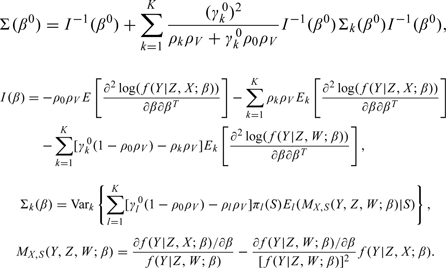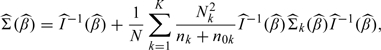Abstract
Two-stage design has long been recognized to be a cost-effective way for conducting biomedical studies. In many trials, auxiliary covariate information may also be available, and it is of interest to exploit these auxiliary data to improve the efficiency of inferences. In this paper, we propose a 2-stage design with continuous outcome where the second-stage data is sampled with an “outcome-auxiliary-dependent sampling” (OADS) scheme. We propose an estimator which is the maximizer for an estimated likelihood function. We show that the proposed estimator is consistent and asymptotically normally distributed. The simulation study indicates that greater study efficiency gains can be achieved under the proposed 2-stage OADS design by utilizing the auxiliary covariate information when compared with other alternative sampling schemes. We illustrate the proposed method by analyzing a data set from an environmental epidemiologic study.
Keywords: Auxiliary covariate, Kernel smoothing, Outcome-auxiliary-dependent sampling, 2-stage sampling design
1. INTRODUCTION
Biomedical studies are often designed to assess the relationship between some exposure X of interest and the corresponding outcome Y of individual adjusted by some confounding covariates Z. In many situations, due to limited budget, the assessment of X is not feasible to be conducted on all subjects under study. One useful approach to accommodating this issue is to use a 2-stage stratified sampling design, originally introduced by Neyman (1938), to enhance the study efficiency while minimizing the costs. At the first stage of a typical 2-stage design, a relatively large random sample is drawn and measurements are conducted on outcome Y and Z, which are easier to measure, while at the second stage, ascertainments on the X are made for a subsample drawn randomly, without replacement.
There is great literature on the variations of 2-stage sampling designs with binary outcomes. For example, White (1982) proposed a stratified case–control design of a rare disease (i.e. Y) and a rare exposure (i.e. X), where a large preliminary random sample is drawn at the first stage, from which strata are identified on the basis of both the disease and the exposure. At the second stage, a subsample is drawn from within the strata identified in the first stage and measurements of the potential confounding variables are made on the subsample. Compared with the simple random sampling at the second stage regardless of either the disease or the exposure status, great efficiency gains can be achieved by selecting the desirable number of cases and controls within each stratum identified in the first stage. Rathouz and others (2002) considered a matched case–control study with binary outcome using the conditional logistic regression method. Recently, Schildcrout and Rathouz (2010) extended this stratified case–control design to a more general case where the response is a longitudinal binary variable.
On the other hand, when there exists an additional auxiliary variable W for the expensive X, which is easily obtained for all subjects under study at the first stage, it is necessary to incorporate the information implied by W into the statistical analysis. For instance, in a lung cancer biomarker study, one of the aims is to assess the epidermal growth factor receptor (EGFR) mutations (X) as a predictive biomarker for whether a subject responds to a greater extent to EGFR inhibitor drugs (Y). Due to high cost of genotyping EGFR genes, it is prohibitive to ascertain the genotype of EGFR genes on all samples at the first stage. However, the likelihood score of EGFR mutations (W) obtained by a designed questionnaire has been shown to relate to the EGFR mutations and can be easily observed for all patients in Paez and others (2004). Wang and Zhou (2010) considered inference of the 2-stage outcome-auxiliary-dependent sampling (OADS) design to increase the study efficiency by utilizing the auxiliary covariate information when the outcome is categorical. Zhang and others (2008) and Lu and Tsiatis (2008) also showed that using the available baseline auxiliary covariate information can achieve more efficient estimators in the analysis of randomized clinical trials and survival data, respectively.
As the scope of biomedical studies inquiry grows, it is important to investigate the relationships between continuous biological outcomes and exposure of interest adjusted by other covariates. It is cost-effective to adopt a 2-stage design when the exposure is hard to obtain. However, most current 2-stage designs have been developed for categorical outcomes, the statistical method for the 2-stage design with continuous outcome is limited. When an auxiliary W does not exist, Chatterjee and others (2003) considered a pseudoscore estimator for regression parameter with a 2-stage sampling. Weaver and Zhou (2005) proposed a 2-stage outcome-dependent sampling (ODS) design for continuous outcome regression models, wherein the subsample was drawn at the second stage within the stratum that was achieved by subdividing the range of continuous outcome variable into class intervals.
In this paper, we proposed a 2-stage OADS design when outcome Y is continuous and there exists auxiliary variable W at the first stage. Specifically, outcome Y, auxiliary variable W for exposure X, and other covariates Z are all observed for all patients at the first stage. Then we selected the subsample within each stratum defined by the partition of the domain of Y×W to ascertain the value of X at the second stage. An estimated likelihood function by estimating its infinite-dimensional nuisance parameter through the kernel smoother is proposed and the estimator maximizing the estimated likelihood is used to estimate the regression parameter. The proposed 2-stage OADS design with continuous outcome is shown to be more efficient than other alternative competing sampling schemes.
The rest of this paper is structured as follows. We describe the 2-stage OADS design, data structure, and the model in Section 2. The estimated likelihood function method and the asymptotic properties of the resulting estimator are presented in Section 3. We conduct a simulation study to assess the small sample approximation under the 2-stage OADS design in Section 4. In Section 5, a real data example is analyzed to illustrate our proposed method. Some conclusions are provided in Section 6, and the proof of the asymptotic properties of proposed estimator is investigated in the supplementary material available at Biostatistics online.
2. DESIGN AND MODEL
2.1. Two-stage OADS design and data structure
To fix notation, let Y denote a continuous outcome variable, {Z,X} be a covariate vector, and W be a continuous auxiliary variable for X. We assume that the conditional distribution of Y given Z and X is known up to a finite vector of unknown parameters, that is,
| (2.1) |
where β0 is the true value of q-vector regression parameter β of interest. Assume that W offers no additional information regarding the outcome Y given covariate X.
