Abstract
The molecular mechanisms by which plants sense their micronutrient status, and adapt to their environment in order to ensure a sufficient micronutrient supply, are poorly understood. Zinc is an essential micronutrient for all living organisms. when facing a shortage in zinc supply, plants adapt by enhancing the zinc uptake capacity. The molecular regulators controlling this adaptation were recently identified. in this mini-review, we highlight recent progress in understanding the adaptation to zinc deficiency in plants and discuss the future challenges to fully unravel its molecular basis.
Key words: adaptation, zinc deficiency, biofortification, molecular regulators, plant nutrition
In an increasingly populated world, agricultural production is an essential element of social development. Agriculture is the primary source of all nutrients required for human life, and nutrient sufficiency is the basis for good health and welfare of the human population.1 Soils with zinc deficiency are widespread in the world, affecting large areas of cultivated soils in India, Turkey, China, Brazil and Australia,2,3 making zinc the most common crop micronutrient deficiency.4 In addition, risk of inadequate zinc diet and zinc malnutrition are estimated to affect one-third of the global human population, i.e., around two billion people.5 Most affected are people living in developing countries, where diets are rich in cereal-based foods. Cereal grains are rich in phytate, which is a potent anti-nutrient, limiting micronutrient bioavailability.6 Zinc deficiency in crop production can be easily ameliorated through zinc fertilization, making agronomic biofortification an important strategy,3 however in the poorer regions, the required infrastructure to provide a reliable supply of zinc fertilizers of sufficient quality, is often not available. In those situations, biofortified crops, in which the zinc status of crops is genetically improved by selective breeding or via biotechnology, offer a rural-based intervention that will more likely reach the population.7 Different traits can be targeted to developing such improved crops, such as plant zinc deficiency tolerance, zinc use efficiency and the accumulation of zinc in edible parts. However, insufficient knowledge on the molecular mechanisms and the regulation of the zinc homeostasis network in plants is a serious bottleneck when pursuing zinc biofortification.
Zinc Homeostasis Network
The zinc homeostasis network comprises the coordinated activities of zinc uptake, transport, trafficking and sequestration, providing an adequate amount of zinc to all cell types, at all stages of development and under different environmental conditions.8 Although zinc is an essential element, it can be toxic when present in excess. Therefore, plants are thought to control zinc homeostasis using a tightly regulated network where the coordinated expression of zinc transporters plays a major role in zinc acquisition from soil, in mobilization between organs and tissues and in intracellular sequestration.8,9
In Arabidopsis, different members of important families of cation transporters have been characterized and found to be relevant for zinc homeostasis.10,11 Members of the ZIP (Zrt/Irt-like Proteins) family, which facilitate the influx of zinc into the cytosol, are good candidates to mediate the uptake of zinc from soil and the unloading of zinc from xylem.12–14 Members of the CDF (Cation Difusion Facilitator) or MTP (Metal Tolerance Protein) family are involved in vacuolar zinc import,15,16 while NRAMP proteins ensure vacuolar zinc unloading.17 Members of the divalent metal cation transporting 1b P-type ATPases (HMA) are responsible for zinc xylem loading,18 the control of plastid zinc contents19 and