Abstract
Genetic studies have suggested that transmembrane proteins CLAVATA1 (CLV1), CLV2, CORYNE (CRN), BAM1 and BAM2 all play a role in relaying the CLV3 signal and thus regulating stem cell homeostasis at the shoot meristem (SM). The extracellular domain of CLV1 was previously shown to bind the CLE peptide derived from CLV3, providing direct evidence that CLV3-CLV1 function as a ligand-receptor pair. How the other putative receptors function in the CLV pathway, however, remained unclear. We demonstrated in a recent Plant Journal article that the receptor-like protein CLV2 and the receptor-kinases BAM1 and BAM2 also bind to the CLV3 CLE peptide ligand with an affinity similar to that of CLV1. Critically, these ligand binding receptors form two distinct complexes in both transient expression in tobacco and in Arabidopsis meristem cells: a CLV2/CRN multimer and a CLV1/BAM multimer. Here we examine in detail the subcellular membrane partitioning for the receptor proteins in transient expression by two-phase partitioning and co-expression with known subcellular markers. All tested proteins measurably accumulate at the plasma membrane. While CLV1 primarily co-localizes with a plasma membrane marker, CLV2 shows greater co-localization with an endoplasmic reticulum (ER) marker.
Key words: CLAVATA2, CLAVATA1, CORYNE, subcellular localization, plasma membrane, endoplasmic reticulum, receptor complex, meristem
The CLV signaling pathway regulates stem cell specification at the SM by regulating the asymmetric nature of L3 stem cell divisions, indirectly promoting differentiation of lateral stem cell daughters in the L1 and L2 layers.1,2 Known components of this pathway include the CLV3 proprotein for the CLE peptide ligand,3–5 the leucine-rich repeat receptor kinase (LRR-RK) CLV1,6,7 and its close homologs BAM1 and BAM2,8,9 the LRR receptor-like protein CLV2,10,11 the transmembrane kinase-related protein CRN,12,13 the type 2C protein phosphatases POLTERGEIST (POL) and PLL1,14–16 and the homeodomain transcription factor WUSCHEL (WUS).17,18 The CLV3 signal is believed to be perceived by the membrane-localized receptors and relayed through POL and PLL1, leading to the suppression in the L3 stem cells of WUS, which functions to promote stem cell identity in neighboring cells. Biochemical mechanisms of CLV signal perception and transduction are largely unknown. Our recent study in Plant Journal19 presents evidence of ligand perception by different receptors and the formation of distinct receptor complexes, providing insights into the mechanisms of signal perception and receptor activation in the CLV pathway.
Using co-immunoprecipitation (co-IP) analysis in both transient expression in tobacco and in vivo in Arabidopsis meristem tissue, we demonstrated that CLV1 forms stable homomultimers and heteromultimers with BAM1 and BAM2, and that CLV2 forms a stable complex with CRN.19 While the CLV2-CRN complex has been reported by two other studies using fluorescent-based association assays in transient expression systems, both of these studies also suggested the formation of a larger CLV1-CRN-CLV2 complex20,21 and one study reported CLV1-CRN interaction,20 raising the question of whether CLV2-CRN and CLV1 function in two distinct complexes or in one large super-complex. By using a quantitative approach to measure the efficiency of co-IPs, we showed that the interactions between components of the two complexes such as CLV1-CLV2, CLV1-CRN, BAM1-CLV2 and BAM2-CLV2 are significantly weaker in transient expression and not detectable in vivo.19 Consistent with a model that the two ligand-binding receptor complexes function independently from each other, we showed that ectopic expression of BAM1, BAM2 and to a less extent, CLV1, rescued the clv2 mutant phenotype, suggesting that BAM and CLV1 can replace CLV2 function in vivo.19
If CLV2 forms a distinct complex with CRN, what then is the role of the large CLV2 extracellular domain? We found that CLV2 binds to the CLV3 CLE ligand with the same affinity as CLV1. Because CLV2 lacks a cytosolic signaling domain, CLV2 perception of CLE is likely relayed through its stable association with the transmembrane kinase-related protein CRN in vivo. CLV1/BAM on the other hand, contains both extracellular CLE-binding domains as well as intracellular protein kinase domains to relay signal. Thus, the CLV pathway features two distinct ligand-binding complexes with similar ligand affinity, a novel mode of signal perception.
