Abstract
The European wild boar Sus scrofa was first introduced into Uruguay, in southern South America during the early decades of the last century. Subsequently, and starting from founder populations, its range spread throughout the country and into the neighbouring Brazilian state Rio Grande do Sul. Due to the subsequent negative impact, it was officially declared a national pest. The main aim in the present study was to provide a more comprehensive scenario of wild boar differentiation in Uruguay, by using mtDNA markers to access the genetic characterization of populations at present undergoing rapid expansion. A high level of haplotype diversity, intermediate levels of nucleotide diversity and considerable population differentiation, were detected among sampled localities throughout major watercourses and catchment dams countrywide. Phylogenetic analysis revealed the existence of two different phylogroups, thereby reflecting two deliberate introduction events forming distantly genetic lineages in local wild boar populations. Our analysis lends support to the hypothesis that the invasive potential of populations emerge from introgressive hybridization with domestic pigs. On taking into account the appreciable differentiation and reduced migration between locales in wild boar populations, management strategies could be effective if each population were to be considered as a single management unit.
Keywords: Uruguayan wild boar
Introduction
The European wild boar Sus scrofa was first introduced into Uruguay, in southern South America, during the early decades of the last century. During the 1920’s, Aaron de Anchorena, an Argentinean landowner, introduced a number of wild boars onto his ranch in the south-western Department of Colonia (one of the administrative divisions in Uruguay) for hunting purposes (Figure 1a). The founder population, on encountering adequate environmental conditions, and through specific dispersal capacity and generalist predator habits, began to increase in numbers and widely expand its range. The present day wild boar population is presumed to comprise a cross-breed with domestic pigs (Figure 1b), thereby giving rise to great variability in phenotypes, albeit with a predominance of wild boar characteristics (Herrero and Fernandez de Luco, 2003). The quick expansion of the animal’s range was facilitated by the locally mild climate, dense network of rivers, forest corridors, and an abundant food source of cultivated crops and vulnerable domestic animals, together with the absence of natural predators. Nowadays, species distribution is widespread, having started in the west, the main agricultural zone, from there later extending to central and eastern parts of the country, and in the neighbouring Brazilian state Rio Grande do Sul.
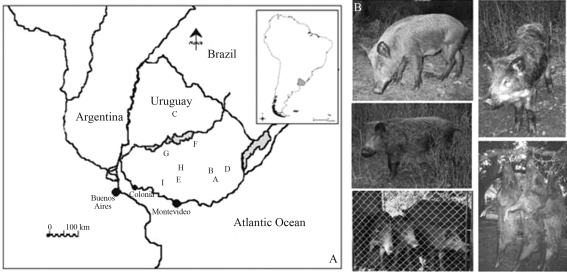
Figure 1-.
A. Distribution map of Uruguayan wild boar collecting sites: A- Neighborhood of Velazquez (Rocha Department); B- Rio Cebollatí (Lavalleja Department); C- Aº Malo (Tacuarembó Department); D- Neighborhood of Lazcano (Rocha Department); E- Aº Aries (Florida Department); F- Aº Sauces (Durazno Department); G- R. Negro (Durazno Department); H- Neighborhood of Sarandí del Yí (Durazno Department); I- Neighborhood of San José (San José Department). B. Various phenotypes of captured Uruguayan wild boars from 9 localities and captives from a farm reserve at Velazquez. Scale bar: 100 km.
The species has been extensively hunted over recent years. Some landowners have a highly negative perception of wild boars, regarding them as being responsible for direct predation of sheeps, even though such was not verified and quantified and only apparent by interviewing farmers (Herrero and Fernandez de Luco, 2003). In 1982, the animal was officially declared a national pest (Decree 463/982).
Feral pigs are also potential reservoirs or `vectors for a number of endemic and exotic diseases capable of affecting domestic livestock, wildlife and even humans. In Australia, for example, besides leptospirosis and brucellosis, they are also capable of transmitting exotic diseases, such as foot-and-mouth disease and Japanese encephalitis (Dexter, 2003; Caley and Hone, 2004).
Along with knowledge of spatial genetic structure, that of population dispersal is also essential for reducing and reversing environmental impacts (Hampton et al., 2004), particularly so in the development of effective control programmes for feral or invasive species, and for obtaining informative and reliable risk analysis (Edwards et al., 2004).
A major genetic paradox in invasive species is resolving how bottlenecked populations, with typically low genetic diversity, low evolutionary potential and, possibly, reproductive fitness, can become invasive (Frankham, 2005). The wild boar from Uruguay thus constitutes an interesting species-model for solving the issue, besides representing an unusual challenge for encountering local management strategies.
New approaches, using contemporary molecular techniques, in conjunction with demographic data, can be extremely useful for a better comprehension of the dynamics, population structure and social biology of many invasive species (Taylor et al., 2000).
