeth. Thereafter, the developing epithelium forms the dental bud and cap structures that mark the onset of tooth morphology. The cap stage is characterised by the appearance of a transient epithelial signalling centre called the enamel knot, which is formed by subsets of cells that once more express BMPs, FGFs, Wnt factors and Shh (Jernvall et al., 1998; Mitsiadis, 2001; Tummers and Thesleff, 2009). The enamel knot regulates dental cusp morphology by controlling epithelial cell proliferation and apoptosis (Jernvall et al., 1998; Kim et al., 2006; Viriot et al., 1997). Subsequent folding and growth of the epithelium give rise to the bell stage, at which cytodifferentiation occurs. Four cell layers form the epithelial component during late odontogenesis: the inner dental epithelium (future ameloblasts), stratum intermedium, stellate reticulum and outer dental epithelium. The dental mesenchyme is also composed of different cell types, such as odontoblasts, sub-odontoblastic layer cells, dental papilla cells and dental follicle cells. Ameloblasts and odontoblasts are highly differentiated cells that synthesize and secrete the organic components of the enamel and dentin, respectively (Bluteau et al., 2008; Mitsiadis and Graf, 2009).
Previous data have shown that components of the Notch signalling pathway are expressed in developing mouse teeth. Expression of Notch1, Notch2, Notch3 (Mitsiadis et al., 1995a), Dll1 (Mitsiadis et al., 1998a), Jag1 (Mitsiadis et al., 1997) and Jag2 (Mitsiadis et al., 2005; Valsecchi et al., 1997) in developing teeth prefigures the subdivision of the epithelium into ameloblastic (capable of enamel-matrix synthesis) and non-ameloblastic regions already at the initiation stage. This becomes obvious during cytodifferentiation, in which Notch receptors and ligands show complementary expression patterns: Notch1 expression is confined to the stratum intermedium, whereas Dll1 and Jag2 are expressed in the adjacent inner dental epithelium layer (Mitsiadis et al., 1998a; Mitsiadis et al., 2005; Valsecchi et al., 1997). Similarly, in dental mesenchyme, Dll1 is expressed in differentiating odontoblasts, whereas the Notch genes are predominantly expressed in the sub-odontoblastic layer (Mitsiadis et al., 1998a). These results suggest that Notch receptors and ligands control tooth morphogenesis and influence differentiation events. However, little information exists about the in vivo biological role of Notch signalling during odontogenesis. This is mainly due to the early embryonic death (at E11-12) of Notch1 (Swiatek et al., 1994), Notch2 (Hamada et al., 1999; McCright et al., 2001), Jag1 (Xue et al., 1999) and Dll1 (Hrabe de Angelis et al., 1997) homozygous mice.
Here, we examined in detail the expression, regulation and function of Jag2 in developing mouse teeth. For the functional analysis we used mice with a targeted mutation that deletes exons encoding the Notch-interacting DSL domain of the Jag2 protein (Jiang et al., 1998).
MATERIALS AND METHODS
Animals and tissue preparation
E10.5-18.5 Swiss mouse embryos were used for in situ hybridisation, tissue recombination and bead implantation experiments. Jag2DDSL mutant mice have been described previously (Jiang et al., 1998). E12.5-18.5 wild-type, Jag2+/− and Jag2−/−embryos were obtained by intercrossing Jag2DDSL/+ mice. Embryonic age was determined according to the appearance of the vaginal plug (day 0) and confirmed by morphological criteria. Animals were sacrificed by cervical dislocation and the embryos removed in Dulbecco's phosphate-buffered saline (PBS). Dissected heads were fixed in 4% paraformaldehyde (PFA) for 24 hours at 4°C and prepared for sectioning.
Probes and in situ hybridisation
Digoxigenin-labelled sense and antisense riboprobes for Jag2, Bmp2, Bmp4, Bmp7, Fgf8, Pitx1, Pitx2, Barx1, Pax9, Tbx1 and Mk were used. Whole-mount in situ hybridisation on explants and in situ hybridisation on cryosections of E12.5-18.5 embryos were performed as described (Mitsiadis et al., 2003; Mitsiadis et al., 1998b; Wilkinson, 1995).
Dental explants, tissue recombination and bead implantation experiments
E11.5-13.5 molars were dissected from the rest of the mandible in PBS. Twenty-four dental explants were incubated for 5 minutes in 2.25% trypsin/0.75% pancreatin on ice. Epithelial and mesenchymal tissues were separated in DMEM supplemented with 15% foetal calf serum (FCS). Isolated mesenchymal tissues were transferred onto pieces of Nuclepore filter (pore size, 0.1 μm) supported by metal grids (Trowell type). Thereafter, isolated epithelia were placed either in contact with the mesenchyme or cultured alone. Eight homochronic recombinants (E11.5 epithelium/E11.5 mesenchyme, E13.5 epithelium/E13.5 mesenchyme), eight heterochronic recombinants (E11.5 epithelium/E13.5 mesenchyme and vice versa) and eight isolated epithelia (E11.5 and E13.5) were cultured for 24 hours in DMEM supplemented with 15% FCS and 20 units/ml penicillin/streptomycin in a humidified atmosphere of 5% CO2 in air at 37°C.
The same procedure was followed for bead implantation experiments, in which 18 E12.5 molar tooth germs were collected. Dental epithelia were separated from mesenchyme and then isolated epithelia were recombined with isolated mesenchyme. Beads were placed on top of dental epithelia and cultured for 24 hours. After culture, explants were fixed in 4% PFA, washed in PBS and analysed by whole-mount in situ hybridisation (Mitsiadis et al., 2003; Mitsiadis et al., 1997; Mitsiadis et al., 1995a).
Recombinant proteins and treatment of beads
Recombinant human BMP2, BMP4, FGF2, FGF8 and FGF4 proteins were used for bead implantation experiments. Affigel agarose beads (75-150 μm diameter) and heparin acrylic beads (100-250 μm diameter) were used as carriers of BMP and FGF proteins, respectively. Recombinant proteins were diluted with 0.1% bovine serum albumin (BSA) in PBS to concentrations of 10-25 (FGFs) and 100-250 (BMPs) ng/μl per 5 μl per 50 beads and incubated for 30 minutes at 37°C. Beads were then placed on top of dental explants. Control beads were treated identically with 0.1% BSA in PBS.
Histology
Mouse embryos were dissected and DNA was prepared from the tails for genotyping by PCR analysis. Heads of embryos were fixed in 4% PFA for 24 hours, then embedded in paraffin, sectioned at 6 μm, and stained according to the Masson's trichrome protocol.
Analysis of apoptosis
Terminal deoxynucleotidyl transferase-mediated dUTP nick end (TUNEL) labelling was used to investigate apoptosis. Briefly, after proteinase K treatment (20 μg/ml at 37°C for 30 minutes), slides were incubated with terminal deoxyribonucleotide transferase at 37°C for 1 hour. Anti-digoxigenin antibody conjugated with horseradish peroxidase was applied and 3,3′-diaminobenzidine (DAB) was used to visualise apoptotic DNA strand breaks (brown). A positive control for TUNEL labelling was prepared by nuclease treatment (5 μg/ml at 37°C for 30 minutes), whereas for the negative control we omitted the terminal transferase from the labelling procedure as described previously (Mitsiadis et al., 2008b).
RESULTS
Jag2 expression during embryonic tooth development
To be able to interpret the effects of Jag2 deletion on tooth development, we first determined its expression in sections of E11.5-18.5 teeth. Jag2 expression was observed in dental epithelium from E11.5 onwards, and persisted in epithelium throughout all stages of embryonic development (Fig. 1). During the bud stage (E12.5-13.5), Jag2 transcripts were observed in cells of the inner and outer dental epithelia (Fig. 1B,C), whereas during the cap (E14.5-15.5; Fig. 1D) and bell (E16.5-18.5; Fig. 1E) stages, Jag2 expression was found in the inner dental epithelium of the molars. A similar pattern was observed in developing incisors: at the bud stage, Jag2 was expressed in cells of the inner and outer dental epithelia (Fig. 1F,G), whereas at more advanced stages expression was seen only in the inner dental epithelium (Fig. 1H-J). The signal was absent in slides hybridised with the Jag2 sense probe (data not shown).
Fig. 1.