vacuolar zinc sequestration.20 In addition to these transporters, proteins involved in the synthesis of metal chelators, such as nicotianamine synthases (NAS), or transport of (metal-) chelates, such as Yellow-Stripe Like (YSL) proteins21 and the MATE type transporter FRD3,22 are important for zinc homeostasis. The recent identification of two transcription factor genes that are essential for the transcriptional regulation of Arabidopsis adaptation to zinc deficiency represents the beginning of our understanding of the molecular control of the zinc homeostasis network.23
Adaptation to Zinc Deficiency in Arabidopsis
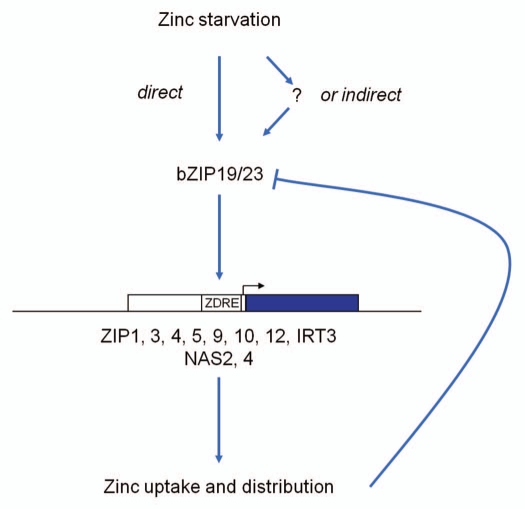
Using a yeast-one-hybrid screening with promoter fragments of the strong zinc-deficiency-induced Arabidopsis ZIP4 zinc transporter gene as bait, two closely related members of the Arabidopsis basic-region leucine-zipper (bZIP) transcription factor gene family, bZIP19 and bZIP23, have been isolated.23 The bZIP19/23 function is essential for a proper zinc deficiency response, allowing Arabidopsis to grow at low zinc supply. Both genes are functionally partially redundant, with single T-DNA insertion mutants showing a weak sensitivity to zinc deficiency, and only the bzip19bzip23 double mutant lines showing a zinc deficiency hypersensitive phenotype.23 This rescue system appears to be zinc specific. Preliminary experiments with the double mutants were unable to display any hypersensitivity for copper, manganese or iron deficiencies (Assunção AGL, unpublished data). The bZIP19 and bZIP23 proteins bind to a palindromic 10-bp ZDRE (Zinc Deficiency Response Element) sequence motif. Transcript profiling revealed only a small set of around 15 genes, most containing ZDRE motifs in their upstream regions, to be induced in wild-type plants in response to zinc deficiency, but not in the bzip19bzip23 double mutants. These genes, which must be pivotal for the zinc deficiency adaptation response, comprise eight (out of 15) members of the ZIP family of cation transporters (Fig. 1). The exact role of these ZIP members in Arabidopsis zinc nutrition is largely unknown, but four of them (ZIP1, ZIP3, ZIP4 and IRT3) have been functionally characterized to mediate zinc uptake in yeast complementation studies.12,14,23 In addition to the ZIPs, the nicotianamine synthetases NAS2 and NAS4 are differentially expressed. This means that all direct targets of bZIP19/bZIP23, the genes containing one or more ZDRE elements in their promoter,23 appear to be essential for the scavenging of zinc from the growth substrate and adequate distribution through the plant.
Figure 1.
Scheme representing the response to zinc starvation in Arabidopsis mediated by the transcription factors bZIP19 and bZIP23.
Recently, the analysis of transcriptional changes upon iron deficiency in Arabidopsis24 suggested that plants respond to avoid imbalances in ion distribution by inducing a suite of transporters that compensates for the surplus of undesired metals. This means that the regulation of these metal transporters is not a consequence of the inadvertent uptake of other metals than iron, which is likely to happen due to the low specificity for other metals of the major Fe uptake transporter IRT1,25 but instead an anticipated response to avoid ion imbalance by promoting prompt ion distribution.24 Four ZIP genes, ZIP2, ZIP3, ZIP4 and ZIP9, were found to be differentially expressed in response to iron deficiency, however, careful examination of the response made clear that these genes are regulated solely by zinc, and their response to iron deficiency is due to a secondary effect.24 This confirms that these genes act primarily in zinc nutrition and homeostasis and that their upstream regulatory mechanisms are zinc-specific.