Among genetic markers, highly polymorphic animal mitochondrial DNA (mtDNA), almost exclusively maternally inherited and without genetic recombination, constitutes a powerful tool for a population genetic approach. The clonal transmission of mtDNA haplotypes facilitates the discrimination of matrilineage within species, the sequence analysis of their most variable regions being useful for investigating the genetic origin of animal populations and breeds, and thus, domestication processes in livestock species themselves (Bradley et al., 1996; Luikart el al., 2001).
The main aim of this investigation is to provide a more comprehensive scenario of wild boar differentiation in Uruguay, using mtDNA population markers and phylogenetic analysis for testing possible hypotheses regarding their rapid expansion. Furthermore, this is a first-time report containing genetic information, with recommendations for more effective control strategies of feral pigs in Uruguay.
Material and Methods
Sampling and DNA extraction
This phylogeographic study of the Uruguayan wild boar included specimens (Appendix 1) mostly from the southern, central, northeastern and eastern regions of the country, covering the geographic range of expansion, starting from the introduction site in the Department of Colonia (Figure 1). The remainder are sequences retrieved from GenBank, and pertaining to Spanish, Italian, central European and Japanese wild boars, as well as commercial pig breeds, viz., Large White, Landrace, Duroc and Pietrain (Supplementary Material, Table S1). Tissues of voucher specimens were deposited in the Sección Genética Evolutiva, Facultad de Ciencias, Uruguay. Outgroup analysis included individuals pertaining to two additional taxa Sus verrucosus and Phacochoerus africanus.
Genomic DNA was isolated from liver and muscle tissues of freshly sacrificed animals (fixed in ethanol 95%), using an extraction with a sodium chloride protein precipitation, followed by ethanol DNA precipitation (modified from Miller et al., 1988).
Mitochondrial cytochrome b (cyt b) sequences
A 661-bp fragment of cyt b gene between sites 14,695 and 15,355 was amplified using forward and reverse primers, as described by Alves et al. (2003), in 30 cycles of 94 °C for 45 s, 53 °C for 45 s, 72 °C for 1 min; 30 cycles. PCR products were cleaned with MARLIGEN Kit (Biosciences inc.) Rapid PCR Purification System, to then undergo sequencing using amplification primers with a Perkin-Elmer ABI Prism 377 automated sequencer. The final sequences for analysis were obtained by reconciling chromatograms for light and heavy DNA strands. Sequence alignment was performed using the CLUSTAL X program (Thompson et al., 1997).
Data analysis and DNA polymorphism
Nucleotide composition and substitution patterns were calculated using the MEGA (Kumar et al., 2004) and DNASP4 (Rozas et al., 2003) computer programs. Corrected estimates of pairwise sequence divergence were obtained using the two-parameter algorithm (K2P) of Kimura (1980) implemented into MEGA. Population DNA polymorphism was measured by calculating the proportion of segregating sites (S), haplotype diversity (Nei, 1987; p. 179), and nucleotide diversity π (Nei, 1987; p.257), using ARLEQUIN (Schneider et al., 2000) and DNASP4 (Rozas et al., 2003) software packages.
In order to evaluate neutrality departure in the data, Tajimas D (Tajima, 1989) was calculated using the DNASP4 (Rozas et al., 2003) software package, as a way of testing any significant excess of low-frequency haplotypes.
Phylogenetic analyses
Two methods of phylogenetic reconstruction viz., maximum-parsimony (MP) and neighbor-joining (NJ), were employed to define phylogeographic association among mitochondrial sequences, using for the purpose PAUP*4.0b8 (Swofford, 1998). Equally weighted maximun-parsimony analysis was undertaken by way of heuristic search (MULPARS option, stepwise addition, tree-bisection-reconnection [TBR] branch swapping, 100 replications). Strict consensus between rival trees was computed to reconcile equally parsimonious topologies. Distance trees were generated using a Hasegawa et al. (1985) model, taking into consideration differences among transversion and transition substitutions, as well as those among base frequencies. The neighbour-joining method (Saitou and Nei, 1987) was employed for phylogenetic reconstruction. In the case of both methods (MP and NJ), the degree of confidence assigned to nodes in trees was assessed by bootstrapping with 1000 replicates. All the trees were rooted by means of the outgroup criterion.