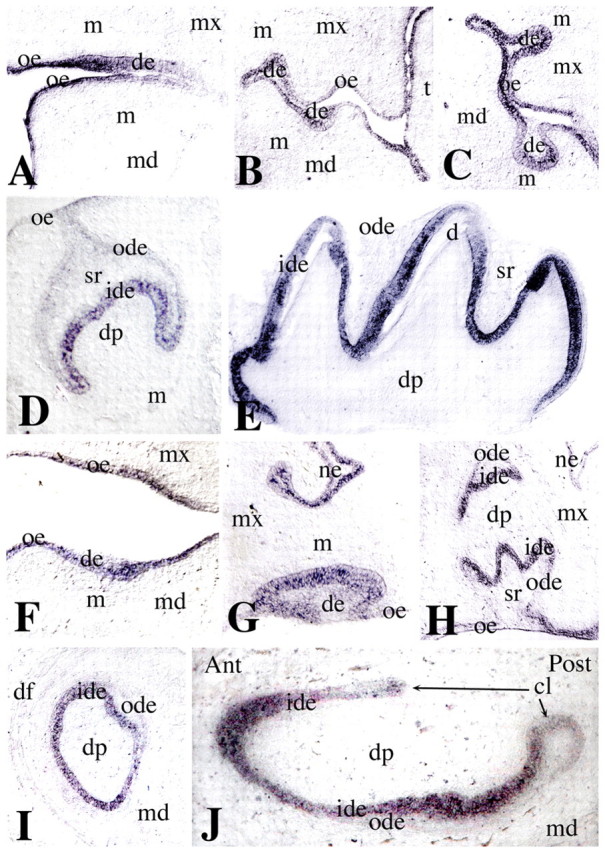
Jag2 expression during embryonic development of molars and incisors. In situ hybridisation on longitudinal (A,E,J) and frontal (B-D,F-I) cryosections of E11.5-18.5 mouse embryos. Shown are the first molar (A-E) and incisor (F-J) tooth germs, and mandibular (F,I,J) and maxillary (G,H) incisors. (A) Jag2 expression in E11.5 dental epithelium (de) and oral epithelium (oe). (B,C) Jag2 transcripts in E12.5 (B) and E13.5 (C) dental bud epithelium (de), in cells juxtaposed to the underlying mesenchyme (m). (D) Jag2 expression in inner dental epithelium (ide) cells of cap stage molars (E15.5). Expression is downregulated in oral epithelium and outer dental epithelium (ode). (E) Jag2 expression in inner dental epithelium of bell stage molars (E18.5). (F,G) Jag2 expression in the epithelium of E12.5 (F) and E13.5 (G) incisors. (H-J) Jag2 expression in E15.5 (H) and E17.5 (I,J) incisors in inner dental epithelium cells. Ant, anterior part of the incisor; cl, cervical loop; d, dentin; df, dental furrow; dp, dental papilla or dental pulp; md, mandibular process; mx, maxillary process; ne, nasal epithelium; ode, outer dental epithelium; Post, posterior part of the incisor; si, stratum intermedium; sr, stellate reticulum; t, tongue.
Jag2 expression in dental epithelium is maintained by mesenchyme-derived signals
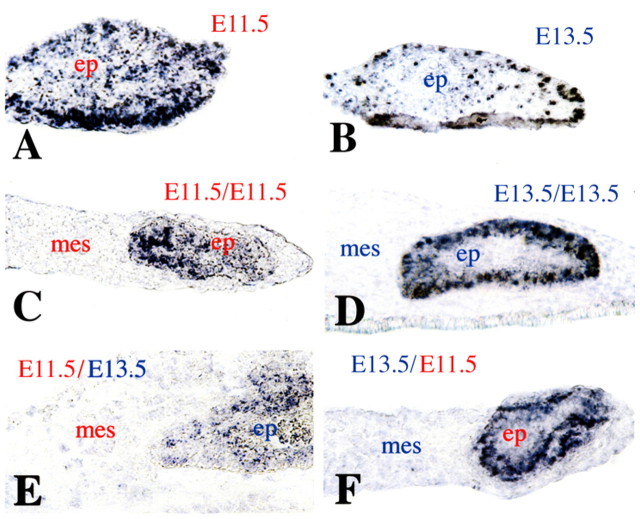
Although the initiation of Jag2 expression in dental epithelium occurs prior to mesenchymal induction (E11.5), its maintenance might depend on mesenchyme-derived signals at later stages (E12.5-13.5), when the mesenchyme possesses odontogenic potential (Mina and Kollar, 1987). To investigate this, we first cultured isolated E11.5 and E13.5 dental epithelia and examined Jag2 expression. Whereas isolated E11.5 dental epithelia strongly expressed Jag2 (Fig. 2A), expression was significantly reduced in E13.5 epithelia cultured alone (Fig. 2B). These findings indicate that Jag2 expression is intrinsic to epithelium at E11.5, whereas expression might require the presence of mesenchyme-derived signals at later stages.
Fig. 2.
Digoxigenin in situ hybridisation on cryosections showing expression of Jag2 in dental epithelial/mesenchymal homochronic and heterochronic recombinants, as well as in dental epithelia cultured alone. (A) Isolated E11.5 dental epithelia cultured in the absence of mesenchyme. (B) Isolated E13.5 dental epithelia cultured alone. (C) E11.5 dental epithelium (ep) recombined with E11.5 dental mesenchyme (mes). (D) E13.5 dental epithelium recombined with E13.5 dental mesenchyme. (E) E13.5 dental epithelium recombined with E11.5 dental mesenchyme. (F) E11.5 dental epithelium recombined with E13.5 dental mesenchyme.
To explore this further, we dissected epithelial and mesenchymal tissues from E11.5-13.5 molar germs and followed Jag2 expression in cultured homochronic and heterochronic tissue recombinants. In homochronic recombinants from E11.5 dental tissues, Jag2 expression was observed in all dental epithelial cells (Fig. 2C). In homochronic recombinants from E13.5 molar germs, Jag2 expression was only observed in dental epithelial cells contacting the mesenchyme (Fig. 2D). These findings reflect the in vivo situation in E13.5 teeth, in which Jag2 is expressed in dental epithelial cells that are in close contact with the mesenchyme and is downregulated in epithelial cells located far from the mesenchyme. To test whether tooth recombinant explants recapitulate the initial processes of tooth development, we also studied the expression of Pitx2, a gene that is exclusively expressed in dental epithelium during odontogenesis (Mucchielli et al., 1997). Expression of Pitx2 was found in the epithelium of dental recombinants (see Fig. S1 in the supplementary material).
Consequently, we tested whether the non-induced E11.5 dental mesenchyme could maintain Jag2 expression in E13.5 epithelia. For this we used heterochronic recombinants, in which E13.5 epithelia were cultured together with E11.5 mesenchyme. Few Jag2 transcripts were found throughout the epithelial cells (Fig. 2E), indicating that E11.5 dental mesenchyme does not have the capacity to maintain Jag2 expression in E13.5 epithelia. By contrast, strong Jag2 expression was observed in recombinants of E13.5 dental mesenchyme with E11.5 epithelia (Fig. 2F). Jag2 transcripts were less abundant (or absent) in epithelial cells separated from the mesenchyme by several cell layers. Together, these data suggest that mesenchyme-derived signals maintain Jag2 expression in epithelium at more advanced stages.
Opposite effects of FGFs and BMPs on Jag2 expression in dental epithelium
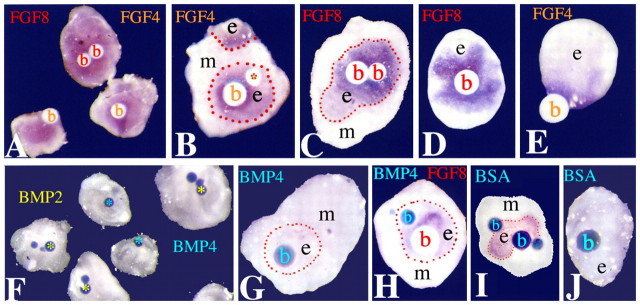
We attempted to elucidate the mesenchyme-derived signals that are responsible for the maintenance of Jag2 expression in dental epithelium. BMPs and FGFs are essential for tooth initiation and morphogenesis and therefore good candidates for such a function. BMP and FGF beads were placed either on top of recombinants of dental epithelium and dental mesenchyme isolated from E12.5 tooth germs or onto isolated E12.5 dental epithelia cultured alone (Fig. 3). Jag2 expression was upregulated by FGF2 (see Fig. S2 in the supplementary material), FGF4 (Fig. 3A,B) and FGF8 (Fig. 3A,C) beads in the epithelium of tooth recombinants, and in isolated dental epithelia (Fig. 3D,E). By contrast, Jag2 expression was downregulated in recombinants cultured with BMP2 (Fig. 3F) or BMP4 (Fig. 3F,G) beads. Jag2 expression was downregulated in dental epithelial cells surrounding BMP4 beads, but upregulated around FGF8 beads, when recombinants were cultured together with both FGF8 and BMP4 beads (Fig. 3H). Control BSA beads did not alter Jag2 expression in the epithelium of tooth recombinants (Fig. 3I) or in epithelia cultured alone (Fig. 3J). Similarly, BSA beads did not affect epithelial Pitx2 expression in tooth recombinants (see Fig. S3 in the supplementary material). These results indicate that FGFs upregulate Jag2 expression in dental epithelium, whereas BMPs have the opposite effect and downregulate its expression.
Fig. 3.
Regulation of Jag2 expression in E12.5 dental epithelium. Whole-mount in situ hybridisation. The red dotted lines indicate the border between dental epithelium and dental mesenchyme. (A) Upregulation of Jag2 in tooth recombinants of epithelium (e) cultured on top of mesenchyme (m) together with FGF4 (orange) or FGF8 (red) beads (b). (B) Tooth recombinants of epithelia cultured in contact, and on top of, a mesenchyme together with FGF4 and BSA (red asterisk) beads. Jag2 expression is seen in epithelial cells surrounding the FGF4 bead. Also note Jag2 induction in epithelial cells contacting the mesenchyme, as expected. (C) Jag2 expression in a recombinant of epithelium cultured on top of mesenchyme together with FGF8 beads. (D) Upregulation of Jag2 expression by FGF8 in dental epithelium cultured alone. (E) Upregulation of Jag2 expression by FGF4 in dental epithelium cultured alone. (F) Downregulation of Jag2 expression in recombinants of epithelium cultured on top of mesenchyme together with BMP2 (yellow) or BMP4 (cyan) beads (asterisks). (G) Downregulation of Jag2 expression in a tooth recombinant of epithelium cultured on top of mesenchyme together with BMP4 beads. (H) Jag2 expression in a tooth recombinant of epithelium cultured on top of mesenchyme together with BMP4 and FGF8 beads. Induction of Jag2 expression in epithelial cells by an FGF8 bead and downregulation by a BMP4 bead are seen. (I) BSA beads do not alter Jag2 expression in the epithelium of tooth recombinants. (J) BSA beads do not induce Jag2 expression in epithelium cultured alone.
