Abstract
Light-harvesting antenna system possesses an inherent property of photoprotection. The single-helix proteins found in cyanobacteria play role in photoprotection and/or pigment metabolism. The photoprotective functions are also manifested by the two- and four-helix proteins. The photoprotection mechanism evolved earlier to the mechanism of light-harvesting of the antenna complex. Here, the light-harvesting complex genes of photosystems I and II from Arabidopsis are enlisted, and almost similar set of genes are identified in rice. Also, the three-helix early light-inducible proteins (ELIPs), two-helix stress-enhanced proteins (SEPs) and one-helix high light-inducible proteins [one-helix proteins (OHPs)] are identified in rice. Interestingly, two independent genomic loci encoding PsbS protein are also identified with implications on additional mode of non-photochemical quenching (NPQ) mechanism in rice. A few additional LHC-related genes are also identified in rice (LOC_Os09g12540, LOC_Os02g03330). This is the first report of identification of light-harvesting complex genes and light-inducible genes in rice.
Key words: Lhca and Lhcb proteins, Lhc proteins evolution, light-inducible proteins, protein alignment, PsbS
The light-harvesting proteins are present in different taxa. The proteins of light-harvesting systems from higher plants, cyano-bacteria, purple bacteria and green sulphur bacteria share no sequence similarity however little structural similarity can be seen.1 Apparently, the light-harvesting systems in these different taxa might have evolved independently from each other.1 To enable efficient transfer of excitation energy into the reaction centers, where charge separation takes place, different proteins are recruited in order to coordinate the photosynthetic pigment molecules. The light-harvesting and light dissipation are tightly coupled processes involving the higher plant light-harvesting antenna. Here, genome-wide analysis of the light-harvesting chlorophyll a/b-binding proteins and light-inducible proteins in Arabidopsis thaliana L. and Oryza sativa L. (rice) is conducted. This study wherein genes coding for antenna proteins are identified and named can be used as a nomenclature guide to the light-harvesting complex gene family members and their relatives in rice.
The Light-harvesting Complexes (LHCs) of Higher Plants
A protein family of 10–12 members, plus a few of related proteins constitutes the light-harvesting chlorophyll a/b-binding (LHC) proteins, the proteins of higher plant light-harvesting antenna. The proteins Lhca1–6 associate with photosystem I (PSI), and the proteins Lhcb1–6 primarily with photosystem II (PSII). The proteins Lhcb1 and Lhcb2 reorient between PSI and PSII to balance energy distribution and thereby the electron flow during a process called state transitions. Other names are proposed to designate the gene products of higher plant Lhcb4, 5 and 6 light-harvesting antenna proteins.2 For instance, Lhcb4, 5 and 6 are also designated as chlorophyll protein (CP) 29, CP26 and CP24 respectively, which suggests the relative mobility of these proteins upon SDS-PAGE electrophoresis. In Arabidopsis, several multiple loci exist for genes of Lhcb1 and Lhcb2. There are at least five (Lhcb1.1–Lhcb1.5) (AT1G29920, AT1G29910, AT1G29930, AT2G34430, AT2G34420), and three (Lhcb2.1–Lhcb2.3) (AT2G05100, AT2G05070, AT3G27690) multiple loci encoding Lhcb1 and Lhcb2 respectively in A. thaliana3–5 (Table 1). The amino acid sequences of these individual Lhcb1 and Lhcb2 proteins are slightly different. These differences are not well conserved in plant species. There are three multiple genes for Lhcb4 (CP29) (Lhcb4.1 to Lhcb4. 3) (AT5G01530, AT3G08940, AT2G40100), and one each for Lhcb3 (AT5G54270), Lhcb5 (CP26) (AT4G10340), and Lhcb6 (CP24) (AT1G15820) (Table 1). There are at least six PSI light-harvesting complex genes in A. thaliana (Lhca1 to Lhca6) (AT3G54890, AT3G61470, AT1G61520, AT3G47470, AT1G45474 and AT1G19150) (Table 1). A few additional LHC-related genes are also identified in A. thaliana (AT1G76570, AT4G17600 and AT5G28450).
Table 1.