Analysis of Molecular Variance and Nested Clade Analysis (NCA)
In order to examine genetic structuring among Uruguayan wild boar populations, variance components among hierarchical partitions in the data set were assessed through the Analysis of Molecular Variance (AMOVA) developed by Excoffier et al. (1992). The Euclidean metric (Excoffier et al., 1992) was used to construct the matrix of pairwise distances. Various grouping hypotheses were proposed for analyzing the hierarchical partitioning of genetic variation. Three among these were retained, viz., 1) all the haplotypes were gathered into a single group, 2) haplotypes from two neighbouring eastern sites and distributed into the corresponding groups (B and D in Figure 1) vs. all the remaining geographic localities, 3) the haplotypes were assigned to three regions (southern, central-northeastern and eastern Uruguay) representing the diverse Uruguayan basins, as a means of recuperating biogeogeographic information.
The existence of geographic association among haplotypes was assessed by NCA (Templeton et al., 1995). The cyt b haplotype network in Uruguayan wild boars was estimated by using the statistical parsimony method, with the algorithm described by Templeton et al. (1992). Accordingly, the cladogram for finding haplotype connections with probabilities above the 0,95 confidence level was contructed using the TCS 1.06 (Clement et al., 2000) program. Statistics related to data distances, viz., internal clade (Dc), between clades (Dn) and between interior and tip clades (I-T), were generated by exact permutation contingency analysis of clades within nested categories as against their respective geographic locality (Templeton et al., 1995), using in the process 10,000 permutations of nesting clades versus sampling localities, and assuming as recommended statistical significance, α = 0.05. Results obtained from GEODIS were then interpreted, using the revised Posada and Templeton (2008) inference key to elucidate alternative historical scenarios of wild boar differentiation.
Population subdivision was measured by assuming the infinite mutation model (Kimura and Crow, 1964) and calculating FST (Slatkin, 1991) for the whole population. Pairwise estimates of FST were calculated using Arlequin (Schneider et al., 2000) to generate pairwise estimates of gene flow levels, as follows: Nfm ≈ 1/2 [(1/ FST) - 1] (Wright, 1951).
Results
Mitochondrial cyt b diversity and population genetic analysis
Mitochondrial cytochrome b sequences pertaining to Uruguayan, European and Japanese wild boars, as well as commercial breeds of Sus scrofa, were included for analysis. Most of those from the Uruguayan samples were new for the wild boar Sus scrofa, the remainder having been retrieved from GenBank (Table S1). Among the 571 bp analyzed from Uruguayan wild boar samples, 47 variables and 28 phylogenetic informative sites were found in the data set, apart from 39 polymorphic segregating sites and 14 haplotypes in the sample itself. Haplotype diversity was high (0.97 - SD = 0.032), whereas intermediate levels of nucleotide diversity were found (0.014 - SD = 0.021). With the exception of haplotypes 1 (shared by 2 individuals), 2 and 11 (both shared by three individuals), the remainder were carried by a single individual.
The estimated average rate of transitional/transversional substitution (r = si/sv) in the wild boar cyt b gene was 2:1 (1st r = 1.7; 2 sd r = 0.5 and 3rd r = 2.8 codon positions). Among the Uruguayan wild boar samples, deduced amino acid sequences showed only 20 of 190 amino acids to be variable and 9 phylogenetically informative sites.
Corrected pairwise K2P sequence divergence among the sampled Uruguayan wild boars is presented in Table 1. The average genetic distance was 1.7% (SD = 0.003), thus remarkably higher than that found among European (0.3% -SD = 0.001), and Japanese and Israeli (1.3% - SD = 0.005) wild boars, as well as the analyzed commercial breeds (0.9% - SD = 0.003). The maximun divergence by including outgroup taxa was 8.4% (SD = 0.009).
Table 1-.
Corrected genetic distances among 14 haplotypes of the Uruguayan wild boar Sus scrofa, according to the Kimura 2-P model (below the diagonal), and Standard Deviation (SD) estimated by the bootstrap method (above the diagonal).