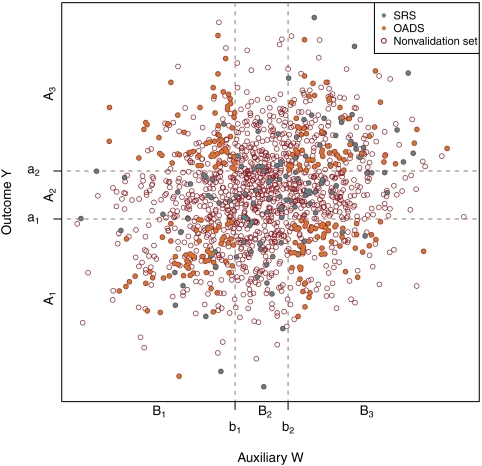
Assume that the domain of (Y,W) is denoted by 𝒴×𝒲. Let 𝒴 be partitioned into J mutually exclusive and exhaustive strata by the known constants − ∞ = a0 < a1 < ⋯ < aJ − 1 < aJ = ∞, and let the jth stratum be denoted by Aj = (aj − 1,aj], for j = 1,…,J. Similarly, let 𝒲 be partitioned into T mutually exclusive and exhaustive strata by the known constants − ∞ = b0 < b1 < ⋯ < bT − 1 < bT = ∞, and let the tth stratum be denoted by Bt = (bt − 1,bt], for t = 1,…,T. For subsequential use, we define B0 = ( − ∞,∞) when T = 0, which indicates that there is no partition on 𝒲. Therefore, we have 𝒴×𝒲 partitioned into J×T mutually exclusive and exhaustive rectangles Aj×Bt, for j = 1,…,J and t = 1,…,T. For simplicity, we rewrite these rectangles as Δk for k = 1,…,K. Hence, {Aj×Bt:j = 1,…,Jandt = 1,…,T} = {Δk:k = 1,⋯,K} and 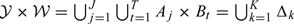
At the first stage, N subjects are sampled at random from a population with (Yi,Zi,Wi)i = 1N being observed. Suppose that there are Nk observations of (Y,W) falling into stratum Δk, then 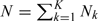 . The second stage, where X is observed, are comprised of 2 components: (i) a simple random sample (SRS) of size n0 and (ii) a supplemental OADS sample of size nk from the kth stratum Δk for k = 1,…,K. Let Ri be an indicator for the ith subject whether Xi is observed (Ri = 1) or not (Ri = 0). Let n0k denote the number of subjects in the SRS falling into the kth stratum Δk. Furthermore, let
. The second stage, where X is observed, are comprised of 2 components: (i) a simple random sample (SRS) of size n0 and (ii) a supplemental OADS sample of size nk from the kth stratum Δk for k = 1,…,K. Let Ri be an indicator for the ith subject whether Xi is observed (Ri = 1) or not (Ri = 0). Let n0k denote the number of subjects in the SRS falling into the kth stratum Δk. Furthermore, let 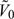 denote all the subjects in the SRS and define Vk = {i:Ri = 1,(Yi,Wi)∈Δk} and
denote all the subjects in the SRS and define Vk = {i:Ri = 1,(Yi,Wi)∈Δk} and 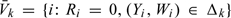 , then nk + n0k = |Vk| and
, then nk + n0k = |Vk| and 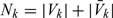 , where and hereafter, we use notation |A| to denote the cardinality of a set A. Let
, where and hereafter, we use notation |A| to denote the cardinality of a set A. Let 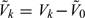 representing the supplemental OADS samples in the stratum Δk, where A − B is defined as the set consisting of elements that are in set A but not in set B. Let V = cupk = 1KVk and
representing the supplemental OADS samples in the stratum Δk, where A − B is defined as the set consisting of elements that are in set A but not in set B. Let V = cupk = 1KVk and 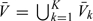 , representing the validation set (set with X observed, i.e. the second-stage set) and nonvalidation set (i.e. the first stage set that are not sampled at the second stage), respectively. Hence, the observed data structure for the proposed 2-stage OADS design with continuous outcome can be summarized as follows: the first stage: {Yi,Zi,Wi} for i = 1,⋯,N; the second stage: (i) the SRS sample: {Yi,Xi,Zi,Wi} for i ∈
, representing the validation set (set with X observed, i.e. the second-stage set) and nonvalidation set (i.e. the first stage set that are not sampled at the second stage), respectively. Hence, the observed data structure for the proposed 2-stage OADS design with continuous outcome can be summarized as follows: the first stage: {Yi,Zi,Wi} for i = 1,⋯,N; the second stage: (i) the SRS sample: {Yi,Xi,Zi,Wi} for i ∈ 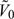 ; (ii) the OADS sample: {Xi|(Yi,Wi)∈Δk,Zi} for
; (ii) the OADS sample: {Xi|(Yi,Wi)∈Δk,Zi} for  and k = 1,…,K; and (iii) the nonvalidation sample: {Yi,Zi,Wi} for
and k = 1,…,K; and (iii) the nonvalidation sample: {Yi,Zi,Wi} for 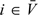 .
.
To better illustrate the proposed OADS design with continuous outcome, we present Figure 1 when J = T = 3. At the second stage, except for the SRS samples, the supplemental OADS samples are selected within strata at the 4 corners Δ1 = A1×B1,Δ2 = A1×B3,Δ3 = A3×B1, and Δ4 = A3×B3 based on the consideration that these combinations of the extreme values of both Y and W contain more information for the relationship of interest between outcome Y and exposure X. Hence, the advantage of such 2-stage OADS design is that, while providing overall information about the population from the SRS samples, it allows the investigator to oversample certain segments of the population that are believed to be more informative.
Fig. 1.
Illustration for the proposed 2-stage OADS design with continuous outcome. Y-axis denotes outcome variable Y. X-axis denotes auxiliary variable W.
The 2-stage ODS design proposed by Weaver and Zhou (2005) assumed that only the outcome variable is observed in the first stage and the covariates are ascertained for a subsample drawn at the second stage from strata defined by the outcome. Our proposed 2-stage OADS design includes this design when T = 0 and the information in Z and W is discarded. We call this design a 2-stage ODS design with only the outcome observed at the first stage. However, in many studies, some covariates such age, gender, and race so forth can be observed for all subjects in the cohort study. To this point, we extended the design by Weaver and Zhou (2005) to this more practical situation. When the auxiliary information is available for all subjects, our proposed 2-stage OADS design can accommodate the 2-stage ODS design with outcome, some covariates, and auxiliary observed at the first stage by letting T = 0. It is worth noting that the subsequential methodology development on the 2-stage OADS design is still valid for the 2-stage ODS design in several abovementioned scenarios.
2.2. Likelihood function
Let G(x|z,w) and g(x|z,w) be the conditional cumulative distribution function and the conditional probability function of X given (Z,W). We will construct the likelihood function based on all the observations under the 2-stage OADS design. First, the contribution from the SRS at the second stage to the full likelihood is proportional to
| (2.2) |
Second, the likelihood for the supplemental OADS sample at the second stage can be shown to be proportional to (Zhou and others, 2002)
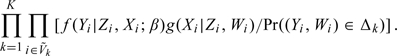 |
(2.3) |
Furthermore, the observations in the nonvalidation sample contribute the following term to the full-information likelihood function:
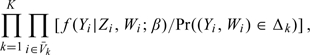 |
(2.4) |
where f(Y|Z,W;β) = ∫𝒳f(Y|Z,x;β)dG(x|Z,W).