This adaptive system of Arabidopsis to cope with growth under zinc limiting conditions is likely present in all plant species as the bZIP19 and bZIP23 transcription factors, their target genes and the characteristic cis elements, are conserved in higher plants, including the major monocot and dicot crops.23 Plants thus appear to have developed their own zinc deficiency response transcriptional regulation system, different from systems found in species of other kingdoms. In yeast, induced transcription of the cytoplasmic zinc uptake transporter genes ZTR1 and ZRT2 depends on the Zn-finger transcription factor Zap1.26 Zap1 binds to zinc-responsive elements in the promoter of target genes, with a consensus sequence (ACC TTN AAG GT)27 that is very different from the ZDRE motif (RTG TCG ACA Y) bound by bZIP19 or bZIP23 in Arabidopsis.23 In vertebrates, including mammals, fish and birds, and in insects, the metal-responsive-element-binding transcription factor-1 (MTF-1) plays a central role in zinc homeostasis.28 This is another Zn-finger transcription factor, binding to metal responsive elements (MRE) with the consensus core sequence TGC RCN C. Initially identified as a transcription factor controlling the response to excess metals, recent work has shown that it is also involved in controlling transcription of zinc uptake transporters upon zinc deficiency.29 Activity of both proteins appears to be controlled by binding of zinc to the Zn-fingers. This zinc sensing ability through Zn-fingers is not directly obvious for the plant zinc deficiency responsive bZIP transcription factors. Nevertheless, an essential question that arises from this newly unraveled zinc deficiency response mechanism in Arabidopsis relates to the regulatory mechanism upstream of the bZIP19/bZIP23 transcription factors: How are plants sensing zinc deficiency? bZIP19 and bZIP23 belong to bZIP family group F,30 which is characterized by containing two histidine-rich motifs. Although the only other member of this family is involved in salt stress response31 and not in zinc homeostasis, a role can be envisaged for these motifs in the binding and sensing of intracellular zinc concentrations. Alternatively, instead of a direct interaction with zinc, an upstream signaling pathway might be responsible for the zinc deficiency signal transduction. In either way these aspects need to be elucidated. Further research on the regulation and adaptation mechanisms to zinc deficiency in plants will contribute to engineering of crops displaying improved acquisition and use of zinc, which will help to overcome serious problems with crop yield and mineral deficiencies among the human population living in areas with low bioavailable zinc in soils.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13469
References
- 1.Welch RM, Combs GF, Duxbury JM. Toward a “Greener” revolution. Issues Sci Tech. 1997;14:50–58. [Google Scholar]
- 2.Broadley MR, White PJ, Hammond JP, Zelko I, Lux A. Zinc in plants. New Phytol. 2007;173:677–702. doi: 10.1111/j.1469-8137.2007.01996.x. [DOI] [PubMed] [Google Scholar]
- 3.Cakmak I. Enrichment of cereal grains with zinc: Agronomic or genetic biofortificaion? Plant Soil. 2008;302:1–17. [Google Scholar]
- 4.White PJ, Broadley MR. Biofortification of crops with seven mineral elements often lacking in human diets—iron, zinc, copper, calcium, magnesium, selenium and iodine. New Phytol. 2009;182:49–84. doi: 10.1111/j.1469-8137.2008.02738.x. [DOI] [PubMed] [Google Scholar]
- 5.Hotz C, Brown KH. Assessment of the risk of zinc deficiency in populations and options for its control. Food Nutr Bull. 2004;25:94–204. [PubMed] [Google Scholar]
- 6.Welch RM, Graham RD. Breeding for micronutrient in staple food crops from a human nutrition perspective. J Exp Bot. 2004;55:353–364. doi: 10.1093/jxb/erh064. [DOI] [PubMed] [Google Scholar]
- 7.Bouis H, Welch RM. Biofortification—A sustainable agricultural strategy for reducing micronutrient malnutrition in the global south. Crop Sci. 2010;50:20–32. [Google Scholar]
- 8.Clemens S. Molecular mechanisms of plant metal tolerance and homeostasis. Planta. 2001;212:475–486. doi: 10.1007/s004250000458. [DOI] [PubMed] [Google Scholar]
- 9.Clemens S, Palmgren MG, Kraemer U. A long way ahead: understanding and engineering plant metal accumulation. Trends Plant Sci. 2002;7:309–315. doi: 10.1016/s1360-1385(02)02295-1. [DOI] [PubMed] [Google Scholar]