| hap1 | hap2 | hap3 | hap4 | hap5 | hap6 | hap7 | hap8 | hap9 | hap10 | hap11 | hap12 | hap13 | hap14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hap1 | - | 0.002 | 0.027 | 0.029 | 0.005 | 0.011 | 0.007 | 0.005 | 0.022 | 0.016 | 0.002 | 0.002 | 0.002 | 0.004 |
| hap2 | 0.002 | - | 0.029 | 0.027 | 0.004 | 0.013 | 0.009 | 0.004 | 0.024 | 0.018 | 0.004 | 0.004 | 0.004 | 0.002 |
| hap3 | 0.007 | 0.008 | - | 0.027 | 0.029 | 0.027 | 0.024 | 0.033 | 0.042 | 0.044 | 0.029 | 0.029 | 0.029 | 0.031 |
| hap4 | 0.007 | 0.007 | 0.007 | - | 0.024 | 0.033 | 0.029 | 0.027 | 0.052 | 0.046 | 0.031 | 0.031 | 0.031 | 0.029 |
| hap5 | 0.003 | 0.002 | 0.008 | 0.007 | - | 0.009 | 0.005 | 0.007 | 0.027 | 0.022 | 0.007 | 0.007 | 0.007 | 0.005 |
| hap6 | 0.004 | 0.005 | 0.007 | 0.008 | 0.004 | - | 0.004 | 0.016 | 0.022 | 0.027 | 0.013 | 0.013 | 0.013 | 0.014 |
| hap7 | 0.003 | 0.004 | 0.007 | 0.008 | 0.003 | 0.002 | - | 0.013 | 0.022 | 0.024 | 0.009 | 0.009 | 0.009 | 0.011 |
| hap8 | 0.003 | 0.002 | 0.008 | 0.007 | 0.003 | 0.005 | 0.005 | - | 0.027 | 0.022 | 0.007 | 0.007 | 0.007 | 0.005 |
| hap9 | 0.006 | 0.007 | 0.010 | 0.011 | 0.007 | 0.006 | 0.006 | 0.007 | - | 0.009 | 0.024 | 0.024 | 0.024 | 0.025 |
| hap10 | 0.005 | 0.006 | 0.010 | 0.010 | 0.006 | 0.007 | 0.007 | 0.007 | 0.004 | - | 0.018 | 0.018 | 0.018 | 0.020 |
| hap11 | 0.002 | 0.002 | 0.008 | 0.008 | 0.003 | 0.005 | 0.004 | 0.003 | 0.007 | 0.006 | - | 0.004 | 0.004 | 0.005 |
| hap12 | 0.002 | 0.002 | 0.008 | 0.008 | 0.003 | 0.005 | 0.004 | 0.003 | 0.007 | 0.006 | 0.002 | - | 0.004 | 0.005 |
| hap13 | 0.002 | 0.002 | 0.008 | 0.008 | 0.003 | 0.005 | 0.004 | 0.003 | 0.007 | 0.006 | 0.002 | 0.002 | - | 0.005 |
| hap14 | 0.002 | 0.002 | 0.008 | 0.008 | 0.003 | 0.005 | 0.004 | 0.003 | 0.007 | 0.006 | 0.003 | 0.003 | 0.003 | - |
No significant excess of low-frequency haplotypes among Uruguayan wild boars (D = −1.418 p > 0.10) was revealed by Tajima D’test.
Phylogenetic analyses
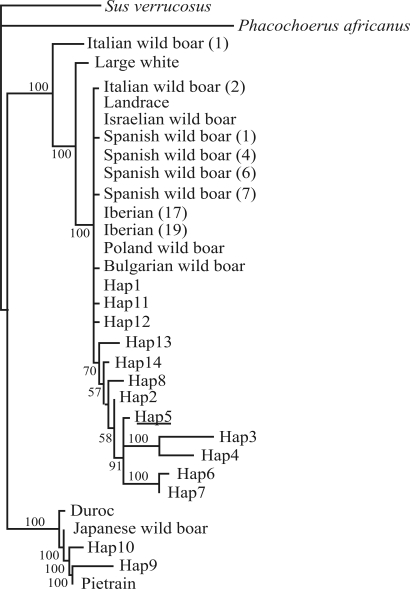
The same two evolutionary sister clades that constitute different phylogroups were identified by maximum parsimony and distance analysis. In maximum parsimony analysis, strict consensus (Figure 2) resulted in the 100 most parsimonious trees (156 steps), thereby showing a major and monophyletic clade integrated by 12 Uruguayan wild boar haplotypes. They collapsed into a basal polytomy joining all the haplotypes belonging to European wild boars, together with Landrace and Large White commercial breeds. A minor clade integrated by the Uruguayan wild boar haplotypes 9 and 10 collapsed together with a Japanese wild boar haplotype and the Duroc and Pietrain commercial breeds. All the major clades received high bootstrap values of 80%–100% in both phylogenetic analyses.
Figure 2-.
Maximum parsimony phylogenetic relationships based on the cytochrome b dataset of haplotypes of Uruguayan, European and Japanese wild boars, as well as sequences from commercial breeds of Sus scrofa. The strict consensus resulted in 100 of the shortest most parsimonious trees (156 steps). All the trees were rooted by using Sus verrucosus and Phacochoerus africanus as outgroups. Bootstrap values above 50% are shown on the relevant nodes.