Light-harvesting complex genes of photosystem I and II, and light-inducible genes in Arabidopsis
| Locus | Annotation | Synonym | A* | B* | C* |
| AT1G15820 | Lhcb6 protein | CP24, Light-harvesting complex PSII subunit 6 | 258 | 27522.3 | 7.6661 |
| AT1G19150 | Lhca2*1 mRNA | LHCA6, PSI Light-harvesting complex gene 6 | 270 | 29939.0 | 6.2432 |
| AT1G29910 | chlorophyll a/b-binding protein | CAB3, Light-harvesting chlorophyll a/b-binding protein 1.2 (LHCB1.2) | 267 | 28226.8 | 5.0852 |
| AT1G29920 | Lhcb1.1 | CAB2, Light-harvesting chlorophyll a/b-binding protein 1.1 (LHCB1.1) | 267 | 28226.8 | 5.0852 |
| AT1G29930 | LHCII | CAB1, Light-harvesting chlorophyll a/b-protein 1.3 (LHCB1.3) | 267 | 28240.9 | 5.3477 |
| AT1G45474 | Lhca5 | LHCA5, PSI Light-harvesting complex gene 5 | 256 | 27801.8 | 7.2572 |
| AT1G61520 | Lhca3*1 | LHCA3, PSI Light-harvesting complex gene 3 | 273 | 29181.2 | 9.0847 |
| AT1G76570 | chlorophyll a/b-binding protein | F14G6.17, F14G6_17 | 327 | 36374.6 | 8.4044 |
| AT2G05070 | Lhcb2.2 | LHCB2.2, PSII Light-harvesting complex gene 2.2 | 265 | 28620.3 | 5.0852 |
| AT2G05100 | Lhcb2.1 | LHCB2.1, PSII Light-harvesting complex gene 2.1 | 265 | 28649.3 | 5.0921 |
| AT2G34420 | chlorophyll a/b-binding protein | PSII Light-harvesting complex gene 1.5 (LHCB1.5), LHB1B2 | 265 | 28053.7 | 5.0913 |
| AT2G34430 | chlorophyll a/b-binding protein | PSII Light-harvesting complex gene 1.4 (LHCB1.4), LHB1B1 | 266 | 28169.8 | 4.9178 |
| AT2G40100 | Lhcb4.3 | LHCB4.3 | 276 | 30211.3 | 4.9911 |
| AT3G08940 | Lhcb4.2 | LHCB4.2 | 287 | 31193.2 | 6.1385 |
| AT3G27690 | Lhcb2.4 | LHCB2.4, PSII Light harvesting complex gene 2.3 (LHCB2.3) | 266 | 28802.5 | 5.6978 |
| AT3G47470 | chlorophyll a/b-binding protein | CAB4, Light-harvesting chlorophyll-protein complex subunit A4 (LHCA4) | 251 | 27733.4 | 6.6847 |
| AT3G54890 | LHCA1 | LHCA1 | 241 | 25995.7 | 6.674 |
| AT3G61470 | LHCA2 | LHCA2 | 257 | 27754.6 | 7.5728 |
| AT4G10340 | LHCB5/CP26 | Light-harvesting complex PSII 5 (LHCB5) | 280 | 30156.4 | 6.2942 |
| AT4G17600 | light-harvesting like | LIL3:1 | 262 | 29403.0 | 4.6688 |
| AT5G01530 | LHCB4/CP29 | LHCB4.1 | 290 | 31139.1 | 6.0326 |
| AT5G28450 | chlorophyll a/b-binding protein | F21B23.110, F21B23_110 | 173 | 18947.7 | 11.2702 |
| AT5G54270 | Lhcb3 | LHCB3*1, Light-harvesting chlorophyll B-binding protein 3 (LHCB3) | 265 | 28706.5 | 4.735 |
| AT1G44575 | PsbS | Nonphotochemical quenching (NPQ) 4, Photosystem II subunit S | 265 | 28007.5 | 9.9957 |
| AT3G22840 | early light-inducible protein 1 | ELIP1 | 195 | 20324.4 | 10.3836 |
| AT4G14690 | early light-inducible protein 2 | ELIP2 | 193 | 20344.5 | 10.3453 |
| AT4G34190 | stress-enhanced protein 1 | SEP1 | 146 | 14858.0 | 11.5119 |
| AT2G21970 | stress-enhanced protein 2 | SEP2 | 202 | 21988.8 | 4.7414 |
| AT5G02120 | One-helix protein | OHP | 110 | 12010.0 | 10.1387 |
| AT1G34000 | One-helix protein 2 | OHP2 | 172 | 18665.1 | 9.9339 |
A, amino acids; B, molecular weight; C, isoelectric point.