Jag2 mutant mice exhibit abnormal tooth morphology and mineral matrix deposition
To explore the role of Jag2 in vivo, we analysed mice deficient in the Notch-interacting domain of Jag2 (Jag2DDSL). Jag2DDSL/Jag2DDSL (Jag2−/−) homozygous mutant mice die shortly after birth from cleft palate caused by fusions of the oral epithelium. Most commonly, the tongue is fused to the palatal shelves, preventing them from elevating. However, we have observed that essentially all the oral epithelial surfaces can fuse with each other in Jag2−/− mice (Casey et al., 2006; Jiang et al., 1998). We observed that the Jag2−/− teeth exhibited an abnormal morphology. Histological analysis of E18.5 Jag2−/− molars revealed that their epithelial compartment was thinner than normal. Furthermore, the crown morphology was affected as small cusps, and possibly changes in cusp number, were evident (Fig. 4A,C). Ameloblasts and odontoblasts are columnar cells that participate in the secretion of enamel and dentin matrix, respectively. Odontoblasts were located at the tip of the cusps of E18.5 Jag2+/− molars (Fig. 4A), but were absent in E18.5 Jag2−/− molars (Fig. 4C). Dentin and enamel were not yet deposited in molars (Fig. 4A,C). In E18.5 Jag2+/− incisors, odontoblasts were fully differentiated and secreted dentin matrix (Fig. 4B, blue arrowhead). Similarly, functional ameloblasts formed a layer of polarised cells that secrete enamel matrix (i.e. black line on top of dentin in Fig. 4B). By contrast, odontoblast differentiation was inhibited and dentin was absent in Jag2−/− incisors (Fig. 4D, blue arrowhead). Likewise, ameloblast differentiation was inhibited, as indicated by their small size and absence of polarity (Fig. 4D).
Fig. 4.
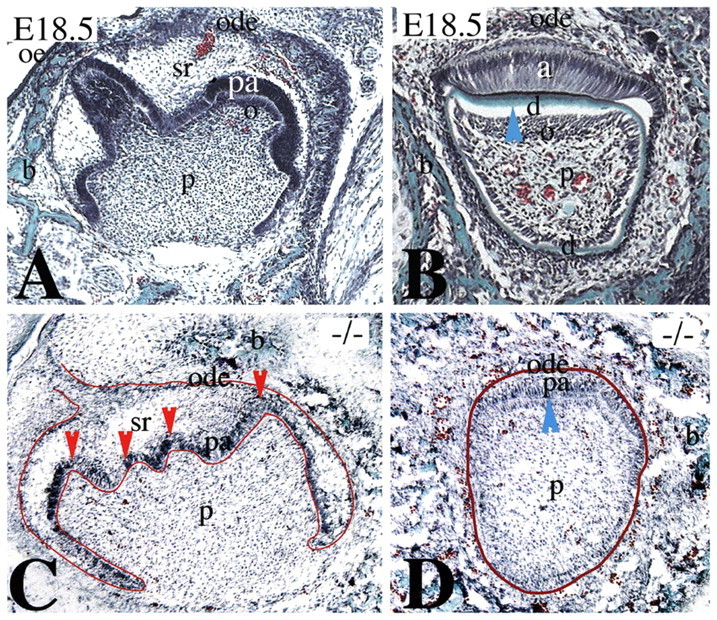
Tooth defects in Jag2 mutant mouse embryos. Masson's trichrome staining in frontal sections. Red lines indicate the border between dental epithelium and mesenchyme. Blue arrowheads indicate the areas of dentin (d) matrix deposition. (A,B) Histology of a lower molar (A) and incisor (B) of E18.5 Jag2 heterozygous embryos. (A) Differentiating odontoblasts (o) are observed at the tip of the cusps, whereas preameloblasts (pa) have not yet differentiated into ameloblasts. Dentin and enamel are absent. (B) In incisors, odontoblasts and ameloblasts are fully differentiated. Dentin is already formed (blue arrowhead). Ameloblasts start to secrete enamel matrix (black line on top of dentin). (C,D) Histology of a lower molar (C) and incisor (D) of E18.5 Jag2−/−embryos. (C) Jag2−/− embryos display defects in molar morphology (small cusps, red arrowheads). Odontoblasts are not seen at the tip of the cusps. (D) Differentiation of ameloblasts and odontoblasts is inhibited and dentin (blue arrowhead) and enamel matrices are absent in incisors. Preameloblasts are small and not yet polarised. b, bone; ode, outer dental epithelium; oe, oral epithelium; p, dental pulp; sr, stellate reticulum.
Correlation of decreased apoptosis and altered BMP expression in the enamel knot of Jag2−/− teeth
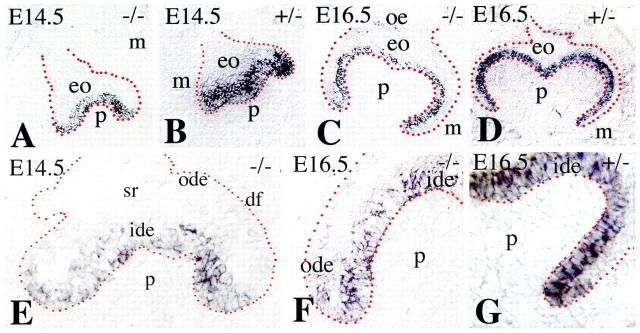
Crown morphology is refined through the controlled elimination of cells in the enamel knot by apoptosis. Using TUNEL staining we visualised apoptosis in E14.5 molars. We noted a substantial reduction of apoptosis in the enamel knot of Jag2−/− (Fig. 5B,D) compared with Jag2+/− (Fig. 5A,C) molars. The enamel knot exerts its activity through the production of BMPs, FGFs, Wnt proteins and Shh (Mitsiadis, 2001; Tummers and Thesleff, 2009). One consequence of the dysregulation of the signalling network in the enamel knot might be the altered Jag2−/− tooth morphology. Previous work has demonstrated that BMP signalling is involved in apoptotic events during the embryonic development of various tissues (Dunn et al., 1997; Hofmann et al., 1996; Macias et al., 1997; Yokouchi et al., 1996; Zou and Niswander, 1996). We examined BMP expression in the enamel knot of E14.5 Jag2−/− and Jag2+/− molars. Bmp2, Bmp4 and Bmp7 were expressed in the enamel knot of Jag2+/− molars (Fig. 6A,D,G). In Jag2−/− embryos, Bmp2 and Bmp7 were expressed in the enamel knot (Fig. 6B,C,H,I), whereas Bmp4 transcripts were detected in tooth mesenchyme (Fig. 6E,F). The absence of Bmp4 expression in the enamel knot of Jag2−/− teeth coincides with the lack of apoptosis (compare Fig. 5B with Fig. 6F) and is accompanied by misshapen tooth crowns.
Fig. 5.
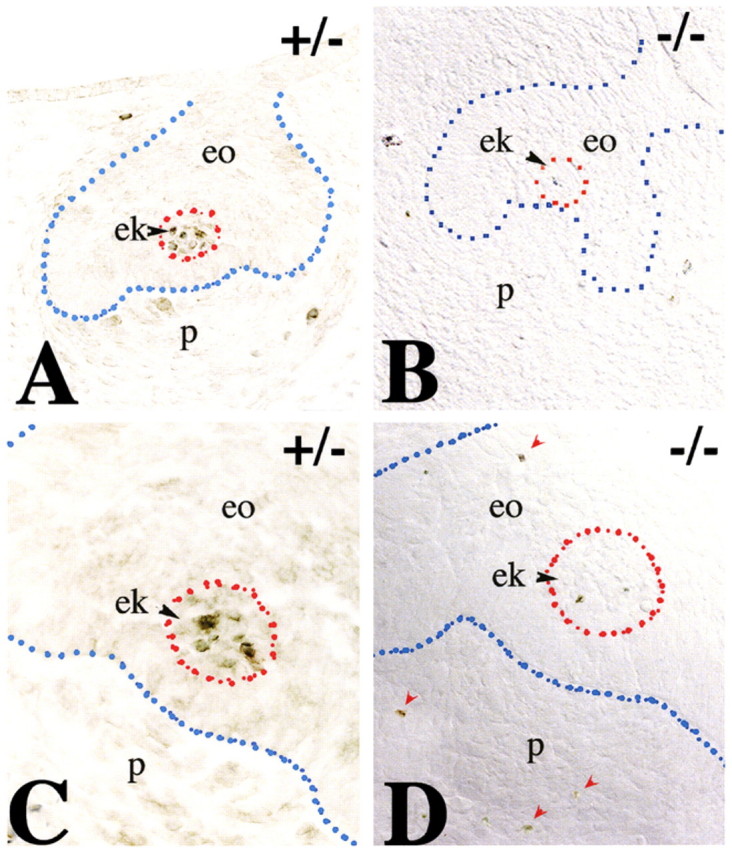
Apoptosis in E14.5 molars of Jag2+/− and Jag2−/− mouse embryos. Frontal sections. Blue dotted lines indicate the border between the enamel organ (eo) and the mesenchyme (m). Red circles and arrowheads indicate the enamel knots. (A) Apoptosis is observed in the enamel knot (ek) of heterozygous littermates, and in dental papilla (p). (B) In Jag2−/− embryos, apoptosis is reduced in the enamel knot. (C,D) Higher magnifications of the enamel knot of Jag2 heterozygous (C) and homozygous (D) embryos. Red arrowheads indicate apoptosis in areas other than the enamel knot. oe, oral epithelium.