Finally, as shown by Weaver and Zhou (2005), conditional on the component size of the OADS being fixed, the kth stratum size for the nonvalidation sample 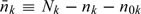 follows a multinomial law such that
follows a multinomial law such that
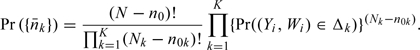 |
(2.5) |
Conditional on the observed size 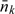 , the observations in the nonvalidation sample are independent of those in the validation sample. After combining and simplifying terms (2.2–2.5), we have derived the full likelihood based on all the observations under the 2-stage OADS design, which is proportional to
, the observations in the nonvalidation sample are independent of those in the validation sample. After combining and simplifying terms (2.2–2.5), we have derived the full likelihood based on all the observations under the 2-stage OADS design, which is proportional to
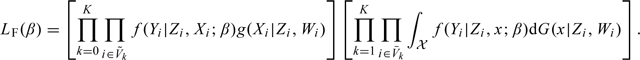 |
(2.6) |
The presence of the nuisance function G(x|z,w) makes the inference for β challenging. Obviously, direct maximization of LF(β) is not feasible since the function G(x|z,w) cannot be factored out. A simple method is to assume a parametric distribution for G(x|z,w), but this could lead to a biased conclusion if the underlying model is misspecified in that, generally, the relationship between W and X may not be known to be specified through a parametric model. A more attractive approach is to model it nonparametrically.
3. AN ESTIMATED LIKELIHOOD METHOD
In the estimated likelihood method, an unspecified nuisance parameter, such as the conditional distribution function G(x|z,w) in (2.6), is replaced by a consistent estimator. When the validation sample is a simple random sample, one could estimate G(x|z,w) using data from validation sample by an empirical imputation method for discrete auxiliary (Pepe and Fleming, 1991) and by kernel smoothing (Carroll and Wand, 1991) for continuous auxiliary. Zhou and Pepe (1995), Zhou and Wang (2000), and Liu and others (2009) applied the estimated likelihood approach to time-to-event data subject to random censoring.
Due to the 2-stage OADS design, the validation sample is not a simple random sample so we cannot use a simple global empirical distribution function to estimate G(x|z,w). Hence, one should account for the sampling mechanism under the 2-stage OADS design to estimate G(x|z,w) nonparametrically. Let S denote the informative components of (Z,W) in the sense that G(X|Z,W) = G(X|S) almost surely. Without loss of generality, assume that S is a continuous variable with dimension d. Note that 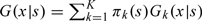 where πk(s) = Pr((Y,W)∈Δk|s) and Gk(x|s) = G(x|s,(Y,W)∈Δk). Then we estimate πk(s) and Gk(x|s), respectively, by
where πk(s) = Pr((Y,W)∈Δk|s) and Gk(x|s) = G(x|s,(Y,W)∈Δk). Then we estimate πk(s) and Gk(x|s), respectively, by 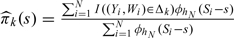 and
and 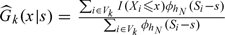 where I(·) is an indicator function and
where I(·) is an indicator function and 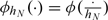 is a d-dimensional kernel function with the bandwidth hN. For simplicity, we suppress the subscript of hN hereafter. Hence, G(x|s) can be subsequently estimated by
is a d-dimensional kernel function with the bandwidth hN. For simplicity, we suppress the subscript of hN hereafter. Hence, G(x|s) can be subsequently estimated by 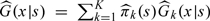 which is a consistent estimator as shown in the supplementary material available at Biostatistics online.
which is a consistent estimator as shown in the supplementary material available at Biostatistics online.
The estimated likelihood function is obtained by substituting G(x|s) in (2.6) with 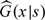 and the corresponding estimated log-likelihood function is denoted by
and the corresponding estimated log-likelihood function is denoted by 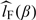 , where
, where
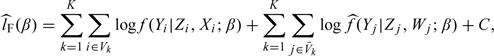 |
with
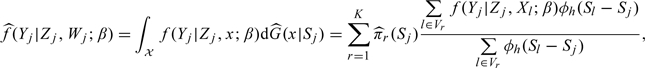 |
and 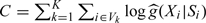 , which is not dependent on β.
, which is not dependent on β.
The solution to the estimated score equations 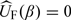 , denoted by
, denoted by 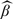 , is used to estimate β0, where
, is used to estimate β0, where
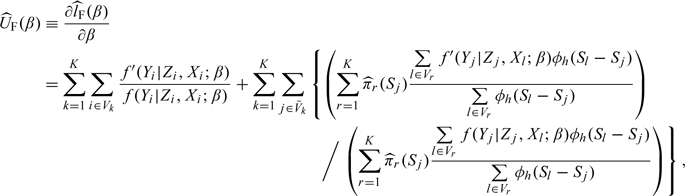 |
with f′(y|z,x;β) = ∂f(y|z,x;β)/∂β. One can adopt the Newton–Raphson iteration method to obtain the estimator 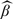 . A simple ad hoc bandwidth selection
. A simple ad hoc bandwidth selection 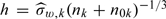 can be used if S = W almost surely, where
can be used if S = W almost surely, where 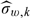 is the sample standard error of {Wi,i∈Vk}.
is the sample standard error of {Wi,i∈Vk}.
The true value of parameters are indicated by superscript “0.” Let Ek denote a conditional expectation given (Y,W)∈Δk, under the true parameters. Assume that |V|/N→ρV > 0 and nk/|V|→ρk ≥ 0 for k = 0,⋯,K, as N→∞. Let γk = Pr{(Y,W)∈Δk}. The regularity conditions needed to derive the asymptotic properties are given in the supplementary material available at Biostatistics online. Then the asymptotic properties of the proposed estimator 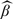 are summarized in the following theorem.
are summarized in the following theorem.
THEOREM 1.
Under the regularity conditions,
converges in probability to β0, while
converges weakly to a normal distribution with mean zero and covariance Σ(β0), where
The proof of Theorem 1 is provided in the separate supplementary material available at Biostatistics online. The consistent variance estimator is stated in the following theorem.
THEOREM 2.
Under the regularity conditions, a consistent estimator for the asymptotic covariance matrix Σ(β0) is
where
and
with
.
4. SIMULATION STUDY
We conducted a simulation study to assess the small sample performance of our proposed estimator. The data were generated from a linear regression model of the form:
where X, Z, and ς were generated independently from standard normal distribution. Thus, the conditional distribution of Y given X and Z is normal with mean β0 + β1X + β2Z and variance 4. Let W = X + ϵ, where ϵ was generated from a zero-mean normal distribution with variance σ2. Note that the value of σ2 indicates the strength of information contained in W for X. We set σ = 1 in simulation, which represents a moderate association between the W and X. Here, we take S = W.
Suppose there are N subjects available at the first stage. Let ai and bi denote the i/3 percentile of Y and W, respectively, for i = 1,2. First, we use the method depicted in Figure 1 to obtain the second stage samples for the 2-stage OADS design. Then the size of the validation set is 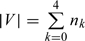 Second, while selecting the same SRS sample of size n0, we also select the 2 supplemental ODS samples in the stratum A1 of size n1 + n2 and stratum A3 of size n3 + n4, respectively, to mimic the design proposed by Weaver and Zhou (2005). Note that the sizes of validation set V obtained at the second stage through the above 2 sampling designs are the same.