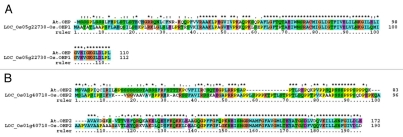
In rice, there are 17 genomic loci which encode for chlorophyll a/b-binding proteins (Table 2). Of these, genes of light-harvesting complex proteins associated with PSI and PSII are annotated during the study. The identified PSI light-harvesting complex genes are Lhca1 to Lhca6 (Table 3). The chlorophyll a/b-binding proteins of PSII light-harvesting complex are Lhcb1.1 to Lhcb1.3, Lhcb2, Lhcb3, Lhcb4, Lhcb5 and Lhcb6 (Table 3). Interestingly, two genomic loci (LOC_Os01g64960, LOC_Os04g59440) encoding PsbS protein are identified in rice as compared to one psbS locus in Arabidopsis (AT1G44575) (Tables 1 and 3). These two loci are hereafter named as psbS1 (LOC_Os01g64960) and psbS2 (LOC_Os04g59440) (Table 3). The rice PsbS1 and PsbS2 shows homology %73/80 and %80/87 (identities/positives) for 270 and 232 amino acids respectively, with the PsbS protein of Arabidopsis. The PsbS proteins are aligned which indicated that the sequences are relatively well conserved in rice and Arabidopsis (Fig. 1). The present finding wherein two PsbS-related loci are identified in rice contradict a previous report where the PSII-S gene, psbS, was suggested to be a single-copy gene in rice.6 A few additional LHC-related genes are also identified in rice (LOC_Os09g12540, LOC_Os02g03330) (Table 3). Some of the Lhc genes that are found in Arabidopsis do not exist in rice (Table 3).
Table 2.
A list of 17 loci which encodes for chlorophyll a/b-binding proteins in rice
| Chromosome | Locus Id | Putative function |
| 1 | LOC_Os01g41710 | Chlorophyll a/b-binding protein, putative expressed |
| 1 | LOC_Os01g52240 | Chlorophyll a/b-binding protein, putative expressed |
| 1 | LOC_Os01g64960 | Chlorophyll a/b-binding protein, putative expressed |
| 2 | LOC_Os02g10390 | Chlorophyll a/b-binding protein, putative expressed |
| 2 | LOC_Os02g52650 | Chlorophyll a/b-binding protein, putative expressed |
| 3 | LOC_Os03g39610 | Chlorophyll a/b-binding protein, putative expressed |
| 4 | LOC_Os04g38410 | Chlorophyll a/b-binding protein, putative expressed |
| 4 | LOC_Os04g59440 | Chlorophyll a/b-binding protein, putative expressed |
| 6 | LOC_Os06g21590 | Chlorophyll a/b-binding protein, putative expressed |
| 7 | LOC_Os07g37240 | Chlorophyll a/b-binding protein, putative expressed |
| 7 | LOC_Os07g37550 | Chlorophyll a/b-binding protein, putative expressed |
| 7 | LOC_Os07g38960 | Chlorophyll a/b-binding protein, putative expressed |
| 8 | LOC_Os08g33820 | Chlorophyll a/b-binding protein, putative expressed |
| 9 | LOC_Os09g12540 | Chlorophyll a/b-binding protein, putative expressed |
| 9 | LOC_Os09g17740 | Chlorophyll a/b-binding protein, putative expressed |
| 9 | LOC_Os09g26810 | Chlorophyll a/b-binding protein, putative expressed |
| 11 | LOC_Os11g13890 | Chlorophyll a/b-binding protein, putative expressed |
Table 3.