- 10.Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraemer U, Talke IN, Hanikenne M. Transition metal transport. FEBS Lett. 2007;581:2263–2272. doi: 10.1016/j.febslet.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Grotz N, Fox T, Connolly E, Park W, Guerinot ML, Eide D. Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc Natl Acad Sci USA. 1998;95:7220–7224. doi: 10.1073/pnas.95.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerinot ML. The ZIP family of metal transporters. Biochim Biophys Acta. 2000;1465:190–198. doi: 10.1016/s0005-2736(00)00138-3. [DOI] [PubMed] [Google Scholar]
- 14.Lin YF, Liang HM, Yang SY, Boch A, Clemens S, Chen CC, et al. Arabidopsis IRT3 is a zinc-regulated plasma membrane localized zinc/iron transporter. New Phytol. 2009;182:392–404. doi: 10.1111/j.1469-8137.2009.02766.x. [DOI] [PubMed] [Google Scholar]
- 15.Van der Zaal BJ, Neuteboom LW, Pinas JE, Chardonnens AN, Schat H, Verkleij JA, et al. Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol. 1999;119:1047–1056. doi: 10.1104/pp.119.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobae Y, Uemura T, Sato MH, Ohnishi M, Mimura T, Nakagawa T, et al. Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol. 2004;45:1749–1758. doi: 10.1093/pcp/pci015. [DOI] [PubMed] [Google Scholar]
- 17.Oomen RJ, Wu J, Lelièvre F, Blanchet S, Richaud P, Barbier-Brygoo H, et al. Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator Thlaspi caerulescens. New Phytol. 2009;181:637–650. doi: 10.1111/j.1469-8137.2008.02694.x. [DOI] [PubMed] [Google Scholar]
- 18.Hussain DM, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, et al. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell. 2004;16:1327–1339. doi: 10.1105/tpc.020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YY, Choi H, Segami S, Cho HT, Martinoia E, Maeshima M, et al. AtHMA1 contributes to the detoxification of excess Zn(II) in Arabidopsis. Plant J. 2009;58:737–753. doi: 10.1111/j.1365-313X.2009.03818.x. [DOI] [PubMed] [Google Scholar]
- 20.Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, Vavasseur A, et al. AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2009;149:894–904. doi: 10.1104/pp.108.130294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curie C, Cassin G, Couch D, Divol F, Higuchi K, Le Jean M, et al. Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann Bot. 2009;103:1–11. doi: 10.1093/aob/mcn207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durrett TP, Gassmann W, Rogers EE. The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 2007;144:197–205. doi: 10.1104/pp.107.097162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Assunção AGL, Herrero E, Lin YF, Huettel B, Talukdar S, Smaczniak C, et al. The Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc Natl Acad Sci USA. 2010;107:10296–10301. doi: 10.1073/pnas.1004788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang TJW, Lin WD, Schmidt W. Transcriptional profiling of the Arabidopsis iron deficiency response reveals conserved transition metal homeostasis networks. Plant Physiol. 2010;152:2130–2141. doi: 10.1104/pp.109.152728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol Biol. 1999;40:37–44. doi: 10.1023/a:1026438615520. [DOI] [PubMed] [Google Scholar]
- 26.Zhao H, Eide D. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:5044–5052. doi: 10.1128/mcb.17.9.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyons TJ, Gasch AP, Gaither LA, Botstein D, Brown PO, Eide DJ. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc Natl Acad Sci USA. 2000;97:7957–7962. doi: 10.1073/pnas.97.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laity JH, Andrews GK. Understanding the mechanisms of zinc-sensing by metal-response element binding transcription factor-1 (MTF-1) Arch Biochem Biophys. 2007;463:201–210. doi: 10.1016/j.abb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 29.Zheng D, Feeney GP, Kille P, Hogstrand C. Regulation of ZIP and ZnT zinc transporters in zebrafish gill: zinc repression of ZIP10 transcription by an intronic MRE cluster. Physiol Genom. 2008;34:205–214. doi: 10.1152/physiolgenomics.90206.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 31.Yang O, Popova OV, Suethoff U, Lueking I, Dietz KJ, Golldack D. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene. 2009;436:45–55. doi: 10.1016/j.gene.2009.02.010. [DOI] [PubMed] [Google Scholar]