Second, while selecting the same SRS sample of size n0, we also select the 2 supplemental ODS samples in the stratum A1 of size n1 + n2 and stratum A3 of size n3 + n4, respectively, to mimic the design proposed by Weaver and Zhou (2005). Note that the sizes of validation set V obtained at the second stage through the above 2 sampling designs are the same.
Having obtained the data under the 2-stage OADS design, we denote the proposed estimator by 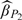 . We also denote the reduced proposed estimator by
. We also denote the reduced proposed estimator by 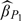 for the 2-stage ODS design with (Y,Z,W) observed at the first stage. We compare estimators
for the 2-stage ODS design with (Y,Z,W) observed at the first stage. We compare estimators 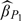 and
and 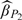 with some competing estimators. The first estimator, denoted by
with some competing estimators. The first estimator, denoted by 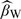 , is the inverse probability weighted estimator (Horvitz and Thompson, 1952) based on the 2-stage OADS design. The second estimators to be compared, as discussed in the Section 2.1, are the estimator
, is the inverse probability weighted estimator (Horvitz and Thompson, 1952) based on the 2-stage OADS design. The second estimators to be compared, as discussed in the Section 2.1, are the estimator 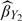 for the 2-stage ODS design with (Y,Z) observed at the first stage and, similarly, the estimator
for the 2-stage ODS design with (Y,Z) observed at the first stage and, similarly, the estimator 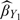 for the 2-stage ODS design with only Y observed at the first stage and (X,Z) observed at the second stage. The bandwidth
for the 2-stage ODS design with only Y observed at the first stage and (X,Z) observed at the second stage. The bandwidth 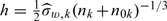 is used for these estimators involving kernel smoothing, where
is used for these estimators involving kernel smoothing, where 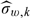 is the sample standard error of {Wi,i∈Vk}. Finally, as a benchmark, we also consider the efficient linear regression estimator, denoted by
is the sample standard error of {Wi,i∈Vk}. Finally, as a benchmark, we also consider the efficient linear regression estimator, denoted by 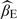 , which is a hypothetical situation in which all subjects at the first stage have X observed, and the ordinary linear regression estimator, denoted by
, which is a hypothetical situation in which all subjects at the first stage have X observed, and the ordinary linear regression estimator, denoted by 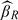 , from a simple random sample of the same size as the validation set at the second stage. Note that the efficiency difference for methods βY1, βY2, βP1, and βP2 should be attributed to the study design instead of estimating procedure. However, βP2 and βW are different estimating procedures under the same 2-stage OADS design.
, from a simple random sample of the same size as the validation set at the second stage. Note that the efficiency difference for methods βY1, βY2, βP1, and βP2 should be attributed to the study design instead of estimating procedure. However, βP2 and βW are different estimating procedures under the same 2-stage OADS design.
For narrative simplicity, we define an allocation function denoted by allocation(μ,ν) to allocate the validation set of size μ + 4ν at the second stage, which means that n0 = μ and n1 = n2 = n3 = n4 = ν under the 2-stage OADS design as illustrated in Figure 1. Under the 2-stage ODS design, allocation(μ,ν) means SRS sample of size μ and 2 supplemental ODS samples in the stratum A1 of size 2ν and in stratum A3 of size 2ν are allocated. We also investigate the impact on the parameter estimation of different allocations of total validation sample size between the SRS sample and the supplemental OADS (ODS) samples at the second stage, with (N,β0,β1,β2) = (1500,0.5,0.3,0.5) fixed.
For each simulation configuration, 1000 replicated samples were generated and the results were presented in Table 1. Under the model studied, we make the following observations on the estimator 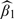 , the parameter of interest. Note that the estimator
, the parameter of interest. Note that the estimator 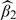 works quite well in all scenarios. First, all the methods in all the scenarios yield consistent estimators, the variance estimators accurately reflect the true variations, and the confidence intervals have proper coverage probabilities. Second, the proposed estimators
works quite well in all scenarios. First, all the methods in all the scenarios yield consistent estimators, the variance estimators accurately reflect the true variations, and the confidence intervals have proper coverage probabilities. Second, the proposed estimators 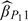 and
and 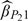 are more efficient than the estimators
are more efficient than the estimators 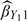 and
and 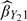 , which indicates that taking auxiliary information into consideration indeed gains substantial estimation efficiency. Furthermore,
, which indicates that taking auxiliary information into consideration indeed gains substantial estimation efficiency. Furthermore, 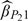 is more efficient than
is more efficient than 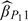 . This fits our expectation since
. This fits our expectation since 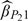 not only utilizes the auxiliary in the stratification (i.e. study design) but also incorporates it into the estimation procedure, while
not only utilizes the auxiliary in the stratification (i.e. study design) but also incorporates it into the estimation procedure, while 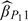 uses it just in the estimation procedure. On the other hand, although the precision of estimator
uses it just in the estimation procedure. On the other hand, although the precision of estimator 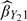 and that of
and that of 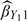 are almost the same in the scenarios considered, the efficiency gains of
are almost the same in the scenarios considered, the efficiency gains of 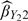 over
over 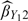 are apparent due to the fact that the covariate Z is observed for all subjects in
are apparent due to the fact that the covariate Z is observed for all subjects in 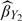 . The estimator
. The estimator 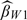 is less efficient than
is less efficient than 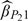 since
since 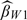 just utilizes the second-stage sample and sampling probability under the 2-stage OADS design. Third, when we increase the size of the validation set from |V| = 240 to |V| = 360, more accurate estimators (including
just utilizes the second-stage sample and sampling probability under the 2-stage OADS design. Third, when we increase the size of the validation set from |V| = 240 to |V| = 360, more accurate estimators (including 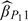 ,
, 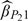 ,
, 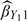 ,
, 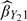 ,
, 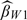 , and
, and 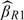 ) are obtained as expected. Here, we consider 3 different ways to add the additional 120 samples to the validation set |V| = 240. It can be seen that more efficiency gains are achievable through the way from allocation(120,30) to allocation(180,45), that is, putting half of the additional 120 samples to the SRS part and the other half to the OADS part averagely, than that from allocation(120,30) to allocation(240,30), that is, putting the additional 120 samples to the SRS part. Efficiency gains are also achieved through the way from allocation(120,30) to allocation(120,60), which puts the additional 120 samples to the OADS part evenly. These different allocation patterns indicate that adding the additional sample to both the SRS part and the supplemental OADS part or the supplemental OADS part is better than to the SRS part only. Finally, under the allocation(120,60), when the cutpoints vary from the
) are obtained as expected. Here, we consider 3 different ways to add the additional 120 samples to the validation set |V| = 240. It can be seen that more efficiency gains are achievable through the way from allocation(120,30) to allocation(180,45), that is, putting half of the additional 120 samples to the SRS part and the other half to the OADS part averagely, than that from allocation(120,30) to allocation(240,30), that is, putting the additional 120 samples to the SRS part. Efficiency gains are also achieved through the way from allocation(120,30) to allocation(120,60), which puts the additional 120 samples to the OADS part evenly. These different allocation patterns indicate that adding the additional sample to both the SRS part and the supplemental OADS part or the supplemental OADS part is better than to the SRS part only. Finally, under the allocation(120,60), when the cutpoints vary from the 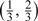 to
to 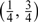 , that is, when the product sample space 𝒴×𝒲 is stratified by more extreme cutpoints, more precise estimators (including
, that is, when the product sample space 𝒴×𝒲 is stratified by more extreme cutpoints, more precise estimators (including 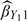 ,
, 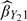 ,
, 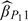 , and
, and 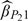 ) are obtained, and the efficiency advantage of
) are obtained, and the efficiency advantage of 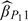 over
over 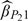 becomes more obvious. We also investigate the effect of the strength of W for X, represented by σ, on the efficiency of estimator
becomes more obvious. We also investigate the effect of the strength of W for X, represented by σ, on the efficiency of estimator 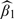 , under the methods considered. Please see Figure A.1 in the supplementary material available at Biostatistics online.