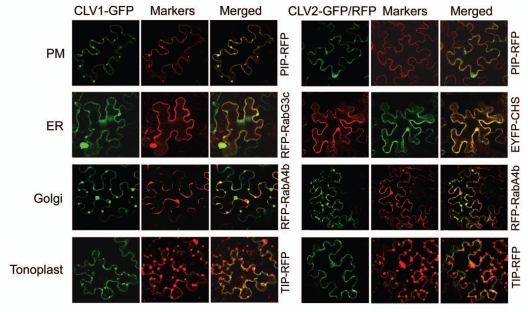
Consistent with the predicted plasma membrane (PM) localization of the receptors, in our recent study we showed that when transiently expressed in tobacco leaves, all tested CLV receptor proteins accumulated in the membrane fraction and GFP/RFP-fused proteins showed fluorescence primarily at the cell periphery.19 To further examine the subcellular localizations of the receptors, we co-expressed CLV1 and CLV2 with fluorescent markers for various membrane localizations, including a trans-Golgi marker RFP-RabA4b,22 a tonoplast marker TIP-RFP,23 a PM marker PIP-RFP,23 and endoplasmic reticulum (ER) markers EYFP-CHS24 and RFP-RabG3c22 (Fig. 1).
Figure 1.
Co-localization of CLV1 and CLV2 with fluorescent markers in N. benthamiana leaves. Shown are confocal images of fluorescent-tagged receptors and co-expressed fluorescent markers two days post-inoculation. Tobacco leaf infiltration and confocal analysis were performed as described19 except that leaves were viewed at a 40x magnification.
We observed that CLV1-GFP co-localized largely with the PM marker PIP-RFP and to a lesser extent with the ER maker RFP-RabG3c. CLV2-RFP co-localized primarily with the ER marker EYFP-CHS. Neither CLV1-GFP nor CLV2-GFP co-localized well with the tonoplast or the Golgi marker (Fig. 1). Fluorescence at ER could be an effect of overexpression from accumulation of newly synthesized receptor proteins as they transport to the PM. ER localization of CLV receptors, on the other hand, could also be a regulatory step during receptor activation and signal transduction. A recent study using the similar tobacco transient system indicated that CLV1, CLV2 and CRN all showed ER localization, CLV2 and CRN were predominantly ER-localized and co-expression of CRN and CLV2 brought both proteins to the PM.21 Interestingly, CLV signaling function requires the ER-resident molecular chaperone SHEPHERD proteins.25 Furthermore, the ER quality control machinery is emerging as an important facet of brassinosteroid receptor function.26,27
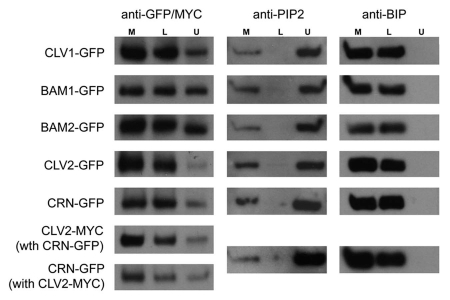
To further test membrane localization of the CLV receptors, we performed two-phase partitioning on membrane fractions (Fig. 2). Protein gel blot analysis showed that CLV1-GFP, CLV2-GFP, CRN-GFP, BAM1-GFP and BAM2-GFP all partially partition in the upper phase which is highly enriched for plasma membranes, as demonstrated by the PM-localized PIP2 control.23 However, we also observed significant accumulation for all receptors in the lower fraction, which is enriched for chloroplast, mitochondrial and ER membranes, as exemplified by the ER-localized BIP control.28
Figure 2.
Two-phase partitioning of membranes isolated from tobacco leaves expressing different receptor proteins. Data from CLV2-GFP alone, CRN-GFP alone and CLV2-MYC/CRN-GFP co-expression are shown. Two-phase partitioning14 and protein gel blot analysis19 were performed as described. The rabbit anti-PIP2 29 and mouse anti-BiP (SPA-818; Stressgen) antibodies were used to detect the PM and ER marker proteins. M, total microsomal fraction; U, upper phase enriched in PM proteins; L, lower phase depleted of PM.
Consistent with data from marker colocalization, CLV1 and the related BAM receptors were detected more strongly in the plasma membrane compared to CLV2 and CRN. In addition, we did not observe significant increase in PM partitioning for CLV2 or CRN when they were co-expressed, which is in contradiction to a previous report21 (Fig. 2). The discrepancy between our results and the Bleckmann et al.21 study could be the result of different experimental approaches and different expression strategies in the tobacco transient system (inducible promoter in their study versus 35S promoter in this study).
In summary, as an interesting addition to our recent study, the confocal and two-phase partitioning data presented here support the idea of partial PM localization of the CLV pathway receptors. However, significant accumulation in interior membranes was also observed, especially for CLV2 and CRN. To what extent these localizations in transient expression reflect in vivo membrane partitioning of these proteins in meristem cells and how this might be affected by signaling activation are critical questions for future studies.