Fig. 6.
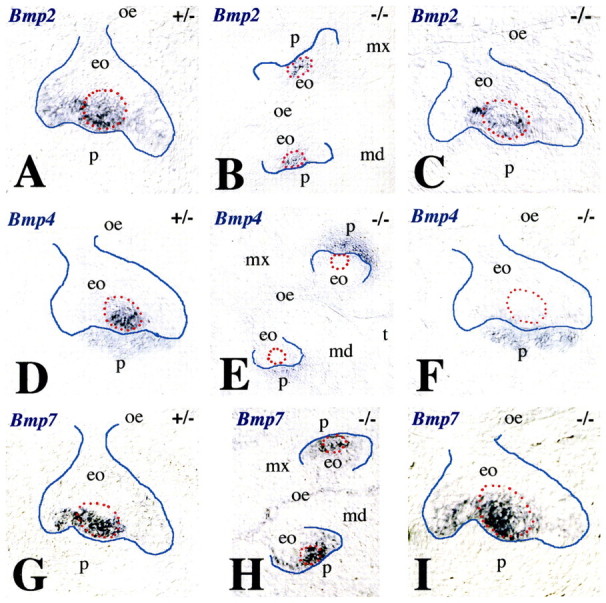
Expression of Bmp2, Bmp4 and Bmp7 in E14.5 molar tooth germs of Jag2+/− and Jag2−/− mouse embryos. In situ hybridisation on frontal cryosections. (A-C) Bmp2 expression in the enamel knot (red circle) of Jag2 heterozygous (A) and homozygous (B,C) littermates. (D-F) Bmp4 expression in the enamel knot and dental papilla (p) of heterozygous mice (D). Bmp4 expression is seen only in the dental papilla of Jag2−/− teeth (E,F). (G-I) Bmp7 expression in Jag2 heterozygous (G) and homozygous (H,I) mutants. Note the fusion of the maxillary (mx) and mandibular (md) oral epithelia (oe) in Jag2−/− mice (B,E,H). eo, enamel organ; t, tongue.
Alteration of the molecular cascade in Jag2−/− mouse embryos during the different stages of tooth development
Pitx1, Pitx2, Pax9, Barx1 and Mk (Mdk – Mouse Genome Informatics) are required for proper tooth formation and represent excellent markers for the dental epithelium (Pitx1, Pitx2), dental mesenchyme (Pax9, Barx1), or both (Mk) (Mitsiadis et al., 2008a; Mitsiadis and Drouin, 2008; Mitsiadis et al., 1998b; Mitsiadis et al., 1995b; Mucchielli et al., 1997; Neubuser et al., 1995; Neubuser et al., 1997; Peters et al., 1998; Tissier-Seta et al., 1995). Similarly, Fgf8 is a marker for dental epithelium during tooth initiation (Heikinheimo et al., 1994; Kettunen and Thesleff, 1998). We examined the expression of these genes in teeth of Jag2-deficient embryos. During dental epithelial thickening (bud stage, E12.5), Fgf8 (Fig. 7A,E), Pitx1 (data not shown) and Pitx2 (Fig. 7B,F) expression was restricted to the dental epithelium of both Jag2−/− and Jag2+/− embryos. However, the domain of Fgf8 expression was reduced in the dental epithelium of Jag2−/− embryos when compared with that of heterozygous littermates (Fig. 7E, red arrowhead).
Fig. 7.
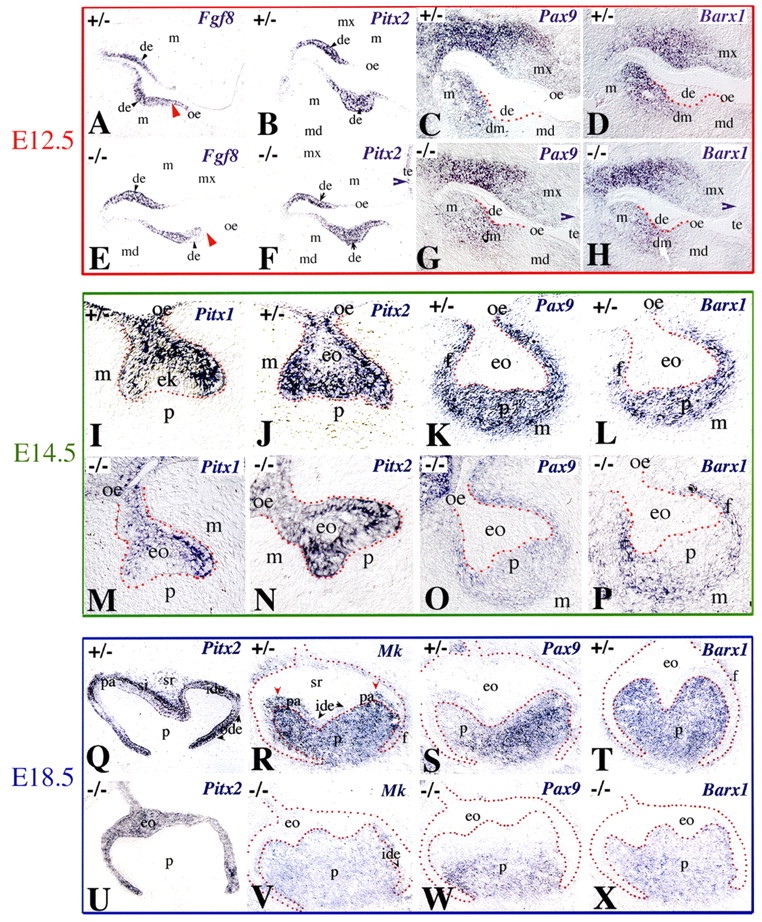
Expression of Fgf8, Pitx1, Pitx2, Pax9, Barx1 and Mk in molars of Jag2+/− and Jag2−/− mouse embryos. In situ hybridisation on frontal cryosections of E12.5 (A-H), E14.5 (I-P) and E18.5 (Q-X) embryos. Red dotted lines indicate borders between dental epithelium (de) and mesenchyme (dm). (A-H) Fgf8 (A,E), Pitx2 (B,F), Pax9 (C,G) and Barx1 (D,H) expression. Fgf8 expression is slightly restricted in Jag2−/− dental epithelium (red arrowheads in A and E). (I-P) Pitx1 (I,M), Pitx2 (J,N), Pax9 (K,O) and Barx1 (L,P) expression. A weak Pax9 signal is seen in dental papilla (p) and follicle (f) and downregulation of Barx1 expression is seen in dental papilla of Jag2−/− teeth. (Q-X) Pitx2 (Q,U), Mk (R,V), Pax9 (S,W) and Barx1 (T,X) expression. Pitx2 expression in inner dental epithelium (ide), stratum intermedium (si) and outer dental epithelium (ode) in Jag2+/− teeth (Q). Downregulation of Pitx2 in preameloblasts (pa) and stellate reticulum (sr). In Jag2−/− embryos (U), Pitx2 is expressed in all epithelial cells. Downregulation of Mk (V), Pax9 (W) and Barx1 (X) expression in dental papilla and follicle of Jag2−/− embryos. Mk transcripts are absent in preameloblasts of Jag2−/− teeth (V), but are present in Jag2+/− teeth (areas indicated by red arrowheads in R). eo, enamel organ; ek, enamel knot; m, mesenchyme; md, mandible; mx, maxilla; oe, oral epithelium; te, tongue epithelium.
We next tested whether Jag2 controls epithelial Pitx1 and Pitx2 expression at later stages of odontogenesis (Fgf8 was not used further as a marker because it is not expressed in dental epithelium during the following stages). No alterations in Pitx1 and Pitx2 expression were observed in the epithelium of E14.5 Jag2−/− molars (Fig. 7M,N) when compared with those of heterozygous mice (Fig. 7I,J). At the bell stage, Pitx1 and Pitx2 were strongly expressed in the epithelium of E18.5 Jag2+/− and Jag2−/− molars (Fig. 7Q,U; data not shown). Pitx2 was downregulated in inner dental epithelial cells that differentiated into preameloblasts (Fig. 7Q) (Mitsiadis et al., 1998b; Mucchielli et al., 1997). The robust expression of Pitx2 in dental epithelium allowed a morphological assessment of mutant teeth, and showed the existence of additional cusps in E18.5 Jag2−/− molars (Fig. 7U). Although a clear distinction between the four different cell layers forming the dental epithelium was evident in E18.5 Jag2+/− molars (Fig. 7Q), such a distinction was impossible in Jag2−/− molars (Fig. 7U). Furthermore, the dental epithelium of Jag2−/− molars appeared thinner than that of Jag2+/− molars (Fig. 7Q,U).