, under the methods considered. Please see Figure A.1 in the supplementary material available at Biostatistics online.
Table 1.
Simulation study for the proposed estimators. Results are based on 1000 replicated data sets with 1500 subjects at the first stage for each data set†
| Cutpoints | |V| | Method |
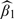 |
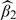 |
|||||||
| Mean | SE | CI | Mean | SE | CI | ||||||
| — | — | βE | 0.299 | 0.050 | 0.052 | 0.966 | 0.497 | 0.052 | 0.052 | 0.946 | |
| — | 240 | βR | 0.298 | 0.124 | 0.130 | 0.962 | 0.500 | 0.126 | 0.130 | 0.958 | |
| — | 360 | βR | 0.297 | 0.107 | 0.106 | 0.943 | 0.496 | 0.105 | 0.106 | 0.943 | |
| allocation(120, 30) | |||||||||||
| 240 | βW | 0.303 | 0.113 | 0.123 | 0.978 | 0.502 | 0.138 | 0.134 | 0.931 | ||
| 0.304 | 0.113 | 0.112 | 0.955 | 0.505 | 0.121 | 0.102 | 0.906 | ||||
| 0.305 | 0.116 | 0.116 | 0.953 | 0.499 | 0.050 | 0.052 | 0.960 | ||||
| 0.305 | 0.072 | 0.068 | 0.941 | 0.500 | 0.050 | 0.052 | 0.957 | ||||
| 0.301 | 0.070 | 0.068 | 0.948 | 0.500 | 0.050 | 0.052 | 0.953 | ||||
| allocation(180, 45) | |||||||||||
| 360 | βW | 0.302 | 0.095 | 0.100 | 0.967 | 0.494 | 0.110 | 0.109 | 0.951 | ||
| 0.299 | 0.094 | 0.093 | 0.951 | 0.502 | 0.092 | 0.087 | 0.941 | ||||
| 0.300 | 0.096 | 0.096 | 0.952 | 0.500 | 0.053 | 0.052 | 0.943 | ||||
| 0.307 | 0.068 | 0.066 | 0.940 | 0.500 | 0.053 | 0.052 | 0.947 | ||||
| 0.303 | 0.064 | 0.065 | 0.954 | 0.500 | 0.053 | 0.052 | 0.945 | ||||
| allocation(240, 30) | |||||||||||
| 360 | βW | 0.303 | 0.091 | 0.099 | 0.971 | 0.496 | 0.103 | 0.105 | 0.952 | ||
| 0.301 | 0.099 | 0.095 | 0.936 | 0.498 | 0.096 | 0.089 | 0.936 | ||||
| 0.305 | 0.098 | 0.098 | 0.947 | 0.500 | 0.053 | 0.052 | 0.939 | ||||
| 0.308 | 0.070 | 0.066 | 0.933 | 0.500 | 0.053 | 0.052 | 0.948 | ||||
| 0.302 | 0.069 | 0.066 | 0.932 | 0.503 | 0.053 | 0.052 | 0.939 | ||||
| allocation(120, 60) | |||||||||||
| 360 | βW | 0.302 | 0.100 | 0.107 | 0.967 | 0.504 | 0.118 | 0.120 | 0.952 | ||
| 0.295 | 0.093 | 0.091 | 0.950 | 0.502 | 0.093 | 0.086 | 0.935 | ||||
| 0.304 | 0.097 | 0.093 | 0.931 | 0.500 | 0.053 | 0.052 | 0.943 | ||||
| 0.308 | 0.069 | 0.065 | 0.928 | 0.502 | 0.053 | 0.052 | 0.943 | ||||
| 0.299 | 0.067 | 0.065 | 0.938 | 0.502 | 0.053 | 0.052 | 0.944 | ||||
| 360 | βW | 0.303 | 0.114 | 0.114 | 0.942 | 0.510 | 0.135 | 0.131 | 0.940 | ||
| 0.303 | 0.085 | 0.085 | 0.954 | 0.505 | 0.086 | 0.082 | 0.942 | ||||
| 0.300 | 0.084 | 0.086 | 0.949 | 0.500 | 0.053 | 0.052 | 0.942 | ||||
| 0.304 | 0.067 | 0.064 | 0.936 | 0.499 | 0.051 | 0.052 | 0.956 | ||||
| 0.293 | 0.061 | 0.063 | 0.963 | 0.499 | 0.051 | 0.052 | 0.958 | ||||
Results are based on the model Y = β0 + β1X + β2Z + 2ς with true values β0 = 0.5, β1 = 0.3, and β2 = 0.5, where X, Z, and ς are mutually independently standard normal variables. The auxiliary variable W is defined to be equal to X plus a standard normal error term. 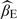 : the regression estimator when X is observed for all subjects at the first stage;
: the regression estimator when X is observed for all subjects at the first stage; 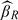 : the regression estimator from a simple random sample of the same size as the validation set at the second stage;
: the regression estimator from a simple random sample of the same size as the validation set at the second stage; 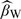 : the inverse probability weighted estimator using the validation set under the 2-stage OADS design;
: the inverse probability weighted estimator using the validation set under the 2-stage OADS design; 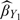 : the estimator for the 2-stage ODS design with only Y observed at the first stage and (X, Z) observed for the second-stage sample;
: the estimator for the 2-stage ODS design with only Y observed at the first stage and (X, Z) observed for the second-stage sample; 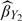 : the estimator for the 2-stage ODS design with (Y, Z) observed at the first stage;
: the estimator for the 2-stage ODS design with (Y, Z) observed at the first stage; 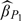 : the estimator for the 2-stage ODS design with (Y, Z, W) observed at the first stage;
: the estimator for the 2-stage ODS design with (Y, Z, W) observed at the first stage; 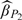 : the estimator for the proposed 2-stage OADS design with (Y, Z, W) observed at the first stage.