AMOVA analysis and geographic distribution of genetic variation
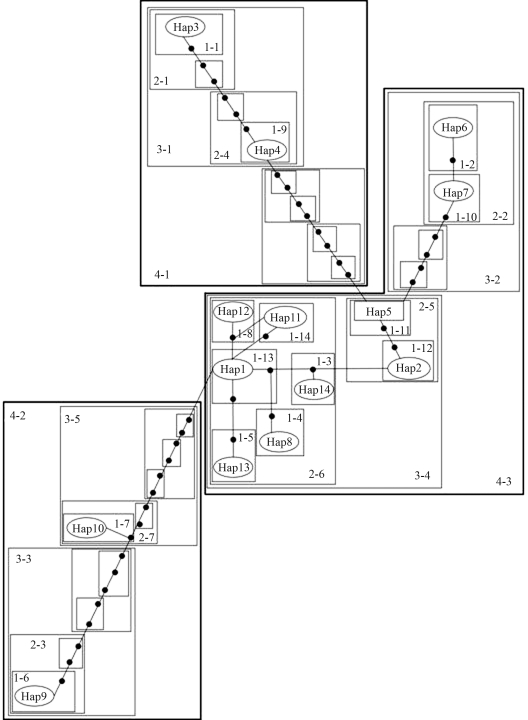
Statistical parsimony network based on the cyt b gene (Figure 3), revealed the strong genetic structuring among haplotypes of the Uruguayan wild boar showing different phylogroups. The total cladogram includes five levels of nested hierarchical clades presenting 11 maximum connection steps at 95%. At a higher level of hierarchy, a major clade (4–3) of haplotypes remains connected by few mutational steps, thereby retaining a central position in the network. This major clade includes those locales in central Uruguay, in the neighborhood of the site in the Department of Colonia (Figure 1, sites I, E, H, G and F), the center of expansion throughout the central region itself, as well as the northeastern (C, Figure 1) and eastern (A, Figure 1). Haplotype 5 relates this clade (4–3) to other more distant haplo-types that integrate clade 4–1. This haplotype corresponds to locale F (Figure 1) in the central region. Furthermore, two other genetically more distant haplotypes (9 and 10) represent a divergent clade (4–2 in Figure 3) in the eastern region, separated by ninety and ten step mutations, respectively, from the remainder. Finally, nested contingency analysis of almost all clade levels revealed no significant association of clades and geographic distances.
Figure 3-.
Statistical parsimony network and the corresponding nested design for cytochrome b haplotypes in Uruguayan wild boars. The cladogram was estimated under 95% statistical limits of parsimony using the algorithm by Templeton et al. (1992). The oval includes haplotype numbers (0-step clades). Solid circles represent hypothetical haplotypes. Thin-lined polygons enclose 1-step to 3-step clades and thick-lined polygons enclose 4-step clades, all within the total cladogram. Specimens and the respective haplotype are listed in Table S1, Supplementary Material.
AMOVA results compiling the three retained hypotheses are shown in Table 2. Under the two-group hypothesis (2), most genetic variation among cyt b haplotypes was distributed among-groups (ΦCT = 0. 673). All the other tested structuring hypotheses failed to provide a more reasonable explanation for maximization of among-group hierarchical molecular variation.
Table 2-.
Analysis of molecular variance (AMOVA) in the Uruguayan wild boar Sus scrofa. Hierarchical partition of genetic variation into three components: among groups (ΦCT), among populations within groups (ΦCS), and among individuals within populations (ΦST), disregarding either their original populations or groups. Among the tested hypotheses, three were selected: a) including all the collecting sites into just one group; b) forming two groups of populations, consisting of all the collection sites vs. Rio Cebollatí and Lascano; c) the separation into three groups of samples pertaining to the various river basins, viz., the eastern, central-northeastern and southern. The highlighted line corresponds to values which maximal among-group differentiation.
| Hypothesis | Source of variation | df | Sum of squares | Variance components | Percentage of variation | Φ statistics |
|---|---|---|---|---|---|---|
| a | Among groups | 8 | 26.140 | 0.35864Va | 11.71 | - |
| Among populations within groups | - | - | - | - | - | |
| Within populations | 8 | 21.625 | 2.70312 Vb | 88.29 | ΦST = 0.11714 | |
| b | Among groups | 1 | 15.731 | 0.01813 Va | 67.39 | ΦCT = 0.67394 |
| Among populations within groups | 7 | 10.408 | −0.76469Vb | −12.86 | ΦSC = −0.39449 | |
| Within populations | 8 | 21.625 | 2.70312 Vc | 45.47 | ΦST = 0.54531 | |
| c | Among groups | 2 | 7.248 | 0.01698Va | 0.55 | ΦCT = 0.00554 |
| Among populations within groups | 6 | 18.892 | 0.34564 Vb | 11.27 | ΦCS = 0.11337 | |
| Within populations | 8 | 21.625 | 2.70312 Vc | 88.17 | ΦST = 0.00554 | |
Indirect estimates of pairwise FST values revealed almost complete genetic isolation among locales, except for site A, with considerable genetic exchange with the remainder. On the other hand, all were genetically isolated from B and D.