Tissue recombination experiments have demonstrated that tooth crown morphology is under the influence of mesenchyme-derived signals (Mina and Kollar, 1987). Thus, the morphological defects observed in Jag2−/− teeth are unlikely to be caused by the lack of Jag2 expression in dental epithelium alone. Consequently, we investigated the eventual molecular consequences of Jag2 deletion on tooth mesenchyme. Expression of Pax9 and Barx1 was restricted to the mesenchyme of developing teeth (Fig. 7C,D,K,L,S,T). Although Mk was also expressed in the mesenchyme during all stages of odontogenesis, differentiating preameloblasts started to express Mk (Fig. 7R; data not shown) (Mitsiadis et al., 2008a; Mitsiadis et al., 1995b). At E12.5, the expression patterns of Pax9 (Fig. 7G), Barx1 (Fig. 7H) and Mk (data not shown) were not altered in the mesenchyme of Jag2−/− teeth. By E14.5, mesenchymal expression of Pax9 (Fig. 7O), Barx1 (Fig. 7P) and Mk (data not shown) was dramatically decreased, although not completely abolished, in Jag2−/− molars. Similarly, in the mesenchyme of E18.5 Jag2−/− molars, Pax9 (Fig. 7W), Barx1 (Fig. 7X) and Mk (Fig. 7V) expression was faint, whereas strong expression was seen in E18.5 Jag2+/− molars (Fig. 7R,S,T). Hence, deletion of Jag2 in dental epithelium contributes indirectly, through the influence of epithelial-mesenchymal interactions, to the downregulation of Pax9, Barx1 and Mk expression in the mesenchyme.
Enamel formation in Jag2−/− teeth was also affected. A hallmark of enamel induction is the expression of Tbx1 in ameloblast progenitors of developing teeth (Mitsiadis and Drouin, 2008; Mitsiadis et al., 2008c; Zoupa et al., 2006), as incisors of Tbx1−/− mice do not form enamel (Caton et al., 2009). We tested whether Jag2 deletion also affects Tbx1 expression in ameloblast progenitors. In situ hybridisation in sections of E14.5 and E16.5 Jag2−/− molars showed that Tbx1 expression was considerably downregulated in inner dental epithelium cells (Fig. 8A,C,E,F) when compared with E14.5 and E16.5 Jag2+/− teeth (Fig. 8B,D,G). Tbx1 was not the only gene altered in the inner dental epithelial cells: their differentiation into preameloblasts correlated with downregulation of Pitx2 expression (Fig. 7Q) (Mitsiadis et al., 1998b; Mucchielli et al., 1997) and upregulation of Mk expression (Fig. 7R, red arrowheads) (Mitsiadis et al., 1995b). Pitx2 expression was not downregulated (Fig. 7U) and nor was Mk upregulated (Fig. 7V) in the inner dental epithelium of E18.5 Jag2−/− molars, indicating a failure or delay in the differentiation process of the ameloblast precursors.
Fig. 8.
Tbx1 expression in developing molars of E14.5 and E16.5 Jag2+/− and Jag2−/− mouse embryos. In situ hybridisation on frontal cryosections. Red dotted lines indicate borders between dental epithelium (de) and mesenchyme (m). (A) Tbx1 in E14.5 Jag2−/− molars. (B) Tbx1 in E14.5 Jag2+/− molars. (C) Tbx1 expression in E16.5 Jag2−/− molars. (D) Tbx1 expression in E16.5 Jag2+/− molars. (E,F) Higher magnifications showing downregulation of Tbx1 expression in inner dental epithelium (ide). (G) Higher magnification showing Tbx1 expression in E16.5 Jag2+/− molars. df, dental follicle; eo, enamel organ; ode, outer dental epithelium; oe, oral epithelium; p, dental papilla; sr, stellate reticulum.
These findings establish that Jag2 participates in the cascade of epithelial-mesenchymal interactions that govern tooth development, and demonstrate that its absence interferes with the regulated expression of several key genes involved in odontogenesis.
DISCUSSION
Teeth develop through sequential and reciprocal interactions between oral epithelium and cranial neural crest-derived mesenchyme (Bluteau et al., 2008; Cobourne and Mitsiadis, 2006; Lumsden, 1988; Mitsiadis, 2001). Dental epithelium contains the progenitors/precursors of ameloblasts, which are responsible for enamel formation. For their terminal differentiation, ameloblast progenitors undergo a specific developmental programme controlled by secreted signalling molecules and transcription factors (reviewed by Mitsiadis, 2001; Mitsiadis and Graf, 2009; Tummers and Thesleff, 2009). How this determination is achieved is not yet understood, but it might occur via Notch-mediated lateral inhibition, whereby inhibitory interactions between adjacent progenitor cells regulate cell fate specification (Artavanis-Tsakonas et al., 1995). Our previous studies have shown that Notch expression in the epithelium of developing teeth correlates with ameloblast fate specification (Mitsiadis et al., 1995a). Jag2 is expressed in prospective ameloblast precursor cells that are adjacent to the Notch1-expressing cells of the stratum intermedium (Mitsiadis et al., 1998a; Mitsiadis et al., 1995a). This well-defined expression pattern suggests that, in the developing dental epithelium, Notch1 signalling is mediated through the Jag2 receptor, as has been shown in previous studies in a variety of mammalian tissues (Francis et al., 2005; Lindsell et al., 1995; Luo et al., 1997). This signalling pair might play a pivotal role in ameloblast lineage commitment from the earliest stages of odontogenesis. As shown in this study, interruption of Jag2-mediated Notch signalling in vivo greatly affects dental epithelial progenitor cells and diminishes their potential to form ameloblasts, culminating in tooth germs with abnormal morphology and lacking enamel.
Regulation of Notch signalling in developing teeth
E11.5 epithelium possesses the inductive capacity for tooth formation (Lumsden, 1988; Mina and Kollar, 1987). Jag2 expression in the E11.5 dental epithelium is independent of mesenchyme-derived signals. This was demonstrated in cultured E11.5 dental explants, in which expression persisted in epithelium after removal of the mesenchyme. By E12.5, a time corresponding to the shift of the odontogenic potential from epithelium to mesenchyme (Mina and Kollar, 1987), epithelial Jag2 expression is dependent on mesenchyme-derived signals. Indeed, Jag2 expression was downregulated in E13.5 dental epithelial explants cultured in the absence of mesenchyme. By contrast, E13.5 dental mesenchyme maintained epithelial Jag2 expression in homochronic tooth recombinants. Interestingly, Jag2 was downregulated in the epithelium of heterochronic recombinants (E13.5 epithelium/E11.5 mesenchyme). This suggests that the E11.5 mesenchyme does not possess the adequate repertoire of signalling molecules necessary for Jag2 maintenance in epithelium. Conversely, in heterochronic recombinants composed of E11.5 epithelium and E13.5 mesenchyme, Jag2 was expressed in epithelial cells contacting the mesenchyme. This suggests that the E11.5 epithelium is competent to respond to E13.5 mesenchyme-derived signals. Taken together, these findings indicate that Notch-mediated decisions in dental epithelium are influenced by epithelial-mesenchymal interactions and thus must be under the control of other signalling pathways.
Members of the BMP and FGF families are essential for odontogenesis (Aberg et al., 1997; Kettunen et al., 1998) (reviewed by Mitsiadis, 2001; Mitsiadis and Graf, 2009). BMPs and FGFs exert opposite effects on the expression of Notch receptors and ligands in dental tissues (Mitsiadis et al., 1997; Mitsiadis et al., 1998a), indicating that dental cell fate choices are under the concomitant control of the Notch and BMP/FGF signalling pathways. Several studies have indicated that FGFs are important for the maintenance of ameloblast progenitors (Klein et al., 2008; Wang et al., 2007). FGFs may have an autocrine (e.g. Fgf4) or a paracrine (e.g. Fgf2) function that affects cell behaviour in dental epithelium, which expresses the FGF receptor Fgfr2b (Kettunen et al., 1998). We found that the in vitro implantation of FGF2, FGF4 and FGF8 beads into E12.5 explants upregulated Jag2 expression in dental epithelium, whereas BMP2 and BMP4 beads exerted the opposite effect. Hence, within the dental epithelium, Notch signalling is regulated by FGFs and BMPs to assure the maintenance of ameloblast precursors. However, there is not yet sufficient information about a genetic interaction between these three signalling pathways during embryogenesis (Hurlbut et al., 2007).
Morphological and cytodifferentiation defects in Jag2−/− teeth
To assess defects in tooth morphology we examined E18.5 Jag2−/− mouse embryos. The overall morphology and structure of the developing teeth were disturbed in mutant embryos. Abnormal crown morphology, as shown by the presence of small cusps and possibly changes in cusp number, is observed in Jag2−/− molars. Unfortunately, more detailed insight into the effects of Jag2 deficiency on late tooth morphology, using kidney capsule experiments, could not be obtained. Unanticipated difficulties were encountered in dissecting out intact tooth germs from Jag2−/− embryos because of fusions occurring as early as E12.5 in developing structures of the oral cavity (tongue, palatal shelves). This technical difficulty should be overcome as soon as a conditional floxed allele for Jag2 is available. We found that Jag2 inactivation significantly reduced apoptosis in the enamel knot, a transient signalling centre (Jernvall et al., 1998; Tummers and Thesleff, 2009), and this event could be responsible for the morphological defects of the crown. A similar effect of Jag2 on apoptosis has been reported for the developing limb (Francis et al., 2005; Jiang et al., 1998). In the developing teeth, the consequence of Jag2 inactivation on apoptosis might be indirect and mediated through BMP signalling, as reduction of apoptosis in the enamel knot of Jag2−/− mutant embryos correlates with downregulation of Bmp4 expression in dental epithelium. BMPs have been suggested to be important regulators of programmed cell death in various embryonic tissues, including the neural tube and limb buds (Dunn et al., 1997; Hofmann et al., 1996; Macias et al., 1997; Yokouchi et al., 1996; Zou and Niswander, 1996). Although Bmp4 expression was downregulated in the enamel knot of Jag2 mutants, expression of Bmp2 and Bmp7 was unaffected, suggesting that Bmp4 alone can act as an apoptotic signal in dental epithelium. However, there is no direct proof that Bmp4 activity is required for apoptosis in the developing teeth. The transcriptional regulation of BMPs is extremely complex, involving large genomic loci that contain multiple enhancer elements upstream and downstream of the coding exons (Pregizer and Mortlock, 2009), and tooth-specific enhancer regions have been reported for Bmp2 and Bmp4 (Chandler et al., 2009; Chandler et al., 2007). Insight into whether Bmp4 directly regulates apoptosis in the tooth could be obtained through applying recombinant BMP4 or BMP antagonists (e.g. noggin) to E14.5 tooth germs and probing for changes in apoptosis in the enamel knot. However, as BMPs often act in a spatially restricted manner, the use of a genetic model would clearly be preferred. Bmp4-deficient mice die at the gastrulation stage (Winnier et al., 1995), making a conditional approach using a floxed Bmp4 allele (Chang et al., 2008) in combination with a suitable (e.g. enamel knot-specific) Cre driver a necessity.