Light-harvesting complex genes of photosystem I and II in rice
| Locus Id | Annotation | Synonym | A* | B* | C* |
| LOC_Os04g38410 | Chlorophyll a/b-binding protein | Lhcb6/CP24 | 253 | 27060 | 7.6727 |
| LOC_Os09g26810 | Chlorophyll a/b-binding protein | LHCA6 | 265 | 28902 | 6.2372 |
| LOC_Os01g41710 | Chlorophyll a/b-binding protein | Lhcb1.2 | 262 | 27552.5 | 5.0852 |
| LOC_Os01g52240 | Chlorophyll a/b-binding protein | Lhcb1.1 | 266 | 27901.8 | 5.085 |
| LOC_Os09g17740 | Chlorophyll a/b-binding protein | Lhcb1.3 | 266 | 28014 | 4.904 |
| LOC_Os02g52650 | Chlorophyll a/b-binding protein | LHCA5 | 262 | 27895 | 6.6016 |
| LOC_Os02g10390 | Chlorophyll a/b-binding protein | LHCA3 | 270 | 29209.5 | 7.8011 |
| LOC_Os09g12540 | Chlorophyll a/b-binding protein | Chl a/b | 322 | 34925.4 | 9.2047 |
| LOC_Os03g39610 | Chlorophyll a/b-binding protein | LHCB2.1 | 264 | 28495.3 | 5.7079 |
| n/f | LHCB2.2 | ||||
| n/f | Lhcb1.5 | ||||
| n/f | Lhcb1.4 | ||||
| n/f | Lhcb4.3 | ||||
| n/f | Lhcb4.2 | ||||
| n/f | Lhcb2.4 | ||||
| LOC_Os08g33820 | Chlorophyll a/b-binding protein | LHCA4 | 245 | 26955.7 | 7.0578 |
| LOC_Os06g21590 | Chlorophyll a/b-binding protein | LHCA1 | 242 | 26185.8 | 6.0177 |
| LOC_Os07g38960 | Chlorophyll a/b-binding protein | LHCA2 | 264 | 28209.1 | 6.2689 |
| LOC_Os11g13890 | Chlorophyll a/b-binding protein | LHCB5 | 284 | 30283.6 | 5.3897 |
| LOC_Os02g03330 | Chlorophyll a/b-binding protein | LiL* | 251 | 27593.4 | 6.5533 |
| LOC_Os07g37240 | Chlorophyll a/b-binding protein | LHCB4.1 | 291 | 31349.5 | 5.1588 |
| n/f | Chla/b | ||||
| LOC_Os07g37550 | Chlorophyll a/b-binding protein | Lhcb3 | 267 | 28777.9 | 6.1247 |
| LOC_Os01g64960 | Chlorophyll a/b-binding protein | PsbS1 | 269 | 27903.3 | 6.9945 |
| LOC_Os04g59440 | Chlorophyll a/b-binding protein | PsbS2 | 255 | 26950.4 | 6.0137 |
A, amino acids; B, molecular weight; C, isoelectric point. n/f = genes not found in rice but exists in Arabidopsis. *LiL, light-harvesting like.
Figure 1.
Amino acid alignment of PsbS proteins in Arabidopsis and rice. Note the presence of two PsbS proteins in rice. Proteins are aligned using ClustalX.14 Gene names are identified on left and amino acid positions on right. Asterisks and dots drawn on top of sequence indicate identical residues and conservative amino acid changes, respectively. Gaps in the amino acid sequences are introduced to improve the alignment. At-Arabidopsis thaliana L.; Os-Oryza sativa L.
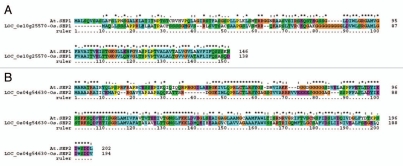
The Table 4 enlists light-inducible genes in rice. These include six loci which encode for early light-inducible proteins (ELIP1 to ELIP6), two loci for stress-enhanced proteins (SEP1 and SEP2), two loci for one-helix proteins (OHP1 and OHP2), and a single loci for light-induced protein 1-like (LIP1-like) (Table 4). In the Rice Genome Annotation Project (RAP) database (http://rice.plantbiology.msu.edu/), the loci LOC_Os04g54630, LOC_Os05g22730 and LOC_Os10g25570 are annotated as “expressed proteins” (Table 4). However, the BLAST search against Arabidopsis protein database identified these loci as SEP1 (LOC_Os10g25570), SEP2 (LOC_Os04g54630) and OHP1 (LOC_Os05g22730) (Table 4). The Arabidopsis SEP1 (AT4G34190) shares homology %48/66 (identities/positives) for 109 amino acids with rice LOC_Os10g25570, and SEP2 (AT2G21970) shares homology %48/68 (identities/positives) for 206 amino acids with rice LOC_Os04g54630. The protein alignment of SEPs in Arabidopsis and rice is shown in Figure 2A and B. While, the Arabidopsis OHP (AT5G02120) shares homology %58/73 (identities/positives) for 110 amino acids with rice LOC_Os05g22730. Also, the Arabidopsis OHP2 (AT1G34000) shares homology %66/76 (identities/positives) with rice LOC_Os01g40710, a high light-inducible protein. The protein alignment of OHPs in Arabidopsis and rice is shown in Figure 3A and B. Thus the present study has attested three of the formerly annotated “expressed proteins” to their respective “functional class”.