: the estimator for the proposed 2-stage OADS design with (Y, Z, W) observed at the first stage.
It should be noted that in above simulation results, the covariate X was generated independently from Z. Therefore, we took S = W and then adopted a univariate kernel smoothing method to estimate the function g(X|Z,W) = g(X|W) nonparametrically. As suggested by one of the referees, here we intend to investigate our proposed estimators when g(X|W) is specified parametrically instead of being estimated by kernel smoothing. Note that in our above simulation setups g(X|W) is a normal density function with mean W and variance 2. The resultant estimate is denoted by 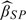 . Furthermore, we also consider this estimate in the misspecified situation in which the X was generated from the model
. Furthermore, we also consider this estimate in the misspecified situation in which the X was generated from the model 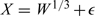 but the working model remains to be X = W + ϵ. The related results are formulated in Table 2. Obviously, when g(X|W) is correctly specified, the estimate
but the working model remains to be X = W + ϵ. The related results are formulated in Table 2. Obviously, when g(X|W) is correctly specified, the estimate 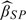 outperforms the nonparametric methods. However, when g(X|W) is misspecified, the estimate
outperforms the nonparametric methods. However, when g(X|W) is misspecified, the estimate 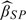 is biased with low coverage probability while the nonparametric smoothing estimates, including our proposed estimates
is biased with low coverage probability while the nonparametric smoothing estimates, including our proposed estimates 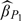 and
and 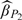 , still work well.
, still work well.
Table 2.
Simulation study for the proposed estimators. Results are based on 1000 replicated data sets with 1500 subjects at the first stage and allocation pattern allocation(120, 60) at the second stage under the cutpoints 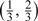 for each data set†
for each data set†
| g(X|W) | Method |
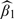 |
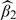 |
|||||||
| Mean | SE | CI | Mean | SE | CI | |||||
| Specified | βW | 0.302 | 0.100 | 0.107 | 0.967 | 0.504 | 0.118 | 0.120 | 0.952 | |
| 0.295 | 0.093 | 0.091 | 0.950 | 0.502 | 0.093 | 0.086 | 0.935 | |||
| 0.304 | 0.097 | 0.093 | 0.931 | 0.500 | 0.053 | 0.052 | 0.943 | |||
| 0.308 | 0.069 | 0.065 | 0.928 | 0.502 | 0.053 | 0.052 | 0.943 | |||
| 0.299 | 0.067 | 0.065 | 0.938 | 0.502 | 0.053 | 0.052 | 0.944 | |||
| βSP | 0.302 | 0.060 | 0.059 | 0.951 | 0.504 | 0.052 | 0.052 | 0.947 | ||
| Misspecified | βW | 0.307 | 0.104 | 0.108 | 0.969 | 0.505 | 0.121 | 0.121 | 0.955 | |
| 0.307 | 0.098 | 0.095 | 0.932 | 0.504 | 0.096 | 0.090 | 0.931 | |||
| 0.310 | 0.099 | 0.096 | 0.925 | 0.508 | 0.059 | 0.057 | 0.941 | |||
| 0.309 | 0.075 | 0.074 | 0.926 | 0.503 | 0.058 | 0.057 | 0.947 | |||
| 0.306 | 0.071 | 0.068 | 0.934 | 0.505 | 0.057 | 0.056 | 0.941 | |||
| βSP | 0.269 | 0.066 | 0.063 | 0.903 | 0.512 | 0.059 | 0.051 | 0.929 | ||
See note for Table 1.
On the other hand, as suggested by another referee, in some practice, d, the dimension of W, could be greater than one, and then multivariate kernel smoothing method would be involved. Hence, it is of practical importance to see how sensitive the resulting inference on the parameters of interest is with regard to the dimension d of kernel smoothing. We explore this issue with some modifications of the simulation models, where we generate Z from model Z = W2 + ϵ2, where W and ϵ2 are both generated independently from a standard normal distribution. We keep the remaining parametric simulation settings unchanged. We use 2 dimensional product standard normal kernels to estimate g(X|Z,W) with bandwidth matrix diag(h1,h2), where 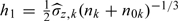 , h2 is defined in a similar pattern, and
, h2 is defined in a similar pattern, and 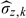 is the sample standard error of {Zi,i∈Vk}. The corresponding estimates are listed in Table 3. It can be seen that when the dimension of kernel smoothing d equals 2, the resultant estimates of β1 of main interest are slightly biased with low coverage probability except for the inverse probability estimate
is the sample standard error of {Zi,i∈Vk}. The corresponding estimates are listed in Table 3. It can be seen that when the dimension of kernel smoothing d equals 2, the resultant estimates of β1 of main interest are slightly biased with low coverage probability except for the inverse probability estimate 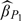 . Even then, our proposed estimators
. Even then, our proposed estimators 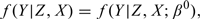 and
and 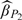 outperform
outperform 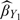 and
and 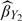 .
.
Table 3.
Simulation study for the proposed estimators. Results are based on 1000 replicated data sets with 1500 subjects at the first stage and allocation pattern allocation(120, 60) at the second stage under the cutpoints 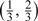 for each data set with S = (Z, W)†
for each data set with S = (Z, W)†
| Method |
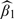 |
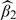 |
|||||||
| Mean | SE | CI | Mean | SE | CI | ||||
| βE | 0.302 | 0.053 | 0.054 | 0.957 | 0.506 | 0.057 | 0.056 | 0.949 | |
| βR | 0.294 | 0.113 | 0.110 | 0.941 | 0.498 | 0.107 | 0.109 | 0.949 | |
| βW | 0.297 | 0.109 | 0.109 | 0.953 | 0.506 | 0.115 | 0.117 | 0.956 | |
| 0.315 | 0.101 | 0.100 | 0.929 | 0.512 | 0.098 | 0.092 | 0.930 | ||
| 0.317 | 0.099 | 0.096 | 0.926 | 0.503 | 0.059 | 0.058 | 0.949 | ||
| 0.315 | 0.076 | 0.074 | 0.927 | 0.505 | 0.059 | 0.058 | 0.946 | ||
| 0.287 | 0.072 | 0.071 | 0.929 | 0.506 | 0.059 | 0.057 | 0.941 | ||
See note for Table 1.