Discussion
Mitochondrial cytochrome b variation in Uruguayan wild boar populations
Present results represent the first population genetic characterization of Uruguayan wide-ranging wild boars, when using the mitochondrial cytochrome b gene. High levels of haplotype diversity, intermediate levels of nucleotide diversity, and considerable population differentiation among sampled localities throughout major watercourses and catchment dams, were detected (Figure 1).
Intermediate nucleotide diversity was similar to that reported for Artiodactyla, taxa, when using mtDNA cyto-chrome b sequences in a comparative analysis of various mammalian orders and families (Nabholz et al., 2008).
According to the present study, levels of corrected sequence divergence among Uruguayan wild boar haplo-types, although higher than in the European wild type and commercial breeds, was similar to the Japanese and Israeli. In previous studies, when considering both cyt b and the mtDNA control region, an appreciable genetic distance (1.2 ± 0.09%) between European wild boars (Italy and Poland) and Asian (Israel), was found (Giuffra et al., 2000), therefore consistent with the divergence found in the Uruguayan wild boar data set. In contrast, on analyzing mitochondrial control region data from domestic pigs, Fang and Andersson (2006) reported low genetic divergence in all the Chinese mtDNA haplotypes (mean ± SD, 0.006 ± 0.001), as well as all the European (mean ± SD, 0.005 ± 0.001).
All accumulated data, plus the average si/sv ratio detected in the cyt b fragment, indicate that this molecular marker, besides revealing no evidence of among-site saturation, represents a useful tool for genetically characterizing the recently introduced feral pig in Uruguay.
Population structuring in Uruguayan wild boar populations
Phylogenetic analysis (Figure 2) revealed the existence of two different phylogroups among the Uruguayan wild boar haplotypes. The major of the two comprises all those, with the exception of 9 and 10, pertaining to the different localities in souththern, central and northeastern Uruguay. This clade collapsed into a basal polytomy with all the sequences from European wild boars, and Landrace and Large White domestic pigs. The minor phylogroup, comprised of haplotypes 9 and 10, joins the sequences from Pietrain and Duroc domestic pigs, as well as the Japanese wild boar. Generally speaking, the statistical parsimony network (Figure 3) showed the same genealogical history, although here the major phylogroup is divided into two clades, 4–1 (haplotypes 3 and 4) and 4–3 (this including the remaining haplotypes of the group). Consequently, clade 4–1 corresponds to a minor derivative monophyletic clade (integrated by haplotypes 3 and 4) in the phylogenetic tree (Figure 2). On the other hand, the minor phylogroup in phylogenetic reconstruction (Figure 2) was consistent with clade 4–2 in the statistical parsimony network (Figure 3).
The present analysis supports the hypothesis of two different deliberate introduction events of distantly related genetic lineages in the Uruguayan wild boar populations. The distribution of the major clade is consistent with the historical introduction of the European wild boar into the Colonia Department, and its dispersal throughout localities in southern, central and northeastern Uruguay, by way of the principal basins and watercourse of the Rio Santa Lucía andthe Río Negro. The second introduction, occurred in southeastern Uruguay near the Cebollatí river in the Merin lagoon basin, the border between Uruguay and the Brazilian state Rio Grande do Sul, may have been a deliberate introduction since southern Brazil. Further analysis, including samples from these neighbouring localities, could possibly clarify the issue.
Moreover, two major hypotheses would explain the association of the Uruguayan wild boar with others populations of different origins, as well as with different strains of domestic pigs, i.e.,the existence of ancestral polymorphisms and/or multiple introgressive hybridization events, as possible, although not mutually exclusive, scenarios. These scenarios are consistent with the level of genetic variation accumulated in the Uruguayan wild boar populations. The evidence of ancestral polymorphism has been a plausible hypothesis since 1920, when the first introduction of wild boars of unknown origin was reported (Herrero and Fernández de Luco, 2003). In this case, various types of European wild boars would be involved, as can be inferred on examining taxa association in the major clade (Figure 2). The presence of lung parasites of the genus Metastrongylus sp., quite common in European wild boars, is notable in the Uruguayan wild boar and could give support to this possible origin. Nevertheless, it is difficult to explain the association with the Japanese wild boar, Duroc and Pietrain sequences in the minor clade, through the lack of additional information regarding neighbouring south Brazilian wild boar populations.
The hypothesis of hybridization and introgression is well supported in phenotypical characterization among current Uruguayan wild boar populations (Herrero and Fernández de Luco, 2003). These authors proposed their emergence from cross-breeding between wild boars and domestic pigs, hence the wide diversity in phenotypes, with a predominance of characteristics from the former. Consequently, some individuals have white hair on the feet, the underside of the neck, tarsus and carpus, drooping ears and jet black or red tails, whereas others present outstanding and fast growth, great intrapopulation morphological variability and considerable accumulation of subcutaneous fat (Herrero and Fernández de Luco, 2003).