In addition to the morphological defects, Jag2 deletion disturbs the differentiation of dental epithelial progenitors into ameloblasts and of dental mesenchyme progenitors into odontoblasts. This is obvious in E18.5 incisors, which exhibit an earlier cytodifferentiation programme than molars. In the E18.5 incisors of wild-type embryos, odontoblasts and ameloblasts are fully differentiated and dentin and enamel deposition is evident, whereas odontoblast and ameloblast differentiation did not occur in the incisors of E18.5 Jag2−/− embryos and, as a consequence, the deposition of dentin and enamel matrices was severely affected. These findings demonstrate that Jag2-mediated Notch signalling is essential for both ameloblastic and odontoblastic fates, and are in accordance with numerous findings showing that the fates of progenitor cells from various tissues, such as the haematopoietic system, thymus and intestine, are under the influence of Notch signalling (Radtke and Clevers, 2005; Wilson and Radtke, 2006).
A major aspect of the tooth phenotype reflects the mutual interaction between Jag2-mediated Notch signalling and transcription factors and growth factors. Analysis of lineage and differentiation marker genes, such as Pitx1 (Mitsiadis and Drouin, 2008), Pitx2 (Mitsiadis and Drouin, 2008; Mitsiadis et al., 1998b; Mucchielli et al., 1997), Pax9 (Peters et al., 1998) and Barx1 (Mitsiadis et al., 1998b; Mucchielli et al., 1997), provides a good indication of the molecular alterations that take place in tooth germs of Jag2−/− mice. Of particular interest is that whereas the expression of the epithelial genes Pitx1 and Pitx2 was not significantly affected in Jag2−/− mutants, expression of both Barx1 and Pax9 was severely diminished in tooth mesenchyme. Thus, the specification of dental mesenchymal cells is partly controlled by Jag2-mediated Notch signalling in the epithelium, indicating an additional role of Notch as a central regulator of the epithelial-mesenchymal interactions that regulate tooth morphogenesis and cytodifferentiation.
The link between the Notch signalling pathway and Tbx1 in developing teeth
It has been shown that Jag2 has survival and proliferative effects on a variety of progenitor cells (e.g. haematopoietic cells) (DeHart et al., 2005; Francis et al., 2005; Radtke et al., 2005; Tsai et al., 2000). It is thus conceivable that Jag2 might exert the same effect on ameloblast progenitors. A key gene for the ameloblastic lineage is that of the transcription factor Tbx1 (Mitsiadis et al., 2008c). In its absence, proliferation of ameloblast precursors and their differentiation into ameloblasts are severely affected (Caton et al., 2009). Tbx1 expression was significantly reduced in the dental epithelium of Jag2−/− teeth, indicating that Notch signalling controls Tbx1 expression. Indeed, the lack of enamel in Jag2−/− incisors (the enamel phenotype) is similar to that observed in Tbx1−/− incisors (Caton et al., 2009). Furthermore, analysis of the mouse Tbx1 promoter using a combination of web-based programmes (Genomatix, Transfac, TFsearch) and phylogenetic examination revealed the existence of three potential Notch binding sites (P. Papagerakis, personal communication). Downregulation of Tbx1 expression could be responsible for the reduction of Fgf8 expression in the epithelium of E12.5 Jag2−/− tooth germs, as we have shown recently that Tbx1 and FGFs form a regulatory loop in dental tissues (Mitsiadis et al., 2008c).
On the basis of our results, we propose the following model of Jag2 function in developing teeth (Fig. 9). During tooth initiation, a group of oral epithelial cells forms a population of dental cells with an as yet unspecified fate. Through the influence of FGF signalling, a subset of cells within this group starts to express Jag2 and adopt the ameloblast fate. Jag2, in turn, activates Bmp4 expression in cells of the enamel knot. Under the influence of Bmp4 signalling, these cells will be eliminated by apoptosis, contributing to normal tooth morphology. Deletion of Jag2 inhibits local Notch signalling, leading to an uncontrolled execution of parallel differentiation programmes, reflected in the increased number of tooth cusps. Furthermore, inhibition of Jag2-mediated Notch signalling causes downregulation of Tbx1 expression in ameloblast progenitors. Through this process, a larger number of unspecified cells remain in the dental epithelium. These cells are unable to interact properly with the underlying dental mesenchyme, resulting in the concomitant downregulation of Barx1 and Pax9 expression in mesenchyme and the disturbance of both cytodifferentiation and mineralisation processes.
Fig. 9.
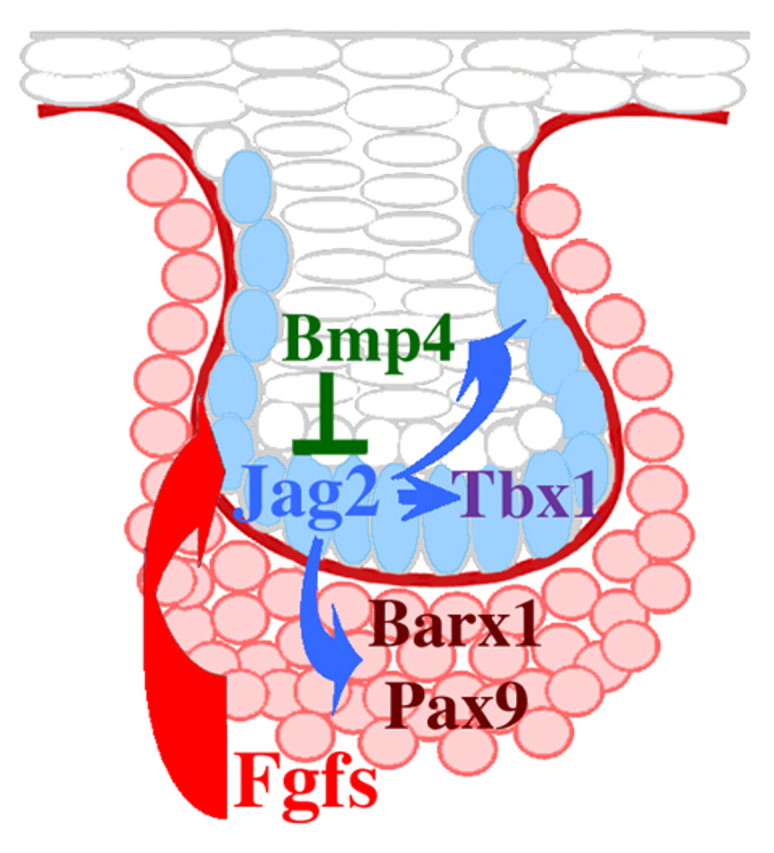
Model of the interactions between Jag2, Bmp4, FGF, Tbx1, Barx1 and Pax9 during early tooth development (E13, bud stage). Mesenchyme-derived FGF signals upregulate Jag2 expression in dental epithelial cells (blue) juxtaposed to mesenchyme (pink). Epithelial-derived Bmp4 signal is responsible for inactivation of Jag2 expression in dental epithelial cells. Jag2 inactivation leads to downregulation of Tbx1 expression in epithelial cells destined to form ameloblasts, and of Barx1 and Pax9 expression in dental mesenchyme.
In conclusion, the present findings show that Jag2-mediated Notch signalling is required for proper tooth formation. Correct regulation of the Notch pathway by FGF and BMP signals is important not only for the control of cell fates, but also for the maintenance of the correct balance of cell proliferation, differentiation and apoptosis.
Supplementary Material
Acknowledgements
We are grateful to Dr Papagerakis (CCMB, University of Michigan, Ann Arbor, USA) for help with the Tbx1 promoter. This work was supported by grants from the University of Zurich and Hartmann-Müller Foundation (Zurich, Switzerland).