5. ANALYSIS OF THE COLLABORATIVE PERINATAL PROJECT DATA
As an illustration, we applied our proposed method to a data set from the Collaborative Perinatal Project (CPP) to evaluate the effect of maternal pregnancy serum level of polychlorinated biphenyls (PCB) of a mother on her children's intelligence quotient (IQ) test performance. Pregnant mothers were enrolled through university-affiliated medical clinics and data were collected on the mothers each prenatal visit. The children born during the study were also followed for various outcomes for up to 8 years. One hypothesis is that PCB levels are related to the performance on the Weschler Intelligence Scale for children at 7 years of age (Longnecker and others, 1997). To investigate the in utero exposure of PCB in relation to neurodevelopmental abnormality, the PCB levels were measured by analyzing the third trimester blood serum specimens that had been preserved from mothers in the CPP study. Due to the expense of conducting the blood serum assay to measure the PCB level, the study investigators decided to assess the PCB levels for an overall simple random sample of 849 subjects from the underlying population. In addition to the PCB level as the exposure variable of interest, other confounding variables available for all subjects under study include socioeconomic status of the child's family (SES), gender (SEX) and race (RACE) of the child indicating for female and black, respectively, the mother's education (EDU) and age (AGE).
To illustrate our methods, we use the simple random sample of 849 subjects as our underlying population. We then construct a 2-stage OADS design for this base population as an illustration. The first stage sample is the 849 subjects, that is, N = 849. We first explore the relationship between SES and PCB based on the first-stage sample data. A linear model fit for PCB given SES yields the estimate of slope 0.154(p < 0.0001), which indicates a linear association between SES and PCB. On the other hand, in terms of practical consideration in environmental epidemiology, higher SES usually leads to higher PCB level. Hence, we use SES as the auxiliary variable for PCB.
The 1/3 and 2/3 sample quantiles of IQ are 3.7 and 5.3, and the 1/3 and 2/3 sample quantiles of SES are 90 and 101, respectively. Hence, we can take a1 = 3.7,a2 = 5.3,b1 = 90, and b2 = 101. With respect to the second-stage samples, assume that 60 SRS samples and 30 supplemental OADS samples in each corner are selected under the allocation pattern allocation(60,30). We use the chi-square statistics to test the independence between IQ and SES, given PCB. In particular, we discretize PCB by dPCB = (PCB > median(PCB)). Under condition dPCB = 0, we can also define dIQ and dSES in a similar pattern, and then use the chi-square test yielding p-value 0.6038. Similarly, under condition dPCB = 1, the chi-square test yields p-value 0.4386. Hence, we think conditioning on PCB level, IQ does not further depend on SES. The fitted model is
where ε is a zero-mean normal variable with unknown variance.
The results for the CPP data analysis are summarized in Table 4. Note that since the other confounding covariates such EDU, SES, AGE, and so on are observed for all subjects, the method βY1 which assumes that only the outcome is observed at the the first stage is not considered in the data analysis. First, we are interested in the estimate for PCB under various methods. It is evident that all the analyses confirm that the PCB level of mother's third-trimester blood serum specimen is not significantly related to the IQ scores for children at 7 years of age. Second, a more precise 95% confidence interval ( − 0.432,1.002) is achieved for the estimate of PCB using method βP2. For example, the 95% confidence intervals for the estimates of PCB are ( − 0.425,1.253), ( − 0.531,1.303), and ( − 0.791,1.149) using methods βW, βY2, and βP1, respectively. Meanwhile, the estimated standard error for the estimate of PCB in the hypothetical case βE is the smallest one among all the methods considered. Also, the method βE yields the most accurate 95% confidence interval ( − 0.190,0.702) for the estimate of PCB. Third, the estimators for the remaining covariates under various methods considered are all almost the same as confirmed in the simulation study. Finally, despite that slightly different conclusions are obtained under methods βR and βW, the methods βE, βY2, βP1, and βP2 all confirm that SES, EDU, and RACE have a positive impact on the IQ scores of children while there is no evidence that both the AGE and SEX have any effect on the IQ scores.
Table 4.
Analysis results for the CPP study†
| Method | Intercept | PCB | EDU | SES | AGE | RACE | SEX | |
| βE | Est. | 80.025∗ | 0.256 | 1.258∗ | 1.078∗ | 0.018 | – 7.942∗ | – 0.590 |
| 2.795 | 0.228 | 0.223 | 0.266 | 0.070 | 0.927 | 0.842 | ||
| LCI | 74.546 | – 0.190 | 0.822 | 0.558 | – 0.118 | – 9.759 | – 2.240 | |
| UCI | 85.504 | 0.702 | 1.694 | 1.599 | 0.155 | – 6.125 | 1.060 | |
| βR | Est. | 77.897∗ | 0.711 | 1.122∗ | 0.847 | 0.131 | – 7.355∗ | – 0.423 |
| 6.912 | 0.496 | 0.534 | 0.619 | 0.167 | 2.071 | 1.934 | ||
| LCI | 64.349 | – 0.262 | 0.076 | – 0.367 | – 0.195 | – 11.414 | – 4.214 | |
| UCI | 91.446 | 1.683 | 2.168 | 2.061 | 0.458 | – 3.296 | 3.368 | |
| βW | Est. | 78.391∗ | 0.414 | 1.322∗ | 0.592∗ | 0.199∗ | – 7.752∗ | – 1.085 |
| 2.732 | 0.428 | 0.207 | 0.245 | 0.061 | 0.876 | 0.779 | ||
| LCI | 73.036 | – 0.425 | 0.916 | 0.112 | 0.079 | – 9.469 | – 2.612 | |
| UCI | 83.746 | 1.253 | 1.728 | 1.072 | 0.319 | – 6.036 | 0.442 | |
| Est. | 79.154∗ | 0.386 | 1.264∗ | 1.102∗ | 0.028 | – 7.841∗ | – 0.611 | |
| 3.015 | 0.468 | 0.222 | 0.263 | 0.068 | 0.917 | 0.839 | ||
| LCI | 73.245 | – 0.531 | 0.830 | 0.586 | – 0.106 | – 9.638 | – 2.255 | |
| UCI | 85.063 | 1.303 | 1.699 | 1.617 | 0.162 | – 6.044 | 1.034 | |
| Est. | 79.759∗ | 0.179 | 1.268∗ | 1.088∗ | 0.031 | – 7.825∗ | – 0.597 | |
| 2.947 | 0.495 | 0.222 | 0.273 | 0.068 | 0.917 | 0.839 | ||
| LCI | 73.982 | – 0.791 | 0.833 | 0.553 | – 0.103 | – 9.623 | – 2.242 | |
| UCI | 85.536 | 1.149 | 1.702 | 1.623 | 0.165 | – 6.027 | 1.048 | |
| Est. | 80.722∗ | 0.285 | 1.269∗ | 1.174∗ | 0.034 | – 7.732∗ | – 0.588 | |
| 2.894 | 0.366 | 0.222 | 0.273 | 0.068 | 0.921 | 0.839 | ||
| LCI | 75.049 | – 0.432 | 0.834 | 0.639 | – 0.100 | – 9.538 | – 2.232 | |
| UCI | 86.395 | 1.002 | 1.703 | 1.709 | 0.168 | – 5.926 | 1.055 |
The outcome is the IQ scores for children at 7 years of age. PCB is the level measured from the third-trimester blood serum specimens, EDU is the mother's education level, SES is the socioeconomic status of the child's family, AGE is the mother's age, and RACE and SEX are the race and gender of the child, respectively. The fitted model is IQ = βint + β1PCB + β2EDU + β3SES + β4AGE + β5RACE + β6SEX + ϵ, where ϵ is zero-mean normal variable with unknown variance. The auxiliary variable is SES, the cutpoints are 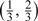 , and the allocation pattern is allocation(60, 30). “Est.” is the estimation of the covariate's effect, “
, and the allocation pattern is allocation(60, 30). “Est.” is the estimation of the covariate's effect, “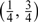 ” is the estimated standard error, “LCI” is the lower bound of the 95% confidence interval, and “UCI” is the upper bound of the 95% confidence interval. The symbol “∗” means the corresponding parameter estimate is significant at 5% level.