According to Lee (2002), the inter- or intraspecific hybridization of an invasive population with native or non-native populations would alleviate the loss of additive genetic variance during founder events, and thus generate novel genotypes. Numerous studies have documented the positive effects of hybridization on invisibility, such as faster growth, greater size and increased aggression. All these characteristics were encountered in Uruguayan wild boar populations (Herrero and Fernández de Luco, 2003). Lee (2002) also proposed successful invasion to be a probable result of advantageous selection among numerous hybrid combinations.
According to Avise (2000), high haplotype and low nucleotide diversities infer rapid population growth from an ancestral population with small Ne. Current results concerning mtDNA polymorphism parameters could conform to this interpretation. Moreover, Frankham (2005) postulated that certain invasive species possess elevated genetic diversity and the enhanced ability to evolve when invading novel localities. All present population genetic data pointed out that hybridization between introduced wild boars and domestic pigs could be a plausible explanation of the invasive potential of these cross-bred populations.
AMOVA analysis (Table 2) confirmed the considerable population differentiation among sampled localities, as well as the existence of two unexpected, different and highly structured evolutionary lineages among Uruguayan wild boars. No genetic exchange was detected among individuals belonging to these different phylogroups. Moreover, indirect estimates of gene flow revealed no homogeneity among all the sampled localities. In fact, with the exception of the neighboring B and D, only locale A (Figure 1) showed any considerable genetic exchange with the remainder. Nevertheless, this is an expected population scenario, seeing that this locality included a mixed group captured from other distant sites in the country, thereby representing a semi-captive wild boar stock.
The levels of differentiation found are consistent with existing knowledge of the regional distribution and biology of the Italian wild boar (Vernesi et al., 2003). There was also a certain similarity with the population structure of feral pigs in southwestern Australia (Hampton et al., 2004), whereby dispersal rates between, but not within, the inferred feral pig populations were relatively low. According to the relatively small home range of feral pigs in this region (Choquenot et al., 1996), a high level of genetic structuring was not unexpected even between populations that were only 25 km apart.
A similar dispersal pattern was encountered in neighbouring wild boar localities (A vs. B and D) which, although only around 60 km apart, remained genetically isolated. Further population genetics analysis, using nuclear markers (i.e. microsatellites, SNPs), could be a means of clarifying aspects of social organization, dispersal and possible asymmetric gene flow among populations in the Uruguayan wild boar. Even so, recent studies on wild boar from Tuscany (Italy) did not reveal that the predicted matrilinearity in wild boar social units. In this study, aggregations of unrelated adult females were detected, thereby indicating a low degree of within-group relatedness (Iacolina et al., 2009).
Population genetics analysis in Uruguayan wild boars and management strategies
Present population genetics has contributed to the strategic management of Uruguayan wild boar populations. Considering both the high levels of differentiation and the normally low migration rates among localities in these populations, management strategies would be more effective if each population were to be considered as a single management unit. A similar modus operandi has already been proposed for other feral pig populations (Hampton et al., 2004). The failure to recognize these units could result in the inevitable recurrent invasion of controlled areas. On the other hand, recurrence from neighbouring pig populations would be relatively slow, due to the low migration rate between discrete or adjacent localities.
Acknowledgments
We wish to thank M. Acosta and N. de los Santos for kindly providing Uruguayan wild boar specimens. The authors are also grateful to the Japanese government for the donation of equipment to the Sección Genética Evolutiva in Facultad de Ciencias, Montevideo, Uruguay.
Footnotes
Associate Editor: João S. Morgante
Supplementary Material
The following online material is available for this article:
List of specimen and haplotypes from cytochrome b sequences.
This material is available as part of the online article from http//www.scielo.br.gmb.
References
- Alves E, Ovilo C, Rodríguez MC, Siló L. Mitochondrial DNA sequence variation and phylogenetic relationships among Iberian pigs and other domestic and wild pigs populations. Anim Genet. 2003;34:319–324. doi: 10.1046/j.1365-2052.2003.01010.x. [DOI] [PubMed] [Google Scholar]
- Avise JC. Phylogeography: The History and Formation of Species. Harvard University Press; Cambridge: 2000. p. 447. [Google Scholar]
- Bradley DG, MacHugh DE, Cunningham P, Loftus RT. Mitochondrial diversity and the origins of African and European cattle. Proc Natl Acad Sci USA. 1996;93:5131–5. doi: 10.1073/pnas.93.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caley P, Hone J. Disease transmission between and within species, and the implications for disease control. J Appl Ecol. 2004;41:94–104. [Google Scholar]
- Choquenot D, McIlroy J, Korn T. Managing Vertebrate Pests: Feral Pigs. Australian Publishing Service; Canberra: 1996. p. 163. [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: A computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Declara plaga nacional al jabalí europeo y autoriza su libre caza en todo el territorio nacional. Diario Oficial. Decreto 463/1982 del Poder Ejecutivo del 15/12/1982. 21388-500-A.