Acknowledgements
We thank Tzvi Tzfira at the University of Michigan for providing fluorescent markers. This work was supported by grants from the National Institute of Health (R01GM62962) and USDA Cooperative Research Extension Service (USDA-2006-35304-17403) to S.E.C.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13359
References
- 1.Fletcher JC. Shoot and Floral Meristem Maintenance in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol. 2002;53:45–66. doi: 10.1146/annurev.arplant.53.092701.143332. [DOI] [PubMed] [Google Scholar]
- 2.Clark SE. Cell signalling at the shoot meristem. Nat Rev Mol Cell Biol. 2001;2:276–284. doi: 10.1038/35067079. [DOI] [PubMed] [Google Scholar]
- 3.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 4.Ni J, Clark SE. Evidence for functional conservation, sufficency and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 2006;140:1–8. doi: 10.1104/pp.105.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- 6.Diévart A, Dalal M, Tax FE, Lacey AD, Huttly A, Li J, et al. CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell. 2003;15:1198–1211. doi: 10.1105/tpc.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 8.DeYoung BJ, Clark SE. BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics. 2008;180:895–904. doi: 10.1534/genetics.108.091108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, Clark SE. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 2006;45:1–16. doi: 10.1111/j.1365-313X.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- 10.Jeong S, Trotochaud AE, Clark SE. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell. 1999;11:1925–1934. doi: 10.1105/tpc.11.10.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kayes JM, Clark SE. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development. 1998;125:3843–3851. doi: 10.1242/dev.125.19.3843. [DOI] [PubMed] [Google Scholar]
- 12.Muller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 2008;20:934–946. doi: 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miwa H, Betsuyaku S, Iwamoto K, Kinoshita A, Fukuda H, Sawa S. The receptor-like kinase SOL2 mediates CLE signaling in Arabidopsis. Plant Cell Physiol. 2008;49:1752–1757. doi: 10.1093/pcp/pcn148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gagne JM, Clark SE. The Arabidopsis stem cell factor POLTERGEIST is membrane localized and phospholipid stimulated. Plant Cell. 2010;22:729–743. doi: 10.1105/tpc.109.068734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song SK, Lee MM, Clark SE. POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells. Development. 2006;133:4691–4698. doi: 10.1242/dev.02652. [DOI] [PubMed] [Google Scholar]
- 16.Song SK, Hofhuis H, Lee MM, Clark SE. Key divisions in the early Arabidopsis embryo require POL and PLL1 phosphatases to establish the root stem cell organizer and vascular axis. Dev Cell. 2008;15:98–109. doi: 10.1016/j.devcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 18.Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 19.Guo Y, Han L, Hymes M, Denver R, Clark SE. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 2010;63:889–900. doi: 10.1111/j.1365-313X.2010.04295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Y, Wang Y, Li R, Song X, Wang Q, Huang S, et al. Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J. 2010;61:223–233. doi: 10.1111/j.1365-313X.2009.04049.x. [DOI] [PubMed] [Google Scholar]
- 21.Bleckmann A, Weidtkamp-Peters S, Seidel CA, Simon R. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 2010;152:166–176. doi: 10.1104/pp.109.149930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preuss ML, Serna J, Falbel TG, Bednarek SY, Nielsen E. The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell. 2004;16:1589–1603. doi: 10.1105/tpc.021634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, et al. pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol. 2005;57:503–516. doi: 10.1007/s11103-005-0340-5. [DOI] [PubMed] [Google Scholar]
- 25.Ishiguro S, Watanabe Y, Ito N, Nonaka H, Takeda N, Sakai T, et al. SHEPHERD is the Arabidopsis GRP94 responsible for the formation of functional CLAVATA proteins. EMBO J. 2002;21:898–908. doi: 10.1093/emboj/21.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong Z, Jin H, Tzfira T, Li J. Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell. 2008;20:3418–3429. doi: 10.1105/tpc.108.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin H, Yan Z, Nam KH, Li J. Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol Cell. 2007;26:821–830. doi: 10.1016/j.molcel.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontes EBP, Shank BB, Wrobel RL, Moose SP, Obrian GR, Wurtzel ET, et al. Characterization of an immunoglobulin binding protein homolog in the maize floury-2 endosperm mutant. Plant Cell. 1991;3:483–496. doi: 10.1105/tpc.3.5.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martre P, Morillon R, Barrieu F, North GB, Nobel PS, Chrispeels MJ. Plasma membrane aquaporins play a significant role during recovery from water deficit. Plant Physiol. 2002;130:2101–2110. doi: 10.1104/pp.009019. [DOI] [PMC free article] [PubMed] [Google Scholar]