” is the estimated standard error, “LCI” is the lower bound of the 95% confidence interval, and “UCI” is the upper bound of the 95% confidence interval. The symbol “∗” means the corresponding parameter estimate is significant at 5% level.
6. CONCLUDING REMARKS
We proposed a new 2-stage OADS design in which the selected supplemental samples at the second stage are allowed to depend on both a continuous outcome variable and a continuous auxiliary variable. This 2-stage OADS design can be easily reduced to the 2-stage ODS design with auxiliary covariate information. An estimated likelihood function based on nonparametric kernel smoothing method is developed to accommodate the 2-stage OADS design with continuous outcome variable. The proposed estimator is shown to be consistent and asymptotically normal. The simulation study suggests that greater efficiency can be gained in estimating the effect of the exposure variable on the outcome using the proposed 2-stage OADS design over the existing or other competing 2-stage ODS designs. Additionally, using the available auxiliary data information can also substantially improve the efficiency of the study. A real data analysis is provided to illustrate our proposed method.
When the dimension d of S is moderately large (e.g. d > = 3), the proposed method will not work well due to the curse of high dimensionality. One possible way is to specify g(X|S) parametrically. However, this parametric method could lead to some biased results when g(X|S) is misspecified. In practice, we suggested using our proposed method when d < = 2 and using the parametric method when d > = 2.
The proposed 2-stage OADS design allows the investigators to focus their attention on the subjects who are more informative for study aims. Generally, the issue of how to appropriately divide the domain of 𝒴×𝒲 to obtain the strata Δk′s may affect the efficiency of estimators. Taking the CPP data as an example, we want to select those subjects with very high or low IQ scores and SES values as much as possible. On the other hand, the number of those subjects that we can sample is decreasing along with higher or lower values of both the IQ scores and SES. Hence, one needs to balance between the 2 above points when using a 2-stage OADS design. Our experience shows that the cutpoints consisting of 1/3 (or 1/4) and 2/3 (or 3/4) quantiles of both the outcome and auxiliary are usually feasible in practice.
SUPPLEMENTARY MATERIAL
Supplementary material is available at http://www.biostatistics.oxfordjournals.org.
FUNDING
National Institutes of Health (R01 CA79949 to H.Z., Y.W.; R01 HL57444 to J.C.); China Postdoctoral Science Foundation (20100480877 to Y.W.); National Nature Science Fund of China (10771163 to Y.L.).
Supplementary Material
Acknowledgments
The authors are very grateful for the valuable comments and suggestions from the editor and the referees. They also thank Ms. Beth Horton for careful reading of the manuscript. Conflict of Interest: None declared.
References
- Carroll RJ, Wand MP. Semiparametric estimation in logistic measurement error models. Journal of the Royal Statistical Society, Series B. 1991;53:573–585. [Google Scholar]
- Chatterjee N, Chen Y-H, Breslow NE. A pseudoscore estimator for regression problems with two-phase sampling. Journal of the American Statistical Association. 2003;98:158–168. [Google Scholar]
- Horvitz DG, Thompson DJ. A generalization of sampling without replacement from a finite universe. Journal of the American Statistical Association. 1952;47:663–685. [Google Scholar]
- Liu Y, Zhou H, Cai J. Estimated pseudo-likelihood method for correlated failure time data with auxiliary covariates. Biometrics. 2009;65:1184–1193. doi: 10.1111/j.1541-0420.2009.01198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker M, Klebanoff M, Zhou H, Wilcox A, Berendes H, Hoffman H. Proposal to Study in Utero Exposure to DDE and PCBs in Relation to Male Birth Defects and Neurodevelopmental Outcomes in the Collaborative Perinatal Project. Study Proposal. Washington, DC: National Institute of Environmental Health Science; 1997. [Google Scholar]
- Lu X, Tsiatis AA. Improving the efficiency of the log-rank test using auxiliary covariates. Biometrika. 2008;95:679–694. [Google Scholar]
- Neyman J. Contribution to the theory of sampling from human populations. Journal of the American Statistical Association. 1938;33:101–116. [Google Scholar]
- Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ. and others. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1450. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pepe MS, Fleming TR. A nonparametric method for dealing with mismeasured covariate data. Journal of the American Statistical Association. 1991;86:108–113. [Google Scholar]
- Rathouz PJ, Satten GA, Carroll RJ. Semiparametric inference in matched case-control studies with missing covariate data. Biometrika. 2002;89:905–916. [Google Scholar]
- Schildcrout JS, Rathouz PJ. Longitudinal studies of binary response data following case-control and stratified case-control sampling: design and analysis. Biometrics. 2010;66:365–373. doi: 10.1111/j.1541-0420.2009.01306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhou H. Design and inference for cancer biomarker study with an outcome and auxiliary-dependent subsampling. Biometrics. 2010;66:502–511. doi: 10.1111/j.1541-0420.2009.01280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver MA, Zhou H. An estimated likelihood method for continuous outcome regression models with outcome-dependent sampling. Journal of the American Statistical Association. 2005;100:459–469. [Google Scholar]
- White JE. A two stage design for the study of the relationship between a rare exposure and a rare disease. American Journal of Epidemiology. 1982;115:119–128. doi: 10.1093/oxfordjournals.aje.a113266. [DOI] [PubMed] [Google Scholar]
- Zhang M, Tsiatis AA, Davidian M. Improving efficiency of inferences in randomized clinical trials using auxiliary covariates. Biometrics. 2008;64:707–715. doi: 10.1111/j.1541-0420.2007.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Pepe MS. Auxiliary covariate data in failure time regression analysis. Biometrika. 1995;82:139–149. [Google Scholar]
- Zhou H, Wang CY. Failure time regression with continuous covariates measured with error. Journal of the Royal Statistical Society, Series B. 2000;62:657–665. [Google Scholar]
- Zhou H, Weaver MA, Qin J, Longnecker MP, Wang MC. A semiparametric empirical likelihood method for data from an outcome-dependent sampling scheme with a continuous outcome. Biometrics. 2002;58:413–421. doi: 10.1111/j.0006-341x.2002.00413.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.