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.049528/-/DC1
References
- Aberg T., Wozney J., Thesleff I. (1997). Expression patterns of bone morphogenetic proteins (Bmps) in the developing mouse tooth suggest roles in morphogenesis and cell differentiation. Dev. Dyn. 210, 383-396 [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Matsuno K., Fortini M. E. (1995). Notch signaling. Science 268, 225-232 [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand M. D., Lake R. J. (1999). Notch signaling: cell fate control and signal integration in development. Science 284, 770-776 [DOI] [PubMed] [Google Scholar]
- Bluteau G., Luder H. U., De Bari C., Mitsiadis T. A. (2008). Stem cells for tooth engineering. Eur. Cell. Mater. 16, 1-9 [DOI] [PubMed] [Google Scholar]
- Casey L. M., Lan Y., Cho E. S., Maltby K. M., Gridley T., Jiang R. (2006). Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development. Dev. Dyn. 235, 1830-1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton J., Luder H. U., Zoupa M., Bradman M., Bluteau G., Tucker A. S., Klein O., Mitsiadis T. A. (2009). Enamel-free teeth: Tbx1 deletion affects amelogenesis in rodent incisors. Dev. Biol. 328, 493-505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler K. J., Chandler R. L., Mortlock D. P. (2009). Identification of an ancient Bmp4 mesoderm enhancer located 46 kb from the promoter. Dev. Biol. 327, 590-602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler R. L., Chandler K. J., McFarland K. A., Mortlock D. P. (2007). Bmp2 transcription in osteoblast progenitors is regulated by a distant 3′ enhancer located 156.3 kilobases from the promoter. Mol. Cell. Biol. 27, 2934-2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W., Lin Z., Kulessa H., Hebert J., Hogan B. L., Wu D. K. (2008). Bmp4 is essential for the formation of the vestibular apparatus that detects angular head movements. PLoS Genet. 4, e1000050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobourne M. T., Mitsiadis T. (2006). Neural crest cells and patterning of the mammalian dentition. J. Exp. Zool. B Mol. Dev. Evol. 306, 251-260 [DOI] [PubMed] [Google Scholar]
- Conlon R. A., Reaume A. G., Rossant J. (1995). Notch1 is required for the coordinate segmentation of somites. Development 121, 1533-1545 [DOI] [PubMed] [Google Scholar]
- Cornell R. A., Eisen J. S. (2005). Notch in the pathway: the roles of Notch signaling in neural crest development. Semin. Cell Dev. Biol. 16, 663-672 [DOI] [PubMed] [Google Scholar]
- D'Souza B., Miyamoto A., Weinmaster G. (2008). The many facets of Notch ligands. Oncogene 27, 5148-5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassule H. R., McMahon A. P. (1998). Analysis of epithelial-mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev. Biol. 202, 215-227 [DOI] [PubMed] [Google Scholar]
- Dassule H. R., Lewis P., Bei M., Maas R., McMahon A. P. (2000). Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 127, 4775-4785 [DOI] [PubMed] [Google Scholar]
- DeHart S. L., Heikens M. J., Tsai S. (2005). Jagged2 promotes the development of natural killer cells and the establishment of functional natural killer cell lines. Blood 105, 3521-3527 [DOI] [PubMed] [Google Scholar]
- Dunn N. R., Winnier G. E., Hargett L. K., Schrick J. J., Fogo A. B., Hogan B. L. (1997). Haploinsufficient phenotypes in Bmp4 heterozygous null mice and modification by mutations in Gli3 and Alx4. Dev. Biol. 188, 235-247 [DOI] [PubMed] [Google Scholar]
- Ellisen L. W., Bird J., West D. C., Soreng A. L., Reynolds T. C., Smith S. D., Sklar J. (1991). TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66, 649-661 [DOI] [PubMed] [Google Scholar]
- Fortini M. E. (2009). Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell 16, 633-647 [DOI] [PubMed] [Google Scholar]
- Fortini M. E., Bilder D. (2009). Endocytic regulation of Notch signaling. Curr. Opin. Genet. Dev. 19, 323-328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis J. C., Radtke F., Logan M. P. (2005). Notch1 signals through Jagged2 to regulate apoptosis in the apical ectodermal ridge of the developing limb bud. Dev. Dyn. 234, 1006-1015 [DOI] [PubMed] [Google Scholar]
- Gridley T. (1997). Notch signaling in vertebrate development and disease. Mol. Cell. Neurosci. 9, 103-108 [DOI] [PubMed] [Google Scholar]
- Gridley T. (2003). Notch signaling and inherited disease syndromes. Hum. Mol. Genet. 12, R9-R13 [DOI] [PubMed] [Google Scholar]
- Hamada Y., Kadokawa Y., Okabe M., Ikawa M., Coleman J. R., Tsujimoto Y. (1999). Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development 126, 3415-3424 [DOI] [PubMed] [Google Scholar]
- Heikinheimo M., Lawshe A., Shackleford G. M., Wilson D. B., MacArthur C. A. (1994). Fgf-8 expression in the post-gastrulation mouse suggests roles in the development of the face, limbs and central nervous system. Mech. Dev. 48, 129-138 [DOI] [PubMed] [Google Scholar]
- Henderson S. T., Gao D., Lambie E. J., Kimble J. (1994). lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120, 2913-2924 [DOI] [PubMed] [Google Scholar]
- Hofmann C., Luo G., Balling R., Karsenty G. (1996). Analysis of limb patterning in BMP-7-deficient mice. Dev. Genet. 19, 43-50 [DOI] [PubMed] [Google Scholar]
- Hrabe de Angelis M., McIntyre J., 2nd, Gossler A. (1997). Maintenance of somite borders in mice requires the Delta homologue DII1. Nature 386, 717-721 [DOI] [PubMed] [Google Scholar]
- Hurlbut G. D., Kankel M. W., Lake R. J., Artavanis-Tsakonas S. (2007). Crossing paths with Notch in the hyper-network. Curr. Opin. Cell Biol. 19, 166-175 [DOI] [PubMed] [Google Scholar]
- Jarriault S., Brou C., Logeat F., Schroeter E. H., Kopan R., Israel A. (1995). Signalling downstream of activated mammalian Notch. Nature 377, 355-358 [DOI] [PubMed] [Google Scholar]
- Jernvall J., Aberg T., Kettunen P., Keranen S., Thesleff I. (1998). The life history of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development 125, 161-169 [DOI] [PubMed] [Google Scholar]
- Jiang R., Lan Y., Chapman H. D., Shawber C., Norton C. R., Serreze D. V., Weinmaster G., Gridley T. (1998). Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 12, 1046-1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joutel A., Corpechot C., Ducros A., Vahedi K., Chabriat H., Mouton P., Alamowitch S., Domenga V., Cecillion M., Marechal E., et al. (1996). Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature 383, 707-710 [DOI] [PubMed] [Google Scholar]
- Kettunen P., Thesleff I. (1998). Expression and function of FGFs-4, -8, and -9 suggest functional redundancy and repetitive use as epithelial signals during tooth morphogenesis. Dev. Dyn. 211, 256-268 [DOI] [PubMed] [Google Scholar]
- Kettunen P., Karavanova I., Thesleff I. (1998). Responsiveness of developing dental tissues to fibroblast growth factors: expression of splicing alternatives of FGFR1, -2, -3, and of FGFR4; and stimulation of cell proliferation by FGF-2, -4, -8, and -9. Dev. Genet. 22, 374-385 [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Cha Y. G., Cho S. W., Kim E. J., Lee M. J., Lee J. M., Cai J., Ohshima H., Jung H. S. (2006). Inhibition of apoptosis in early tooth development alters tooth shape and size. J. Dent. Res. 85, 530-535 [DOI] [PubMed] [Google Scholar]
- Klein O. D., Lyons D. B., Balooch G., Marshall G. W., Basson M. A., Peterka M., Boran T., Peterkova R., Martin G. R. (2008). An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development 135, 377-385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., Ilagan M. X. (2009). The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., Schroeter E. H., Weintraub H., Nye J. S. (1996). Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl. Acad. Sci. USA 93, 1683-1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. (2008). From signals to patterns: space, time, and mathematics in developmental biology. Science 322, 399-403 [DOI] [PubMed] [Google Scholar]
- Li L., Krantz I. D., Deng Y., Genin A., Banta A. B., Collins C. C., Qi M., Trask B. J., Kuo W. L., Cochran J., et al. (1997). Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat. Genet. 16, 243-251 [DOI] [PubMed] [Google Scholar]
- Limbourg F. P., Takeshita K., Radtke F., Bronson R. T., Chin M. T., Liao J. K. (2005). Essential role of endothelial Notch1 in angiogenesis. Circulation 111, 1826-1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsell C. E., Shawber C. J., Boulter J., Weinmaster G. (1995). Jagged: a mammalian ligand that activates Notch1. Cell 80, 909-917 [DOI] [PubMed] [Google Scholar]
- Louvi A., Artavanis-Tsakonas S. (2006). Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 7, 93-102 [DOI] [PubMed] [Google Scholar]
- Louvi A., Arboleda-Velasquez J. F., Artavanis-Tsakonas S. (2006). CADASIL: a critical look at a Notch disease. Dev. Neurosci. 28, 5-12 [DOI] [PubMed] [Google Scholar]
- Lumsden A. G. (1988). Spatial organization of the epithelium and the role of neural crest cells in the initiation of the mammalian tooth germ. Development 103, 155-169 [DOI] [PubMed] [Google Scholar]