Table 4.
A list of light-inducible genes in rice
| Locus Id | Annotation | Synonym | A* | B* | C* |
| LOC_Os01g01340 | light-induced protein 1-like | LIP1-like | 129 | 13317 | 4.1426 |
| LOC_Os01g14410 | early light-inducible protein | ELIP1 | 203 | 20217 | 10.2911 |
| LOC_Os01g40710 | high light-inducible protein | HLIP/OHP2* | 191 | 19884.8 | 10.303 |
| LOC_Os02g16560 | early light-inducible protein | ELIP2 | 241 | 25611.6 | 12.8165 |
| LOC_Os03g30400 | early light-inducible protein | ELIP3 | 129 | 12828.5 | 10.4496 |
| LOC_Os04g54630 | expressed protein | SEP2* | 195 | 20347.1 | 4.5945 |
| LOC_Os05g08110 | early light-inducible protein | ELIP4 | 205 | 20856.3 | 11.7767 |
| LOC_Os05g22730 | expressed protein | OHP1* | 113 | 12109.5 | 11.1885 |
| LOC_Os07g08150 | early light-inducible protein | ELIP5 | 201 | 19840.6 | 9.5072 |
| LOC_Os07g08160 | early light-inducible protein | ELIP6 | 193 | 19352 | 6.9273 |
| LOC_Os10g25570 | expressed protein | SEP1* | 164 | 13625.6 | 10.9811 |
SEP, stress-enhanced protein.
OH P, one-helix protein.
A, amino acids; B, molecular weight; C, isoelectric point.
Figure 2.
Protein alignment of stress-enhanced proteins (SEPs) in Arabidopsis and rice. (A) At.SEP1 and Os.SEP1 protein alignment. (B) At.SEP2 and Os.SEP2 protein alignment. The figure was generated as described in legend to Figure 1. At-Arabidopsis thaliana L.; Os-Oryza sativa L.
Figure 3.
Protein alignment of one-helix proteins (OHPs) in Arabidopsis and rice. (A) At.OHP and Os.OHP1 protein alignment. (B) At.OH P2 and Os.OHP2 protein alignment. The figure was generated as described in legend to Figure 1. At-Arabidopsis thaliana L.; Os-Oryza sativa L.
One, Two, Three and Four-helix Proteins
In higher plants, there are proteins which are related to the LHC proteins. The ELIPs are first to be described which accumulate during early thylakoid development and light stress.7 The ELIPs has three, and the related PsbS protein has four membrane spanning helices. The additional helix of PsbS protein occurs at N-terminus and is homologous to helix-2. The related one-helix and two-helix proteins are also been described in Arabidopsis.8,9 With the exception of PsbS,10 the concrete functions of the other proteins are not well defined.
The Evolution of LHC Proteins
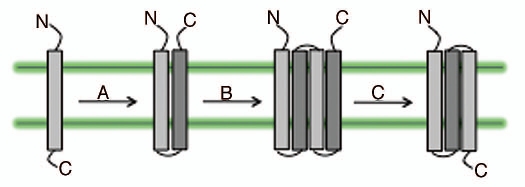
The main light-harvesting structure of cyanobacteria is the phycobilisome. It is a soluble antenna system that coordinates linear tetrapyrroles and shares no homology with the higher plant LHC proteins. However, the high light-inducible proteins (HLIPs) which constitute a group of cyanobacterial proteins are possible ancestors of the LHC proteins.11 The HLIPs are also named as small cab-like proteins (SCPs).12 Green and Pichersky (1994),13 proposed the early events in the evolution of the LHC proteins from the cyanobacterial one-helix proteins (Fig. 4).
Figure 4.
LHC proteins and their evolution. One-helix proteins (like HLIP) acquired a second helix resulting in two-helix proteins (like SEPs). The two-helix proteins underwent internal gene duplication event leading to a four-helix protein (like PsbS). From the loss of the fourth-helix, the three-helix proteins (like ELIPs and LHCs) are evolved. The arrow marks indicate (A), Gain of helix 2; (B), internal duplication; (C), loss of helix 4. At-Arabidopsis thaliana L.; Os-Oryza sativa L.