- Dexter N. Stochastic models of foot and mouth disease in feral pigs in the. Australian semi-arid rangelands. J Appl Ecol. 2003;40:293–306. [Google Scholar]
- Edwards GP, Pople AR, Saalfeld K, Caley P. Introduced mammals in Australian rangelands: Future threats and the role of monitoring programmes in management strategies. Austral Ecol. 2004;29:40–50. [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Andersson L. Mitochondrial diversity in European and Chinese pigs is consistent with population expansions that occurred prior to domestication. Proc R Soc Lond B Biol Sci. 2006;273:1803–1810. doi: 10.1098/rspb.2006.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. Resolving the genetic paradox in invasive species. Heredity. 2005;94:385. doi: 10.1038/sj.hdy.6800634. [DOI] [PubMed] [Google Scholar]
- Giuffra E, Kijas JMH, Amarger LV, Carlborg O, Jeon JT, Andersson L. The origin of the domestic pig: Independent domestication and subsequent introgression. Genetics. 2000;154:1785–1791. doi: 10.1093/genetics/154.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton JO, Spencer PBS, Alpers DL, Twigg LE, Woolnough AP, Doust J, Higgs T, Pluske J. Molecular techniques, wildlife management and the importance of genetic population structure and dispersal: A case study with feral pigs. J Appl Ecol. 2004;41:735–743. [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Herrero J, Fernández de Luco D. Wild boars (Sus scrofa L.) in Uruguay: Scavengers or predators? Mammalia. 2003;67:485–491. [Google Scholar]
- Iacolina L, Scandura M, Bongi P, Apollonio M. Non kin associations in wild boar social units. J Mammal. 2009;90:666–674. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kimura M, Crow JF. The number of alleles that can be maintained in a finite population. Genetics. 1964;49:725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lee CE. Evolutionary genetics of invasive species. Trends Ecol Evol. 2002;17:386–391. [Google Scholar]
- Luikart G, Gielly L, Excoffier L, Vigne JD, Bouvet J, Taberlet P. Multiple maternal origins and weak phylogeographic structure in domestic goats. Proc Natl Acad Sci USA. 2001;98:5927–5932. doi: 10.1073/pnas.091591198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dikes DD, Polesky HF. A simple salting out procedure for extracting DNA for human nucleated cells. Nucleic Acids Res. 1988;16:215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabholz B, Mauffrey JF, Bazin E, Galtier N, Glemin S. Determination of mitochondrial genetic diversity in mammals. Genetics. 2008;178:351–361. doi: 10.1534/genetics.107.073346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York: 1987. p. 512. [Google Scholar]
- Rozas J, Sánchez-Delbarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbour-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Arlequin: A software for population genetics data analysis. University of Geneva; Switzerland: 2000. [Google Scholar]
- Slatkin M. Inbreeding coefficients and coalescence times. Genet Res. 1991;58:167–175. doi: 10.1017/s0016672300029827. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP* (Phylogenetic Analysis Using Parsimony) v 4. Sinauer; Sunderland: 1998. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AC, Cowan PE, Fricke BL, Cooper DW. Genetic analysis of the mating system of the brushtail possum (Trichosurus vulpecular) in New Zealand farmland. Mol Ecol. 2000;9:869–879. doi: 10.1046/j.1365-294x.2000.00941.x. [DOI] [PubMed] [Google Scholar]
- Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Routman E, Phillips CA. Separating population structure from population history: A cladistic analysis of the geographical distribution of mitochondrial DNA haplotypes in the Tiger Salamander, Ambystoma tigrinurn. Genetics. 1995;140:767–782. doi: 10.1093/genetics/140.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernesi C, Crestanello B, Pecchioli E, Tartari D, Carnelli D, Hauffe H, Bertorelle G. The genetic impact of demographic decline and reintroduction in the wild boar (Sus scrofa): A microsatellite analysis. Mol Ecol. 2003;12:585–595. doi: 10.1046/j.1365-294x.2003.01763.x. [DOI] [PubMed] [Google Scholar]
- Wright S. The genetical structure of populations. Ann Eugenics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
Internet Resources
- Posada D, Templeton AR. Inference Key for the Nested Haplotype Tree Analysis of Geographical Distance. 2008. http://darwin.uvigo.es/software/geodis.html (December, 2008).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of specimen and haplotypes from cytochrome b sequences.