- Luo B., Aster J. C., Hasserjian R. P., Kuo F., Sklar J. (1997). Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol. Cell. Biol. 17, 6057-6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias D., Ganan Y., Sampath T. K., Piedra M. E., Ros M. A., Hurle J. M. (1997). Role of BMP-2 and OP-1 (BMP-7) in programmed cell death and skeletogenesis during chick limb development. Development 124, 1109-1117 [DOI] [PubMed] [Google Scholar]
- McCright B., Gao X., Shen L., Lozier J., Lan Y., Maguire M., Herzlinger D., Weinmaster G., Jiang R., Gridley T. (2001). Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development 128, 491-502 [DOI] [PubMed] [Google Scholar]
- Mina M., Kollar E. J. (1987). The induction of odontogenesis in non-dental mesenchyme combined with early murine mandibular arch epithelium. Arch. Oral Biol. 32, 123-127 [DOI] [PubMed] [Google Scholar]
- Mitsiadis T. (2001). Bases moléculaires du développement dentaire. In La Dent Normale et Pathologique (ed. Piette E., Goldberg M.), pp. 19-38 Brussels: De Boeck-Université Press; [Google Scholar]
- Mitsiadis T. A., Drouin J. (2008). Deletion of the Pitx1 genomic locus affects mandibular tooth morphogenesis and expression of the Barx1 and Tbx1 genes. Dev. Biol. 313, 887-896 [DOI] [PubMed] [Google Scholar]
- Mitsiadis T. A., Graf D. (2009). Cell fate determination during tooth development and regeneration. Birth Defects Res. C Embryo Today 87, 199-211 [DOI] [PubMed] [Google Scholar]
- Mitsiadis T. A., Lardelli M., Lendahl U., Thesleff I. (1995a). Expression of Notch 1, 2 and 3 is regulated by epithelial-mesenchymal interactions and retinoic acid in the developing mouse tooth and associated with determination of ameloblast cell fate. J. Cell Biol. 130, 407-418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsiadis T. A., Muramatsu T., Muramatsu H., Thesleff I. (1995b). Midkine (MK), a heparin-binding growth/differentiation factor, is regulated by retinoic acid and epithelial-mesenchymal interactions in the developing mouse tooth, and affects cell proliferation and morphogenesis. J. Cell Biol. 129, 267-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsiadis T. A., Henrique D., Thesleff I., Lendahl U. (1997). Mouse Serrate-1 (Jagged-1): expression in the developing tooth is regulated by epithelial-mesenchymal interactions and fibroblast growth factor-4. Development 124, 1473-1483 [DOI] [PubMed] [Google Scholar]
- Mitsiadis T. A., Hirsinger E., Lendahl U., Goridis C. (1998a). Delta-notch signaling in odontogenesis: correlation with cytodifferentiation and evidence for feedback regulation. Dev. Biol. 204, 420-431 [DOI] [PubMed] [Google Scholar]
- Mitsiadis T. A., Mucchielli M. L., Raffo S., Proust J. P., Koopman P., Goridis C. (1998b). Expression of the transcription factors Otlx2, Barx1 and Sox9 during mouse odontogenesis. Eur. J. Oral. Sci. 106, 112-116 [DOI] [PubMed] [Google Scholar]
- Mitsiadis T. A., Angeli I., James C., Lendahl U., Sharpe P. T. (2003). Role of Islet1 in the patterning of murine dentition. Development 130, 4451-4460 [DOI] [PubMed] [Google Scholar]
- Mitsiadis T. A., Regaudiat L., Gridley T. (2005). Role of the Notch signalling pathway in tooth morphogenesis. Arch. Oral Biol. 50, 137-140 [DOI] [PubMed] [Google Scholar]
- Mitsiadis T. A., Caton J., De Bari C., Bluteau G. (2008a). The large functional spectrum of the heparin-binding cytokines MK and HB-GAM in continuously growing organs: the rodent incisor as a model. Dev. Biol. 320, 256-266 [DOI] [PubMed] [Google Scholar]
- Mitsiadis T. A., De Bari C., About I. (2008b). Apoptosis in developmental and repair-related human tooth remodeling: a view from the inside. Exp. Cell Res. 314, 869-877 [DOI] [PubMed] [Google Scholar]
- Mitsiadis T. A., Tucker A. S., De Bari C., Cobourne M. T., Rice D. P. (2008c). A regulatory relationship between Tbx1 and FGF signaling during tooth morphogenesis and ameloblast lineage determination. Dev. Biol. 320, 39-48 [DOI] [PubMed] [Google Scholar]
- Mucchielli M. L., Mitsiadis T. A., Raffo S., Brunet J. F., Proust J. P., Goridis C. (1997). Mouse Otlx2/RIEG expression in the odontogenic epithelium precedes tooth initiation and requires mesenchyme-derived signals for its maintenance. Dev. Biol. 189, 275-284 [DOI] [PubMed] [Google Scholar]
- Muskavitch M. A. (1994). Delta-notch signaling and Drosophila cell fate choice. Dev. Biol. 166, 415-430 [DOI] [PubMed] [Google Scholar]
- Neubuser A., Koseki H., Balling R. (1995). Characterization and developmental expression of Pax9, a paired-box-containing gene related to Pax1. Dev. Biol. 170, 701-716 [DOI] [PubMed] [Google Scholar]
- Neubuser A., Peters H., Balling R., Martin G. R. (1997). Antagonistic interactions between FGF and BMP signaling pathways: a mechanism for positioning the sites of tooth formation. Cell 90, 247-255 [DOI] [PubMed] [Google Scholar]
- Nye J. S., Kopan R., Axel R. (1994). An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development 120, 2421-2430 [DOI] [PubMed] [Google Scholar]
- Oda T., Elkahloun A. G., Pike B. L., Okajima K., Krantz I. D., Genin A., Piccoli D. A., Meltzer P. S., Spinner N. B., Collins F. S., et al. (1997). Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat. Genet. 16, 235-242 [DOI] [PubMed] [Google Scholar]
- Peters H., Neubuser A., Kratochwil K., Balling R. (1998). Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 12, 2735-2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pregizer S., Mortlock D. P. (2009). Control of BMP gene expression by long-range regulatory elements. Cytokine Growth Factor Rev. 20, 509-515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F., Clevers H. (2005). Self-renewal and cancer of the gut: two sides of a coin. Science 307, 1904-1909 [DOI] [PubMed] [Google Scholar]
- Radtke F., Wilson A., MacDonald H. R. (2005). Notch signaling in hematopoiesis and lymphopoiesis: lessons from Drosophila. BioEssays 27, 1117-1128 [DOI] [PubMed] [Google Scholar]
- Robey E. (1997). Notch in vertebrates. Curr. Opin. Genet. Dev. 7, 551-557 [DOI] [PubMed] [Google Scholar]
- Swiatek P. J., Lindsell C. E., del Amo F. F., Weinmaster G., Gridley T. (1994). Notch1 is essential for postimplantation development in mice. Genes Dev. 8, 707-719 [DOI] [PubMed] [Google Scholar]
- Thesleff I., Hurmerinta K. (1981). Tissue interactions in tooth development. Differentiation 18, 75-88 [DOI] [PubMed] [Google Scholar]
- Tissier-Seta J. P., Mucchielli M. L., Mark M., Mattei M. G., Goridis C., Brunet J. F. (1995). Barx1, a new mouse homeodomain transcription factor expressed in cranio-facial ectomesenchyme and the stomach. Mech. Dev. 51, 3-15 [DOI] [PubMed] [Google Scholar]
- Tsai S., Fero J., Bartelmez S. (2000). Mouse Jagged2 is differentially expressed in hematopoietic progenitors and endothelial cells and promotes the survival and proliferation of hematopoietic progenitors by direct cell-to-cell contact. Blood 96, 950-957 [PubMed] [Google Scholar]
- Tummers M., Thesleff I. (2009). The importance of signal pathway modulation in all aspects of tooth development. J. Exp. Zool. B Mol. Dev. Evol. 312, 309-319 [DOI] [PubMed] [Google Scholar]
- Valsecchi C., Ghezzi C., Ballabio A., Rugarli E. I. (1997). JAGGED2: a putative Notch ligand expressed in the apical ectodermal ridge and in sites of epithelial-mesenchymal interactions. Mech. Dev. 69, 203-207 [DOI] [PubMed] [Google Scholar]
- Viriot L., Peterkova R., Vonesch J. L., Peterka M., Ruch J. V., Lesot H. (1997). Mouse molar morphogenesis revisited by three-dimensional reconstruction. III. Spatial distribution of mitoses and apoptoses up to bell-staged first lower molar teeth. Int. J. Dev. Biol. 41, 679-690 [PubMed] [Google Scholar]
- Wang X. P., Suomalainen M., Felszeghy S., Zelarayan L. C., Alonso M. T., Plikus M. V., Maas R. L., Chuong C. M., Schimmang T., Thesleff I. (2007). An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 5, e159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmaster G. (1997). The ins and outs of notch signaling. Mol. Cell. Neurosci. 9, 91-102 [DOI] [PubMed] [Google Scholar]
- Wilkinson D. G. (1995). RNA detection using non-radioactive in situ hybridization. Curr. Opin. Biotechnol. 6, 20-23 [DOI] [PubMed] [Google Scholar]
- Wilson A., Radtke F. (2006). Multiple functions of Notch signaling in self-renewing organs and cancer. FEBS Lett. 580, 2860-2868 [DOI] [PubMed] [Google Scholar]
- Winnier G., Blessing M., Labosky P. A., Hogan B. L. (1995). Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 9, 2105-2116 [DOI] [PubMed] [Google Scholar]
- Xue Y., Gao X., Lindsell C. E., Norton C. R., Chang B., Hicks C., Gendron-Maguire M., Rand E. B., Weinmaster G., Gridley T. (1999). Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum. Mol. Genet. 8, 723-730 [DOI] [PubMed] [Google Scholar]
- Yokouchi Y., Sakiyama J., Kameda T., Iba H., Suzuki A., Ueno N., Kuroiwa A. (1996). BMP-2/-4 mediate programmed cell death in chicken limb buds. Development 122, 3725-3734 [DOI] [PubMed] [Google Scholar]
- Zou H., Niswander L. (1996). Requirement for BMP signaling in interdigital apoptosis and scale formation. Science 272, 738-741 [DOI] [PubMed] [Google Scholar]
- Zoupa M., Seppala M., Mitsiadis T., Cobourne M. T. (2006). Tbx1 is expressed at multiple sites of epithelial-mesenchymal interaction during early development of the facial complex. Int. J. Dev. Biol. 50, 504-510 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.