To conclude, this study has identified evolutionarily occurring one, two, three and four-helix Lhc-related proteins in rice. The present collection of Lhc family members forms a basis that can be rapidly accessible, reliable and up-to-date. This information will provide a solid base for the interpretation of new results, and a starting point for planning further experiments on Lhc proteins in the monocot model plant, rice. The identification of two independent PsbS-related loci offers an open challenge to unravel their functions in rice.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13410
References
- 1.Green BR. Was “molecular opportunism” a factor in the evolution of different photosynthetic light-harvesting pigment systems? Proc Natl Acad Sci USA. 2001;98:2119–2121. doi: 10.1073/pnas.061023198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jansson S. The light-harvesting chlorophyll a/b-binding proteins. Biochim Biophys Acta. 1994;1184:1–19. doi: 10.1016/0005-2728(94)90148-1. [DOI] [PubMed] [Google Scholar]
- 3.Leutweiler LS, Meyerowitz EM, Tobin EM. Structure and expression of three light-harvesting chlorophyll a/b-binding protein genes in Arabidopsis thaliana. Nucleic Acids Res. 1986;14:4051–4064. doi: 10.1093/nar/14.10.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Legen J, Misera S, Herrmann RG, Meurer J. Map positions of 69 Arabidopsis thaliana genes of all known nuclear encoded constituent polypeptide and various regulatory factors of the photosynthetic membrane: a case study. DNA Res. 2001;8:53–60. doi: 10.1093/dnares/8.2.53. [DOI] [PubMed] [Google Scholar]
- 5.Andersson J, Wentworth M, Walters RG, Howard C, Ruban A, Horton P, et al. Absence of the main light-harvesting complex of photosystem II affects photosynthesis and fitness but not grana stacking. Plant J. 2003;35:350–361. doi: 10.1046/j.1365-313x.2003.01811.x. [DOI] [PubMed] [Google Scholar]
- 6.Iwasaki T, Saito Y, Harada E, Kasai M, Shoji K, Miyao M, et al. Cloning of cDNA encoding the rice 22 kDa protein of Photosystem II (PSII-S) and analysis of light-induced expression of the gene. Gene. 1997;185:223–229. doi: 10.1016/s0378-1119(96)00646-4. [DOI] [PubMed] [Google Scholar]
- 7.Adamska I, Ohad I, Kloppstech K. Synthesis of the early light-inducible protein is controlled by the blue light and related to light stress. Proc Natl Acad Sci USA. 1992;89:2610–2623. doi: 10.1073/pnas.89.7.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jansson S. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 1999;4:236–240. doi: 10.1016/s1360-1385(99)01419-3. [DOI] [PubMed] [Google Scholar]
- 9.Heddad M, Adamska I. The evolution of light stress proteins in photosynthetic organisms. Comp Funct Genom. 2002;3:504–510. doi: 10.1002/cfg.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li XP, Bjorkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK. A pigment binding protein essential for regulation of photosynthetic light-harvesting. Nature. 2000;40:391–395. doi: 10.1038/35000131. [DOI] [PubMed] [Google Scholar]
- 11.Dolganov NAM, Bhaya D, Grossman AR. Cyanobacterial protein with similarity to the chlorophyll a/b binding proteins of higher plants: Evolution and regulation. Proc Natl Acad Sci USA. 1995;92:636–640. doi: 10.1073/pnas.92.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funk C, Vermaas WFJ. Expression of cyanobacterial genes coding for single-helix polypeptides resembling regions of light-harvesting proteins from higher plants. Biochem. 1999;38:9397–9404. doi: 10.1021/bi990545+. [DOI] [PubMed] [Google Scholar]
- 13.Green BR, Pichersky E. Hypothesis for the evolution of three-helix Chl a/b and Chl a/c light-harvesting antenna proteins from two-helix and four-helix ancestors. Photosynth Res. 1994;39:149–162. doi: 10.1007/BF00029382. [DOI] [PubMed] [Google Scholar]
- 14.Umate P, Tuteja R, Tuteja N. Genome-wide analysis of helicase gene family from rice and Arabidopsis: a comparison with yeast and human. Plant Mol Biol. 2010;73:449–465. doi: 10.1007/s11103-010-9632-5. [DOI] [PubMed] [Google Scholar]